Abstract
The environment hosts a diversity of microorganisms whose potential for biotechnological applications has not yet been exhausted. The quest of our study was to find isolates of Pichia kudriavzevii from the environment that could be used as new biotechnological agents. Moreover, we aimed to explore the resource efficiency for microbial cultivation, in particular the efficiency of spent coffee grounds (SCG), an easily accessible waste coffee product with a high unutilized organic content. In this study, Pichia kudriavzevii strain ZMUM_K002, a yeast strain isolated from a grape pomace compost, was investigated. Antifungal susceptibility, particularly fluconazole susceptibility, was assessed, and the strain’s biotechnological potential by comparing its ability to utilize low-cost carbon sources, including SCG, with a natural isolate of Saccharomyces cerevisiae (strain ZMUM_K003) was assessed. The P. kudriavzevii strain ZMUM_K002 exhibited higher fluconazole susceptibility and yielded more than 30% more biomass in optimized media formulations compared to S. cerevisiae ZMUM_K003. These findings demonstrate that P. kudriavzevii ZMUM_K002 has the potential for efficient biomass production in sustainable industrial biotechnology, particularly in processes requiring high biomass yields on alternative substrates.
1. Introduction
Yeasts are at the forefront of the food industry, with growing importance in biotechnology. They exhibit superior stress tolerance, including resistance to high temperatures, osmotic stress, and nutrient limitation—properties that are crucial for industrial applications such as bioethanol production, biosurfactant synthesis, and bioremediation [1,2]. The key role among yeasts used in biotechnology is still played by the yeast Saccharomyces cerevisiae, which has not only been domesticated but also fine-tuned to play essential roles in fermentation, baking, and brewing, as well as in other (bio)processes [3,4,5,6,7,8,9]. However, to expand the desired (bio)products and market demands, new biotechnologically important non-Saccharomyces yeasts from different environments are currently being sought [10]. In the vast and diverse biosphere, certain natural habitats harbor unique (micro)organisms that have adapted to survive and thrive in conditions that are considered hostile to most life forms. Not only extreme habitats (hot springs, acidic lakes, deep-sea hydrothermal vents, and arid deserts), but also unexpected environments close to inhabited areas, such as compost heaps, saline soils, and wastewater treatment plants, represent unexplored environments with great potential for new biotechnological agents [1,11,12]. Recently, the yeast Pichia kudriavzeii has been identified as one such potential new agent for biotechnological applications due to its high tolerance to extreme pH values, high temperatures, high osmotic pressure, and fermentation inhibitors. P. kudriavzeii is ubiquitous in the environment, which has meant historic isolates recovered from diverse settings have been assigned alternative species names. These include: Issatchenkia orientalis, Candida glycerinogenes, and Candida krusei. I. orientalis and C. glycerinogenes are known as yeasts used in industry, and C. krusei is known as an opportunistic human pathogen found in clinical samples. Genomic analyses confirmed that these names correspond to the same species (≥99.6% genetic identity), and nowadays the assigned taxonomic name is P. kudriavzevii [13,14]. In industry, P. kudriavzevii is widely used due to its tolerance to extreme conditions. It plays an important role in food fermentation, feed production, chemical biosynthesis, and environmental engineering [15]. In particular, it is widely found in fermented foods and is used in the production of glycerol and succinate [16,17]. On the other hand, P. kudriavzevii (still named C. krusei in medical microbiology) is responsible for ~3% of yeast infections in humans and is known to be intrinsically resistant to fluconazole, the most-used antifungal drug in clinical treatment of yeast infections [18]. As in the absence of appropriate safety precautions, fluconazole resistance could be a source of public health concern [19,20], and fluconazole susceptibility has to be evaluated prior to any biotechnological application.
After isolating environmental non-Saccharomyces yeast isolates, it is important to optimize conditions to maximize biomass production and/or promote the synthesis of desired metabolites, especially since it has been reported that nutrient requirements are often strain-specific and require individual optimization [11,21,22]. In addition to achieving optimal biomass yields, the cultivation of these non-Saccharomyces yeasts using lignocellulosic and disposable agricultural wastes, such as spent coffee grounds (SCG), is becoming increasingly important due to their sustainability and cost-effectiveness [23,24].
SCG, a waste product of one of the world’s most popular beverages, coffee, still has a high unutilized organic content of polysaccharides, including cellulose, hemicellulose and lignin, as well as proteins, lipids, and important minerals to support microbial growth [25]. Due to its high carbohydrate content, SCG can be a valuable source of carbon. In addition, its nitrogen content is beneficial for the synthesis of microbial proteins and nucleic acids. However, as SCG contains bioactive compounds such as caffeine and phenols that can inhibit the growth of microorganisms, its use as an efficient and cost-effective nutrient supplement could be compromised [26,27].
With the aim of finding new P. kudriazevii isolates with potential for efficient biotechnological applications, we sampled a large array of different natural and anthropogenic environments and subjected the samples to harsh selection conditions to obtain isolates with the properties needed for industrial applications. Among all isolated yeasts, only one P. kudriavzevii strain was found (ZMUM_K002 strain), which was subsequently evaluated in comparison to S. cerevisiae ZMUM_K003. Its fluconazole susceptibility and optimization of cultivation media, including the use of SGC, were investigated. With our approach, we aimed to explore the potential of the P. kudriavzevii strain ZMUM_K002 for sustainable biotechnological applications, focusing on resource recovery and valorization of organic waste streams.
2. Materials and Methods
2.1. Collection of Samples and Selective Isolation of Yeasts
Samples were collected using sterile cotton swab tubes (Labortehnika Golias, Kranj, Slovenia) from various, randomly selected, supposedly non-pathogenic, natural and anthropogenic sources in Slovenia. Sampling sites included compost from grape pomace; fermentation broth from sauerkraut; surfaces from various fruits, including citrus fruits; and swabs from a dishwasher, a coffee machine, and a discarded wine bottle. Each sample was subjected to an enrichment–selection protocol designed to favor the growth of yeasts: In the laboratory, to each swab tube 10 mL of YM liquid medium containing 3 g/L malt extract, 3 g/L yeast extract, 5 g/L peptone, 10 g/L glucose, 0.1 g/L chloramphenicol, and 10% (v/v) ethanol was added and incubated anaerobically in an Oxoid AnaeroJarTM 2.5 L (ThermoScientific, Waltham, MA, USA) for 7 days at 30 °C. After incubation, 100 µL of the liquid broth was transferred onto MEA agar plates (20 g/L malt extract, 1 g/L peptone, 20 g/L glucose, and 20 g/L agarose, with a pH 5.3) and incubated aerobically for 7 days at 30 °C in an incubator (Cool incubator 23l, Domel, Železniki, Slovenia). Single colonies (white to cream, round-shaped colonies with a smooth margin, slightly convex colony profile and smeared) were picked and repeatedly subcultured on MEA agar plates and incubated aerobically for 3–4 days at 30 °C to obtain pure yeast cultures. The pure cultures were preserved in medium with 20% glycerol and kept at −80 °C in the microbial collection of the Chair for Microbial and Molecular Biosciences, Department of Biology, Faculty of Natural Sciences and Mathematics, University of Maribor, Slovenia. All media were autoclaved (121 °C, 15 min, 1.1 bar), and all media components were from Duchefa Biochemie (Haarlem, The Netherlands).
2.2. Yeast Identification Using ITS rDNA Region
Strains were grown in MEA broth for 16 h at 30 °C with agitation (180 rpm). Extraction of genomic DNA was performed using a GeneJET Genomic DNA Purification Kit (ThermoFisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol for yeast’s DNA extraction. DNA quality and concentration were determined spectrophotometrically (Biospectrometer, Eppendorf, Hamburg, Germany). The DNA concentration was diluted to 10 ng/µL and used for PCR amplification. DNA was amplified using the primer set (5′-TCCTCCGCTTATTGATATGC-3′ (ITS4)/5′-AGGTTTCCGTAGGTGAACCT-3′ (18SF1)) targeting the internal transcribed spacer region (ITS). The ITS region, which is located between the 18S and 26S rDNA, is a commonly used genetic marker for identification of yeasts [28]. PCR reactions were carried out in 50 µL reactions consisting of the following: 0.5 µL (100 pmol/μL) primers, 5 µL (2 mM) dNTP (New England Biolabs, Ipswich, MA, USA), 5 µL of 10 × Taq buffer+KCl, 5 µL of 10 mM MgCl2, 0.25 µL of Taq polymerase (Thermoscientific), 3 µL of DNA template, and 30.75 µL of ddH2O. The following PCR program was used: an initial denaturation for 3 min at 95 °C, 30 cycles of denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 45 s, and a final extension for 10 min at 72 °C. The PCR amplification products were separated by gel electrophoresis, cleaned with a PCR Clean-Up System (ThermoFisher Scientific), and Sanger sequenced (Microsynth, Vienna, Austria). The obtained sequences were used to identify the yeast species by BLAST (https://blast.ncbi.nlm.nih.gov/; URL accessed 28 December 2024) [29] and deposited in the GenBank database. The S. cerevisiae strain ZMUM_K003, isolated from a discarded wine bottle in Kamnica, Slovenia (Acc. No.: PQ812486), was used for comparison in all subsequent experiments.
2.3. Sporulation and Spore Stain
Single colonies from MEA agar plates were transferred to liquid acetate medium for sporulation containing 0.5 g/L glucose, 10 g/L sodium acetate, and 1 g/L yeast extract, and also to sodium acetate agar plates for sporulation [30] containing 9.8 g/L sodium acetate, 11 g/L glucose monohydrate, 12 g/L NaCl, 7 g/L MgSO4 × 7H2O, 2.5 g/L yeast extract, and 20 g/L agar and incubated aerobically at 30 °C for 7 days. Salts (NaCl, MgSO4 × 7H2O, and sodium acetate) and glucose monohydrate were from Sigma Aldrich, Saint Louis, MO, USA. Single colonies grown on this plate were transferred to objective slides and stained according to Ziehl–Neelsen with Carbol Fuchsin for 3 min over water vapor, rinsed with 70% ethanol and decolorized with acidic ethanol (96% ethanol: 3M HCl, 20:1, by vol.). After washing, Malachite green was applied for 1 min over water vapor, rinsed, dried, and examined under a light microscope.
2.4. Fluconazole Susceptibility Testing
Yeast isolates were grown on MEA agar plates at 30 °C overnight. Next day, a yeast suspension was prepared in sterile saline (0.85% NaCl), adjusted to 0.5 McFarland and with a sterile swab evenly streaked on the surface of an RPMI agar plate. A fluconazole E-test strip (Biomérieux, Marcy-l’Étoile, France) was placed on the agar surface, and plates were incubated aerobically at 30 °C till visible growth (24 h for P. kudriavzevii ZMUM_K002 and 72 h for S. cerevisiae ZMUM_K003) in an inverted position. After incubation, the elliptical zones of inhibition around the E-test strips were observed. The minimum inhibitory concentration (MIC) was recorded as the point where the inhibition zone intersected the scale on the E-test strip [31,32,33,34]. The E-test was performed on three biological replicates (N = 3) per yeast strain.
2.5. Evaluation of Growth on Different Carbon Sources
Evaluation of growth on different carbon sources was performed in 20 mL tubes with Durham tubes using 1 mL of a carbon-source stock solution and 9 mL of a base medium containing 4.5 g/L yeast extract, 5 g/L peptone, and bromothymol blue as a pH indicator. The initial pH was adjusted to 7.0. Stock solutions of monosaccharides (glucose, fructose, galactose, D-mannose, and xylose) were prepared at 100 g/L in ddH2O, while those of disaccharides (sucrose, lactose, and maltose), polysaccharides (starch and methylcellulose) and sugar alcohols (D-sorbitol, D-mannitol, and glycerol) were prepared at 50 g/L. All carbon sources were from Sigma Aldrich. All solutions of the carbon sources were sterilized by autoclaving (121 °C, 15 min, 1.1 bar) without observable caramelization. Each tube was inoculated with 100 µL of yeast culture grown overnight in MEA broth, followed by incubation for at least 48 h at 30 °C. The utilization of the different carbon sources was evaluated by acid production, which was indicated by a color change of the bromothymol blue pH indicator added in the media. After incubation, the tubes were visually inspected for color shifts of the bromothymol blue from blue (pH ~ 7) to yellow (pH < 6.0), indicating carbohydrate metabolism and subsequent acidification of the medium. The results were recorded as follows: positive (+): a clear color change from blue to yellow, indicating the production of acidic products; negative (−): no visible color change, indicating no significant metabolism of the tested sugar; weak (+/−): a slight change in color (greenish), indicating weak or delayed utilization. The presence of gas formation in the Durham tubes was visually inspected and recorded as follows: positive (+) if a gas bubble was present and negative (−) if no gas was detected. Tests were performed in duplicate (N = 2). Results were recorded for all carbon sources after 48 h, except for glycerol, which was recorded after 120 h for both yeast strains inoculated.
2.6. Evaluation of Growth in Chemically Defined Medium Supplemented with Selected Carbon Sources
Preculture conditions are as follows: in order to reduce the lag phase, yeast cultures were precultured to ~1 × 106 viable cells/mL in 20 mL tubes with 10 mL of YP liquid media containing 5 g/L yeast extract, 10 g/L peptone, and 10 g/L glucose, with a pH of 7.0 for 16 h at 30 °C with agitation (180 rpm).
Culture conditions included the following: Cultivations were carried out in 100 mL Erlenmeyer flasks with metal caps and semi-globular internal deformations on the bases of the flasks. Precultures were 1:100 diluted into 50 mL of liquid chemically defined media (CDM) and cultivated for up to 120 h at 30 °C with agitation (180 rpm). CDM was prepared by combining the following components: 5 mL of the salt stock solution (for composition see Supplementary Table S1), 1 mL of ammonium sulphate stock solution (47.2 mg/mL (NH4)2SO4), 50 µL of the trace element stock solution (for composition see Supplementary Table S1), 38 mL of double distilled water (ddH2O), and 5 mL of either a glucose, fructose, or sucrose stock solution prepared at a concentration of 100 g/L and sterilized by filtration (0.22 µm). Needed controls were performed—no growth was observed when no yeasts were inoculated in any media, and no growth was observed in inoculated CDM without a carbon source. Experiments were performed in three biological replicates (N = 3) for each tested carbon source (glucose, fructose, and sucrose) per yeast strain.
2.7. Evaluation of Growth in YP Medium Supplemented with Glucose or Spent Coffee Grounds
Preculture conditions were the same as described above.
Culture conditions included the following: YP medium (10 g/L yeast extract, 20 g/L peptone in ddH2O) was supplemented either with 20 g/L of glucose or with SCG (4 g/L, 20 g/L and 40 g/L) as a carbon source. The SCG was collected at a local cafeteria, dried for 24 h at 60 °C, and homogenized using a laboratory homogenizer to obtain a uniform particle size (Precellys Evolution, Bertin Technologies, Montigny le Bretonneux, France) and used to supplement the YP medium. Cultivations were carried out in 100 mL Erlenmeyer flasks with metal caps and semi-globular internal deformations on the bases of the flasks. Precultures were 1:100 diluted into 50 mL of liquid media (described above) and cultivated up to 48 h at 30 °C with agitation (180 rpm). Experiments were performed in three biological replicates (N = 3), except for the cultivation of P. kudriavzevii ZMUM_002 in YP medium, which was performed in 6 (N = 6), and in YP medium supplemented with 20 g/L glucose, which was performed in 10 biological replicates (N = 10).
2.8. Measurement of Optical Density (OD600)
For optical measurements, 1 mL of liquid culture was used. The optical density at 600 nm (OD600) was measured with a spectrophotometer (Biospectrometer, Eppendorf) using 1 cm pathlength cuvettes, blanked with the corresponding sterile growth medium. The OD600 measurements were performed at different time points in CDM supplemented with different carbon sources (glucose, fructose, and sucrose), as well as in YP medium supplemented with glucose. Measurements were performed in three technical replicates (N = 3) for each tested carbon source (glucose, fructose, and sucrose) per yeast strain.
However, no OD600 measurements could be performed for the YP medium supplemented with SCG, due to the black color of SCG.
2.9. Measurement of Wet Cell Weight
To determine the wet cell weight (WCW), 1 mL of the grown yeast culture was transferred to a microcentrifuge tube and centrifuged at 10,000× g for 10 min using a MiniSpin centrifuge (Eppendorf). The supernatant was carefully removed, and the sediment was weighed. The WCW was expressed as the weight of sediment normalized to the sample volume, in grams per liter (g/L). WCW was measured in CDM supplemented with different carbon sources (glucose, fructose, and sucrose) and in YP medium supplemented with glucose and in YP medium with SCG. All measurements were performed in three technical replicates.
2.10. Statistical Analysis
Descriptive statistical analyses—means and standard deviations—were calculated to assess the variability and reliability of the data. To evaluate the statistical significance of determined differences in the WCW, a Student’s t-test was used. The threshold for statistical significance was set at p values of <0.05. The calculations and graphical presentations were obtained in MS Office 365 Excel.
3. Results
3.1. Sampling, Isolation, Purification, and Molecular Identification of the Yeast Strains
In order to isolate and identify P. kudriavzevii with potential for biotechnological applications, we collected a series of samples from different environments. After enrichment and stringent selection of all isolates, only one, isolated from grape pomace compost in Kamnica, Slovenia, was found to be P. kudriavzevii. The found P. kudriavzevii isolate was designated as ZMUM_K002, and its ITS sequence was deposited in GeneBank (Acc. No.: PQ812485). Additional yeast isolates collected from 30 sampling sites included: two isolates of Pichia membranifaciens (Candida valida)—one isolated from a kombucha tank in Kamnik, Slovenia (Acc. No.: PQ812484—strain ZMUM_K001) and one from fermentation broth of a sauerkraut in Velenje, Slovenia (Acc. No.: PQ812490—strain ZMUM_K007); one isolate of Saccharomyces cerevisiae from a discarded wine bottle in Kamnica, Slovenia (Acc. No.: PQ812486—strain ZMUM_K003); two isolates of Candida parapsilosis—one isolated from a household dishwasher in Pernica, Slovenia (Acc. No.: PQ812487—strain ZMUM_K004) and one isolated from a public coffee machine in Maribor, Slovenia (Acc. No.: PQ812488—strain ZMUM_K005); and one isolate of Meyerozyma guilliermondii (Pichia guilliermondii) from the surface of a lemon (from a local grocery store) in Ptuj, Slovenia (Acc. No.: PQ812489—strain ZMUM_K006).
3.2. Morphological Characterization
The morphological characterization of P. kudriavzevii ZMUM_K002 in comparison with S. cerevisiae ZMUM_K003 showed different morphological and growth characteristics of both yeasts (Figure 1). On sporulation media used to induce spore formation and evaluate reproductive traits under nutrient-limiting conditions, P. kudriavzevii ZMUM_K002 showed large, diffuse colonies, while S. cerevisiae ZMUM_K003 formed smooth, compact colonies (Figure 1a,c). Microscopic examination after Ziehl–Neelsen staining reveled a species- and genus-specific spore and cell morphology: P. kudriavzevii ZMUM_K002 formed spherical spores, while S. cerevisiae ZMUM_K003 produced round, smooth-walled spores. In liquid base medium with glucose, the growth dynamics were different; P. kudriavzevii ZMUM_K002 formed a thin, climbing pellicle, while S. cerevisiae ZMUM_K003 formed a sediment on the bottom. Both isolates actively produced gas during growth, confirming their fermentative abilities (Figure 1c,d).
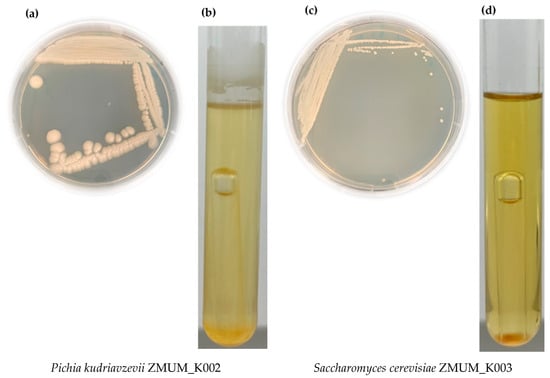
Figure 1.
Morphological comparison of the yeast isolate Pichia kudriavzevii ZMUM_K002 (left panel) in comparison with Saccharomyces cerevisiae ZMUM_K003 (right panel). (a,c) Colony morphology on sporulation media after 120 h at 30 °C. (b,d) Growth in liquid base medium with glucose with a Durham tube after 24 h incubation at 30 °C.
3.3. Fluconazole Susceptibility
In the E-test, the MIC of fluconazole for P. kudriavzevii ZMUM_K002 was 16 mg/L of the fluconazole concentration, causing a fully clear inhibition zone (Figure 2a). The fluconazole MIC for S. cerevisiae ZMUM_K003 could not be determined, as no clear inhibition zone was visible; however, reduced growth could be observed at 2 mg/L (Figure 2b).

Figure 2.
E-test with fluconazole (FL) strips for (a) Pichia kudriavzevii ZMUM_K002 and (b) Saccharomyces cerevisiae ZMUM_K003 on RPMI agar plates. The E-test was performed in triplicate, and the figure shows exemplary results.
3.4. Utilization of Different Carbon Sources
The utilization of different carbon sources was screened (Table 1). We observed growth of P. kudriavzevii ZMUM_K002 in the medium supplemented with glucose, fructose, and mannose, while in medium supplemented with sucrose and glycerol, only partial utilization (blue to green coloration, +/−) was observed. Gas production was observed only in the medium supplemented with glucose and in the medium supplemented with fructose. In comparison, the yeast S. cerevisiae ZMUM_K003 could grow also on galactose, maltose, and starch as a carbon source.

Table 1.
Carbon source utilization and gas production by yeast strains P. kudriavzevii ZMUM_K002 and S. cerevisiae ZMUM_K003. Tests were performed in duplicates. The results were recorded for all carbon sources after 48 h, apart from glycerol, for which the result was recorded after 120 h.
3.5. Growth in Chemically Defined Medium Using Selected Carbon Sources
We tested the growth of P. kudriavzevii ZMUM_K002 using CDM supplemented with three different biotechnologically important carbon sources: glucose, fructose, and sucrose (Figure 3).
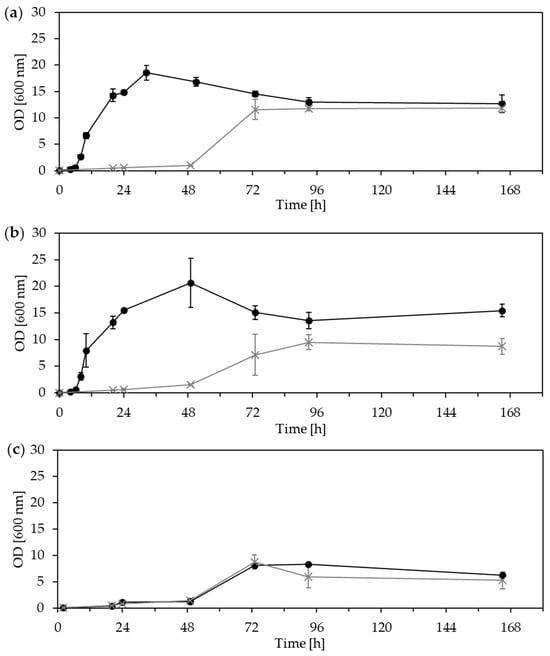
Figure 3.
Growth curves of the yeast isolates Pichia kudriavzevii ZMUM_K002 (black line, ●) and Saccharomyces cerevisiae ZMUM_K003 (gray line, ×) in chemically defined media supplemented by different carbon sources: (a) glucose, 10 g/L; (b) fructose, 10 g/L; (c) sucrose, 10 g/L. The assay was performed in triplicate.
In glucose-supplemented media, both yeast strains demonstrated growth, but the lag phase of P. kudriavzevii ZMUM_K002 was shorter (at about 8 h after inoculation). This strain reached the highest cell accumulation, up to 48 g wet cell weight (WCW) per liter, compared to S. cerevisiae ZMUM_K003, which produced 30% less. Growth patterns in fructose-supplemented media were comparable to those observed with glucose. P. kudriavzevii ZMUM_K002 initiated exponential growth within the first few hours. In contrast, S. cerevisiae ZMUM_K003 exhibited delayed growth requiring two days to reach the exponential phase. In sucrose-supplemented media, both strains showed similar growth patterns, entering exponential growth after about two days. However, the OD600 values were only half as high as when supplemented with glucose or fructose in CDM.
3.6. Growth in YP Medium Supplemented with Glucose or Spent Coffee Grounds
The growth of P. kudriavzevii ZMUM_K002 and S. cerevisiae ZMUM_K003 after 48 h of incubation, expressed as wet cell weight (WCW), varied depending on the supplementation of YP medium (Figure 3). In the absence of a carbon source, both strains exhibited minimal growth, with WCW values of 14.33 ± 2.06 g/L for P. kudriavzevii ZMUM_K002 and 14.53 ± 0.81 g/L for S. cerevisiae ZMUM_K003. The addition of glucose (20 g/L) resulted in the highest growth for both strains, with P. kudriavzevii ZMUM_K002 reaching 49.54 ± 4.21 g/L and S. cerevisiae ZMUM_K003 reaching 33.23 ± 2.46 g/L. When spent coffee ground solution was used as a carbon source, the highest WCW was observed at 40 g/L SCG, with P. kudriavzevii ZMUM_K002 and S. cerevisiae reaching 41.05 ± 2.60 g/L and 34.30 ± 4.80 g/L, respectively. Reducing the SCG concentration by half (to 20 g/L) resulted in moderate growth with WCW values of 27.12 ± 1.20 g/L for P. kudriavzevii ZMUM_K002 and 24.93 ± 1.13 g/L for S. cerevisiae ZMUM_K003, while the lowest SCG concentration (4 g/L) resulted in reduced growth, but not a linear decrease in growth with WCW values of 24.63 ± 1.10 g/L and 22.77 ± 1.83 g/L, respectively. The above stated differences were statistically significant, as shown by Student’s t-test (Figure 4).
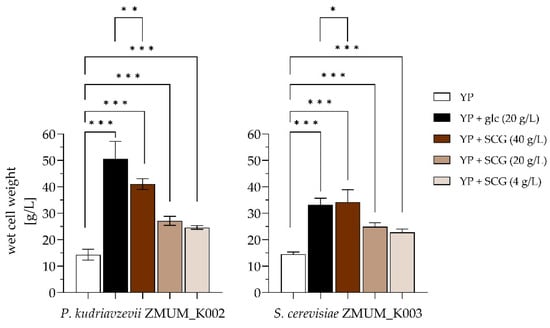
Figure 4.
The wet cell weights (WCWs) of the yeast isolates Pichia kudriavzevii ZMUM_K002 (left) and Saccharomyces cerevisiae ZMUM_K003 (right) were evaluated in complex media containing yeast extract and peptone (YP, white fill) or supplemented as follows: (YP + glc (20 g/L), black fill) YP supplemented with 20 g/L glucose; (YP + SCG (40 g/L), dark brown fill) YP supplemented with 40 g/L spent coffee ground (SCG) solution; (YP + SCG (20 g/L), brown fill) YP supplemented with 20 g/L SCG solution; and (YP + SCG (4 g/L), light brown fill) YP supplemented with 4 g/L SCG solution. The biomass yield (WCW) of each condition is presented as the mean ± standard deviation. Experiments were carried out in at least triplicate. The p-value (Student’s t-test) is indicated by three asterisks (***), if p < 0.001; by two asterisks (**), if p < 0.01; and by one asterisk (*), if p < 0.05.
4. Discussion
Environmental yeast isolates often exhibit unique physiological and metabolic adaptations that distinguish them from food-derived and clinical strains. It is believed we can harness new biotechnological agents from different environments for usage in sustainable and cost-effective industrial processes. In this study, we investigated the biotechnological potential of Pichia kudriavzevii, strain ZMUM_K002, from grape pomace compost in comparison to Saccharomyces cerevisiae, strain ZMUM_K003, isolated from a discarded wine bottle.
The yeast P. kudriavzevii demonstrates strong adaptability to harsh conditions, including extreme pH, elevated temperatures, high osmotic pressure, and fermentation inhibitors, making it a promising candidate for various biotechnological applications [15]. Alas, it is known to have intrinsic fluconazole resistance and is classified as a third-ranked, medium-priority fungal pathogen capable of causing mucosal infections or invasive candidiasis, which could hinder its usage [19]. Using E-tests, we observed that P. kudriavzevii ZMUM_K002 exhibited a fluconazole MIC of 16 mg/L. The clinical cut-off MIC for fluconazole is set at 128 mg/L of fluconazole [35], and hence the P. kudriavzevii ZMUM_K002 can be considered more fluconazole-susceptible as clinical samples. Interestingly, Douglass et al. reported that P. kudriavzevii strains isolated from food showed similar resistance to fluconazole as strains from clinical samples [13], so the P. kudriavzevii ZMUM_K002 can be considered more fluconazole-susceptible as natural food samples. However, as the yeast P. kudriavzevii is a potential pathogen, especially to immunocompromised people in some geographic regions [20], prior to its introduction into biotechnological processes, an in vivo assessment of virulence traits is required, for example with assays on larvae of Tenebrio molitor or Galleria mellonella [36,37] or human cell cultures [38].
In the context of modern industrial sustainability, it is becoming increasingly important to develop processes that utilize efficient and sustainable sources for the production of biomass, including waste materials and by-products. To address this challenge, we conducted experiments to evaluate the utilization of different carbon sources and their corresponding effects on the growth of yeast cells. In all growth experiments, the growth performance of P. kudriavzevii ZMUM_K002 was compared with S. cerevisiae ZMUM_K003, and the main objective was to determine whether P. kudriavzevii showed better growth characteristics.
We selected different carbon sources to represent a range of monosaccharides, disaccharides, polysaccharides, and polyols to provide insight into the utilization capabilities and carbon source preferences of the yeast strain. Our results showed that glucose, fructose, mannose, and sucrose are best fitted as carbon sources.
In growth experiments, using CDM supplemented with different biotechnology commonly used carbon sources (glucose, fructose, and sucrose), and the effects of these carbon sources on the growth of P. kudriavzevii ZMUM_K002 were revealed. The maximum OD600 in CDM experiments was reached after approx. 48 h for all tested carbon sources. In glucose-enriched media, P. kudriavzevii ZMUM_K002 exhibited a shorter lag phase (8 h) compared to S. cerevisiae ZMUM_K003 (24 h). Fructose-enriched media showed similar trends, with P. kudriavzevii ZMUM_K002 rapidly initiating exponential growth, while S. cerevisiae ZMUM_K003 required 48 h for initiating the lag phase. Both strains utilized sucrose less efficiently, with OD600 values approximately 50% lower than for glucose or fructose.
However, we were interested in the utilization of several other carbohydrates, as they may be found in agro-industrial by-products or disposable agricultural wastes and hence can be potentially utilized in line with the principles of circular economy and waste valorization. Galactose utilization was not observed for P. kudriavzevii ZMUM_K002. However, improved galactose utilization in yeasts can also be achieved via metabolic engineering [5], as efficient galactose metabolism is important for the utilization of some agro-industrial residues. Numerous agro-industrial by-products or wastes (e.g., sugar cane and soy molasses) are rich in galactose, which makes them valuable components for utilization by yeasts [39].
Our results showed that P. kudriavzevii ZMUM_K002 also cannot grow on xylose, which is unfortunate, as xylose is a promising carbon source as it is commonly found in lignocellulosic materials. However, it has been reported that certain yeast strains can grow on xylose when lignocellulosic substrates pretreated with acid at 210 °C were added in the grown media [40,41]. Testing the ZMUM_K002 strain for xylose utilization from preheated lignocellulosic substrates in future studies could provide valuable insights.
Partial utilization of glycerol was observed. The assimilation of glycerol by yeasts is of great interest as it is an important by-product in the production of biodiesel, offering potential for biotechnological utilization [42]. As reported by Díaz-Nava et al. for P. kudriavzevii strain ITV-S42 [43], and also our strain ZMUM_002, showed utilization of glycerol but was unable to ferment it. However, further studies on glycerol utilization are needed, as glycerol is a main by-product in bioethanol production [44]. Gallardo et al. have shown that certain P. kudriavzevii strains can effectively utilize glycerol at elevated temperatures (up to 40 °C) [45]. However, this was not investigated in our study, as we prioritized the economic consideration of maintaining lower temperatures in bioprocesses, which we consider more practical for industrial applications. The observation that most S. cerevisiae strains can grow on glycerol as the sole carbon source only when supplemented with complex additives, such as yeast extract or amino acid mixtures [46], and that preadaptation in glucose–glycerol media could activate a specific low-affinity transport system [47,48], provides ground knowledge for the optimization of glycerol utilization in P. kudriavzevii also. These findings will guide our further research to improve the efficiency of glycerol utilization in this species.
To extend the cultivation of P. kudriavzevii ZMUM_K002 beyond CDM, complex YP medium was used, as complex media formulations can support robust yeast growth by providing a balanced nutrient profile [49,50]. Supplementing the YP medium with glucose increased the production of biomass, and the highest OD600 was reached in 48 h. P. kudriavzevii ZMUM_K002 achieved higher biomass (48 g/L WCW) than S. cerevisiae ZMUM_K003, outperforming S. cerevisiae ZMUM_K003 by 30%. Since the addition of glucose in a biotechnological process is not economically viable, while the use of disposable agricultural waste is economically feasible and also contributes to sustainability through the reuse of agro-industrial waste, we explored the effect of supplementing the YP medium with a disposable agricultural waste product—the SCG [25]. Our results revealed a slight decrease in the biomass concentration of P. kudriavzevii ZMUM_K002, when grown in YP medium supplemented with 40 g/L SCG compared to YP medium with 20 g/L glucose. However, when the biomass concentration obtained in YP medium supplemented with 40 g/L SCG was compared to the biomass concentration (determined by WCW) obtained in YP medium with no supplement, the performance of P. kudriavzevii ZMUM_K002 was significant, approximately a 3-fold increase. Moreover, P. kudriavzevii showed remarkable tolerance to SCG-derived inhibitory compounds. These results position P. kudriavzevii ZMUM_K002 as a new potential biotechnological agent and SCG as a potential sustainable replacement for glucose in the microbial medium for industrial biotechnological applications and sustainable production.
5. Conclusions
The quest of our study was based on principles of modern industrial biotechnology and aimed to improve resource efficiency. Our study revealed the potential of P. kudriavzevii ZMUM_K002 for efficient biomass production in biotechnological applications. Successful supplementation of YP medium with SCG, a low-cost, nutrient-rich substrate, underlined the strain’s compatibility with sustainable biotechnological practices. Future research should further investigate the applicability of P. kudriavzevii in various biotechnological processes, paving the way for sustainable and innovative solutions in industrial microbiology.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/applmicrobiol5010030/s1, Table S1: detailed composition of the salt stock solution and the trace element stock solution used for preparation of chemically defined media; Table S2: raw OD600 measurements for Pichia kudriavzevii ZMUM_K002, grown in chemically defined media (CDM) supplemented with different carbon sources; Table S3: raw OD600 measurements for Saccharomyces cerevisiae ZMUM_K003, grown in chemically defined media (CDM) supplemented with different carbon sources.
Author Contributions
T.V.: Conceptualization, Methodology, Validation, and Writing—original draft. M.S.E.: Funding acquisition, Conceptualization, Supervision, and Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The study was financially supported by the project “Development of Research Infrastructure For The International Competitiveness Of The Slovenian RRI Space- RI-SI -LifeWatch”, co-financed by the Republic of Slovenia, the Ministry of Education, Science and Sport and the European Union from the European Regional Development Fund.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author (M.S.E.).
Acknowledgments
We would like to acknowledge Enja Hribernik, Zala Svetec, Nick Leben, Ina Slemnik, and Tajda Bratec for their help in performing experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Segal-Kischinevzky, C.; Romero-Aguilar, L.; Alcaraz, L.D.; López-Ortiz, G.; Martínez-Castillo, B.; Torres-Ramírez, N.; Sandoval, G.; González, J. Yeasts Inhabiting Extreme Environments and Their Biotechnological Applications. Microorganisms 2022, 10, 794. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.K.; Srivastava, R.K. A Review on Sustainable Yeast Biotechnological Processes and Applications. Microbiol. Res. 2018, 207, 83–90. [Google Scholar] [CrossRef]
- Martinić Cezar, T.; Marđetko, N.; Trontel, A.; Paić, A.; Slavica, A.; Teparić, R.; Žunar, B. Engineering Saccharomyces cerevisiae for the Production of Natural Osmolyte Glucosyl Glycerol from Sucrose and Glycerol through Ccw12-Based Surface Display of Sucrose Phosphorylase. J. Biol. Eng. 2024, 18, 69. [Google Scholar] [CrossRef] [PubMed]
- Procópio, D.P.; Lee, J.W.; Shin, J.; Tramontina, R.; Ávila, P.F.; Brenelli, L.B.; Squina, F.M.; Damasio, A.; Rabelo, S.C.; Goldbeck, R.; et al. Metabolic Engineering of Saccharomyces cerevisiae for Second-Generation Ethanol Production from Xylo-Oligosaccharides and Acetate. Sci. Rep. 2023, 13, 19182. [Google Scholar] [CrossRef]
- Lee, K.S.; Hong, M.E.; Jung, S.C.; Ha, S.J.; Yu, B.J.; Koo, H.M.; Park, S.M.; Seo, J.H.; Kweon, D.H.; Park, J.C.; et al. Improved Galactose Fermentation of Saccharomyces cerevisiae through Inverse Metabolic Engineering. Biotechnol. Bioeng. 2011, 108, 621–631. [Google Scholar] [CrossRef]
- Goddard, M.R.; Greig, D. Saccharomyces cerevisiae: A Nomadic Yeast with No Niche? FEMS Yeast Res. 2015, 15, fov009. [Google Scholar] [CrossRef]
- Ostergaard, S.; Olsson, L.; Nielsen, J. Metabolic Engineering of Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2000, 64, 34–50. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Santos, A.; Van Wyk, N.; Pretorius, I.S. Saccharomyces Cerevisiae. Trends Genet. 2019, 35, 956–957. [Google Scholar] [CrossRef]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and Its Industrial Applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Hampsey, M. A Review of Phenotypes in Saccharomyces cerevisiae. Yeast 1997, 13, 1099–1133. [Google Scholar] [CrossRef]
- Schnierda, T.; Bauer, F.F.; Divol, B.; van Rensburg, E.; Görgens, J.F. Optimization of Carbon and Nitrogen Medium Components for Biomass Production Using Non-Saccharomyces Wine Yeasts. Lett. Appl. Microbiol. 2014, 58, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Tiago, F.C.P.; Martins, F.S.; Rosa, C.A.; Nardi, R.M.D.; Cara, D.C.; Nicoli, J.R. Physiological Characterization of Non-Saccharomyces Yeasts from Agro-Industrial and Environmental Origins with Possible Probiotic Function. World J. Microbiol. Biotechnol. 2009, 25, 657–666. [Google Scholar] [CrossRef]
- Douglass, A.P.; Offei, B.; Braun-Galleani, S.; Coughlan, A.Y.; Martos, A.A.R.; Ortiz-Merino, R.A.; Byrne, K.P.; Wolfe, K.H. Population Genomics Shows No Distinction between Pathogenic Candida krusei and Environmental Pichia kudriavzevii: One Species, Four Names. PLoS Pathog. 2018, 14, e1007138. [Google Scholar] [CrossRef] [PubMed]
- Kurtzman, C.P.; Robnett, C.J.; Basehoar-Powers, E. Phylogenetic Relationships among Species of Pichia, Issatchenkia and Williopsis Determined from Multigene Sequence Analysis, and the Proposal of Barnettozyma Gen. Nov., Lindnera Gen. Nov. and Wickerhamomyces Gen. Nov. FEMS Yeast Res. 2008, 8, 939–954. [Google Scholar] [CrossRef]
- Chu, Y.; Li, M.; Jin, J.; Dong, X.; Xu, K.; Jin, L.; Qiao, Y.; Ji, H. Advances in the Application of the Non-Conventional Yeast Pichia kudriavzevii in Food and Biotechnology Industries. J. Fungi 2023, 9, 170. [Google Scholar] [CrossRef]
- Jiang, D.; Wang, M.; Zhao, X.; Lu, X.; Zong, H.; Zhuge, B. Glycerol Production from Undetoxified Lignocellulose Hydrolysate by a Multiresistant Engineered Candida glycerinogenes. J. Agric. Food Chem. 2024, 72, 1630–1639. [Google Scholar] [CrossRef]
- Xi, Y.; Zhan, T.; Xu, H.; Chen, J.; Bi, C.; Fan, F.; Zhang, X. Characterization of JEN Family Carboxylate Transporters from the Acid-Tolerant Yeast Pichia kudriavzevii and Their Applications in Succinic Acid Production. Microb. Biotechnol. 2021, 14, 1130–1147. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species from 1997–2016. Open Forum Infect. Dis. 2019, 6, S79–S94. [Google Scholar] [CrossRef]
- World Health Organization (WHO). WHO Fungal Priority Pathogens List to Guide Research, Development and Public. Health Action; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Nguyen, T.A.; Kim, H.Y.; Stocker, S.; Kidd, S.; Alastruey-Izquierdo, A.; Dao, A.; Harrison, T.; Wahyuningsih, R.; Rickerts, V.; Perfect, J.; et al. Pichia kudriavzevii (Candida krusei): A Systematic Review to Inform the World Health Organisation Priority List of Fungal Pathogens. Med. Mycol. 2024, 62, myad132. [Google Scholar] [CrossRef]
- Sokchea, H.; Thi Hang, P.; Dinh Phung, L.; Duc Ngoan, L.; Thu Hong, T.; Borin, K. Effect of Time, C/N Ratio and Molasses Concentration on Saccharomyces cerevisiae Biomass Production. J. Vet. Anim. Res. 2018, 1, 1–7. [Google Scholar] [CrossRef]
- Ferndahl, C.; Bonander, N.; Logez, C.; Wagner, R.; Gustafsson, L.; Larsson, C.; Hedfalk, K.; Darby, R.A.; Bill, R.M. Increasing Cell Biomass in Saccharomyces cerevisiae Increases Recombinant Protein Yield: The Use of a Respiratory Strain as a Microbial Cell Factory. Microb. Cell Fact. 2010, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, J.; Jiang, H. Microbial Production of Value-Added Bioproducts and Enzymes from Molasses, a by-Product of Sugar Industry. Food Chem. 2021, 346, 128860. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, G.M.; Doran-Peterson, J. A Strain of Saccharomyces cerevisiae Evolved for Fermentation of Lignocellulosic Biomass Displays Improved Growth and Fermentative Ability in High Solids Concentrations and in the Presence of Inhibitory Compounds. Biotechnol. Biofuels 2011, 4, 49. [Google Scholar] [CrossRef]
- Ballesteros, L.F.; Teixeira, J.A.; Mussatto, S.I. Chemical, Functional, and Structural Properties of Spent Coffee Grounds and Coffee Silverskin. Food Bioprocess. Technol. 2014, 7, 3493–3503. [Google Scholar] [CrossRef]
- Calheiros, D.; Dias, M.I.; Calhelha, R.C.; Barros, L.; Ferreira, I.C.F.R.; Fernandes, C.; Gonçalves, T. Antifungal Activity of Spent Coffee Ground Extracts. Microorganisms 2023, 11, 242. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Gabriel, C.; Cerqueira, F.; Manso, M.C.; Vinha, A.F. Coffee Industrial Waste as a Natural Source of Bioactive Compounds with Antibacterial and Antifungal Activities. In The Battle Against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; FORMATEX: Guadalajara, Mexico, 2015; pp. 131–136. [Google Scholar]
- Makimura, K.; Mochizuki, T.; Hasegawa, A.; Uchida, K.; Saito, H.; Yamaguchi, H. Phylogenetic Classification of Trichophyton Mentagrophytes Complex Strains Based on DNA Sequences of Nuclear Ribosomal Internal Transcribed Spacer 1 Regions. J. Clin. Microbiol. 1998, 36, 2629–2633. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Neiman, A.M. Sporulation in the Budding Yeast Saccharomyces cerevisiae. Genetics 2011, 189, 737–765. [Google Scholar] [CrossRef]
- Arendrup, M.C.; Friberg, N.; Mares, M.; Kahlmeter, G.; Meletiadis, J.; Guinea, J.; Andersen, C.T.; Arikan-Akdagli, S.; Barchiesi, F.; Chryssanthou, E.; et al. How to Interpret MICs of Antifungal Compounds According to the Revised Clinical Breakpoints v. 10.0 European Committee on Antimicrobial Susceptibility Testing (EUCAST). Clin. Microbiol. Infect. 2020, 26, 1464–1472. [Google Scholar] [CrossRef]
- Rodriquez Tudela, J.L.; Donnelly, J.P.; Arendrup, M.C.; Arikan, S.; Barchiesi, F.; Bille, J.; Chryssanthou, E.; Cuenca-Estrella, M.; Dannaoui, E.; Denning, D.; et al. EUCAST Technical Note on Fluconazole: The European Committee on Antimicrobial Susceptibility Testing—Subcommittee on Antifungal Susceptibility Testing (EUCAST-AFST). Clin. Microbiol. Infect. 2008, 14, 193–195. [Google Scholar] [CrossRef][Green Version]
- Cuesta, I.; Bielza, C.; Cuenca-Estrella, M.; Larrañaga, P.; Rodríguez-Tudela, J.L. Evaluation by Data Mining Techniques of Fluconazole Breakpoints Established by the Clinical and Laboratory Standards Institute (CLSI) and Comparison with Those of the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Antimicrob. Agents Chemother. 2010, 54, 1541–1546. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, I.; Bielza, C.; Larrañaga, P.; Cuenca-Estrella, M.; de Laguna, F.B.; Rodríguez-Pardo, D.; Almirante, B.; Pahissa, A.; Rodríguez-Tudela, J.L. Data Mining Validation of Fluconazole Breakpoints Established by the European Committee on Antimicrobial Susceptibility Testing. Antimicrob. Agents Chemother. 2009, 53, 2949–2954. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.R.; Verweij, P.E.; Castanheira, M.; Dannaoui, E.; White, P.L.; Arendrup, M.C. Molecular Mechanisms of Acquired Antifungal Drug Resistance in Principal Fungal Pathogens and EUCAST Guidance for Their Laboratory Detection and Clinical Implications. J. Antimicrob. Chemother. 2022, 77, 2053–2073. [Google Scholar] [CrossRef] [PubMed]
- De Souza, P.C.; Morey, A.T.; Castanheira, G.M.; Bocate, K.P.; Panagio, L.A.; Ito, F.A.; Furlaneto, M.C.; Yamada-Ogatta, S.F.; Costa, I.N.; Mora-Montes, H.M.; et al. Tenebrio Molitor (Coleoptera: Tenebrionidae) as an Alternative Host to Study Fungal Infections. J. Microbiol. Methods 2015, 118, 182–186. [Google Scholar] [CrossRef]
- Wojda, I.; Staniec, B.; Sułek, M.; Kordaczuk, J. The Greater Wax Moth Galleria Mellonella: Biology and Use in Immune Studies. Pathog. Dis. 2020, 78, ftaa057. [Google Scholar] [CrossRef]
- Last, A.; Maurer, M.; Mosig, A.S.; Gresnigt, M.S.; Hube, B. In Vitro Infection Models to Study Fungal-Host Interactions. FEMS Microbiol. Rev. 2021, 45, fuab005. [Google Scholar] [CrossRef]
- Wang, Z.P.; Zhang, X.Y.; Ma, Y.; Ye, J.R.; Jiang, J.; Wang, H.Y.; Chen, W. Whole Conversion of Agro-Industrial Wastes Rich in Galactose-Based Carbohydrates into Lipid Using Oleaginous Yeast Aureobasidium namibiae. Biotechnol. Biofuels 2021, 14, 181. [Google Scholar] [CrossRef]
- Behera, S.; Arora, R.; Nandhagopal, N.; Kumar, S. Importance of Chemical Pretreatment for Bioconversion of Lignocellulosic Biomass. Renew. Sustain. Energy Rev. 2014, 36, 91–106. [Google Scholar] [CrossRef]
- Nweze, J.E.; Ndubuisi, I.; Murata, Y.; Omae, H.; Ogbonna, J.C. Isolation and Evaluation of Xylose-Fermenting Thermotolerant Yeasts for Bioethanol Production. Biofuels 2021, 12, 961–970. [Google Scholar] [CrossRef]
- Dobson, R.; Gray, V.; Rumbold, K. Microbial Utilization of Crude Glycerol for the Production of Value-Added Products. J. Ind. Microbiol. Biotechnol. 2012, 39, 217–226. [Google Scholar] [CrossRef]
- Díaz-Nava, L.E.; Montes-Garcia, N.; Domínguez, J.M.; Aguilar-Uscanga, M.G. Effect of Carbon Sources on the Growth and Ethanol Production of Native Yeast Pichia kudriavzevii ITV-S42 Isolated from Sweet Sorghum Juice. Bioprocess. Biosyst. Eng. 2017, 40, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, M.; Eraky, M.; Osman, A.I.; Wang, J.; Farghali, M.; Rashwan, A.K.; Yacoub, I.H.; Hanelt, D.; Abomohra, A. Sustainable Valorization of Waste Glycerol into Bioethanol and Biodiesel through Biocircular Approaches: A Review. Environ. Chem. Lett. 2024, 22, 609–634. [Google Scholar] [CrossRef]
- Gallardo, J.C.M.; Souza, C.S.; Cicarelli, R.M.B.; Oliveira, K.F.; Morais, M.R.; Laluce, C. Enrichment of a Continuous Culture of Saccharomyces cerevisiae with the Yeast Issatchenkia Orientalis in the Production of Ethanol at Increasing Temperatures. J. Ind. Microbiol. Biotechnol. 2011, 38, 405–414. [Google Scholar] [CrossRef]
- Merico, A.; Ragni, E.; Galafassi, S.; Popolo, L.; Compagno, C. Generation of an Evolved Saccharomyces cerevisiae Strain with a High Freeze Tolerance and an Improved Ability to Grow on Glycerol. J. Ind. Microbiol. Biotechnol. 2011, 38, 1037–1044. [Google Scholar] [CrossRef]
- Lages, F.; Lucas, C. Contribution to the Physiological Characterization of Glycerol Active Uptake in Saccharomyces cerevisiae. Biochim. Biophys. Acta (BBA)-Bioenerg. 1997, 1322, 8–18. [Google Scholar] [CrossRef]
- Oliveira, R.; Lucas, C. Expression Studies of GUP1 and GUP2, Genes Involved in Glycerol Active Transport in Saccharomyces cerevisiae, Using Semi-Quantitative RT-PCR. Curr. Genet. 2004, 46, 140–146. [Google Scholar] [CrossRef]
- Zhang, J.; Reddy, J.; Buckland, B.; Greasham, R. Toward Consistent and Productive Complex Media for Industrial Fermentations: Studies on Yeast Extract for a Recombinant Yeast Fermentation Process. Biotechnol. Bioeng. 2003, 82, 640–652. [Google Scholar] [CrossRef]
- Abelovska, L.; Bujdos, M.; Kubova, J.; Petrezselyova, S.; Nosek, J.; Tomaska, L. Comparison of Element Levels in Minimal and Complex Yeast Media. Can. J. Microbiol. 2007, 53, 533–535. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).