De Novo Leaf Transcriptome Assembly and Metagenomic Studies of Coast Live Oak (Quercus agrifolia)
Abstract
1. Introduction
2. Materials and Methods
2.1. Field Sampling and Logistics
2.2. RNA Extraction and Sequencing
2.3. De Novo Assembly, Assessment, and Quantification of mRNA Transcripts
2.4. Soil DNA Extraction
2.5. 16S Microbial Community Diversity Analysis
2.6. Soil Shotgun Metagenomics
3. Results
3.1. Plant Transcriptome Analysis
3.2. Microbial Community Profiling
4. Discussion
4.1. Transcriptome Analysis
4.2. Bacterial Community Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Calscape. Coast Live Oak. California Native Plant Society. 2024. Available online: https://calscape.org/Quercus-agrifolia-(Coast-Live-Oak) (accessed on 14 October 2024).
- Senn, S.; Bhattacharyya, S.; Presley, G.; Taylor, A.E.; Nash, B.; Enke, R.A.; Barnard-Kubow, K.B.; Ford, J.; Jasinski, B.; Badalova, Y. The Functional Biogeography of eDNA Metacommunities in the Post-Fire Landscape of the Angeles National Forest. Microorganisms 2022, 10, 1218. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chris, L. Sudden Oak Death and Fire in California. 2009. Available online: https://www.suddenoakdeath.org/wp-content/uploads/pdf/summary%20of%20fire%20and%20p%20ramorum%20issues%20v5.3.pdf (accessed on 18 October 2024).
- McPherson, B.A.; Mori, S.R.; Wood, D.L.; Storer, A.J.; Svihra, P.; Kelly, N.M.; Standiford, R.B. Sudden oak death in California: Disease progression in oaks and tanoaks. For. Ecol. Manag. 2005, 213, 71–89. [Google Scholar] [CrossRef]
- McPherson, B.A.; Mori, S.R.; Wood, D.L.; Kelly, M.; Storer, A.J.; Svihra, P.; Standiford, R.B. Responses of oaks and tanoaks to the sudden oak death pathogen after 8y of monitoring in two coastal California forests. For. Ecol. Manag. 2010, 259, 2248–2255. [Google Scholar] [CrossRef]
- Parke, J.L.; Rizzo, D.M. Phytophthora ramorum. For. Phytophthoras 2011, 1, 10. [Google Scholar] [CrossRef]
- CEC-500-2012-026; Fire and Climate Change in California. California Energy Commission: Sacramento, CA, USA, 2012.
- Pang, Z.; Chen, J.; Wang, T.; Gao, C.; Li, Z.; Guo, L.; Xu, J.; Cheng, Y. Linking plant secondary metabolites and plant microbiomes: A review. Front. Plant Sci. 2021, 12, 621276. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, Z. State-of-the-Art RNA Sequencing for Drug Discovery. White Paper, BGI Global Genomics Services. 31 May 2018. Available online: www.bgi.com/wp-content/uploads/sites/2/2018/06/BGI-RNA-Transcriptome-Public-white-paper.pdf (accessed on 8 November 2024).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience 2018, 7, gix120. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Bryant, D.M.; Johnson, K.; DiTommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.-H.; Davis, F.G.; et al. A Tissue-Mapped Axolotl De Novo Transcriptome Enables Identification of Limb Regeneration Factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [PubMed]
- Bairoch, A.; Apweiler, R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000, 28, 45–48. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764, Erratum in Bioinformatics 2008, 24, 2942. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Hall, M.; Beiko, R.G. 16S rRNA gene analysis with QIIME2. In Microbiome Analysis: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 113–129. [Google Scholar]
- Hilgert, U.; McKay, S.; Khalfan, M.; Williams, J.; Ghiban, C.; Micklos, D. DNA Subway: Making Genome Analysis Egalitarian. In Proceedings of the 2014 Annual Conference Extreme Science and Engineering Discovery Environment (XSEDE ’14), Association for Computing Machinery, Atlanta, GA, USA, 13–18 July 2014; Article 70. pp. 1–3. [Google Scholar]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Baeza, Y.; Pirrung, M.; Gonzalez, A.; Knight, R. EMPeror: A tool for visualizing high throughput microbial community data. GigaScience 2013, 2, 16. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.G.; Doan, D.; Subramanian, P.; Quiñones, M.; Dolan, M.; Hurt, D.E. WGSA2 Workflow—A Tutorial. 2023. Available online: https://www.protocols.io/view/wgsa2-workflow-a-tutorial-n92ldm98xl5b/v1 (accessed on 23 October 2024).
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Arndt, D.; Xia, J.; Liu, Y.; Zhou, Y.; Guo, A.C.; Cruz, J.A.; Sinelnikov, I.; Budwill, K.; Nesbø, C.L.; Wishart, D.S. METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Res. 2012, 40, W88–W95. [Google Scholar] [CrossRef] [PubMed]
- Dodd, R.; Marimuthu, M.P.A.; Nguyen, O.; Fairbairn, C.W.; Seligmann, W.E.; Escalona, M.; Shaffer, H.B. Whole Genome Shotgun Sequence of Q. agrifolia. Available online: https://www.ncbi.nlm.nih.gov/nucleotide/JARQAE010000006.1 (accessed on 14 November 2024).
- Beals, K.K.; Scearce, A.E.; Swystun, A.T.; Schweitzer, J.A. Belowground mechanisms for oak regeneration: Interactions among fire, soil microbes, and plant community alter oak seedling growth. For. Ecol. Manag. 2022, 503, 119774. [Google Scholar] [CrossRef]
- António, C.B.S.; Obieze, C.; Jacinto, J.; Maquia, I.S.A.; Massad, T.; Ramalho, J.C.; Ribeiro, N.S.; Máguas, C.; Marques, I.; Ribeiro-Barros, A.I. Linking Bacterial Rhizosphere Communities of Two Pioneer Species, Brachystegia boehmii and B. spiciformis, to the Ecological Processes of Miombo Woodlands. Forests 2022, 13, 1840. [Google Scholar] [CrossRef]
- Hart, S.C.; DeLuca, T.H.; Newman, G.S.; MacKenzie, M.D.; Boyle, S.I. Post-fire vegetative dynamics as drivers of microbial community structure and function in forest soils. For. Ecol. Manag. 2005, 220, 166–184. [Google Scholar] [CrossRef]
- Zhang, X.-C.; Gassmann, W. Alternative Splicing and mRNA Levels of the Disease Resistance Gene RPS4 Are Induced during Defense Responses. Plant Physiol. 2007, 145, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Gunjan, S.; Xie, C.H.; Littleton, J.; Yuan, L. Promoter analysis of the Catharanthus roseus geraniol 10-hydroxylase gene involved in terpenoid indole alkaloid biosynthesis. Biochim. Biophys. Acta-Gene Struct. Expr. 2007, 1769, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Collu, G.; Unver, N.; Peltenburg-Looman, A.M.; van der Heijden, R.; Verpoorte, R.; Memelink, J. Geraniol 10-hydroxylase1, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 2001, 508, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Frezza, C.; de Vita, D.; Toniolo, C.; Ventrone, A.; Tomassini, L.; Foddai, S.; Nicoletti, M.; Guiso, M.; Bianco, A.; Serafini, M. Harpagide: Occurrence in plants and biological activities—A review. Fitoterapia 2020, 147, 104764. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M.; Ghassemian, M. Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol. Biol. 2003, 51, 925–948. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Asami, T.; Wu, X.; Tsang, E.W.; Cutler, A.J. A putative hydroxysteroid dehydrogenase involved in regulating plant growth and development. Plant Physiol. 2007, 145, 87–97. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ebadzad, G.; Cravador, A. Quantitative RT-PCR analysis of differentially expressed genes in Quercus suber in response to Phytophthora cinnamomi infection. SpringerPlus 2014, 3, 613. [Google Scholar] [CrossRef] [PubMed] [PubMed Central][Green Version]
- Varma, R.S.; George, K.J.; Balaji, S.; Parthasarathy, V. Differential induction of chitinase in Piper colubrinum in response to inoculation with Phytophthora capsici, the cause of foot rot in black pepper. Saudi J. Biol. Sci. 2009, 16, 11–16. [Google Scholar] [CrossRef][Green Version]
- Malolepszy, A.; Kelly, S.; Sørensen, K.K.; James, E.K.; Kalisch, C.; Bozsoki, Z.; Panting, M.; Andersen, S.U.; Sato, S.; Tao, K.; et al. A plant chitinase controls cortical infection thread progression and nitrogen-fixing symbiosis. eLife 2018, 7, e38874. [Google Scholar] [CrossRef] [PubMed]
- Santos-Sánchez, N.F.; Salas-Coronado, R.; Hernández-Carlos, B.; Villanueva-Cañongo, C. Shikimic acid pathway in biosynthesis of phenolic compounds. In Plant Physiological Aspects of Phenolic Compounds; IntechOpen: Rijeka, Croatia, 2019; Volume 1, pp. 1–15. [Google Scholar]
- Albuquerque, T.G.; Nunes, M.A.; Bessada, S.M.; Costa, H.S.; Oliveira, M.B.P. Biologically active and health promoting food components of nuts, oilseeds, fruits, vegetables, cereals, and legumes. In Chemical Analysis of Food; Academic Press: Cambridge, MA, USA, 2020; pp. 609–656. [Google Scholar]
- Muir, R.M. Analysis of Gallic Acid Production by the Bi-Functional Enzyme Shikimate-5-Dehydrogenase in Higher Plants and Bacteria; University of California: Davis, CA, USA, 2005. [Google Scholar]
- Youn, B.; Moinuddin, S.G.; Davin, L.B.; Lewis, N.G.; Kang, C. Crystal Structures of Apo-form and Binary/Ternary Complexes of Podophyllum secoisolariciresinol dehydrogenase, an Enzyme Involved in Formation of Health-protecting and Plant Defense Lignans. J. Biol. Chem. 2005, 280, 12917–12926. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Geron, C.; Daly, R.; Harley, P.; Rasmussen, R.; Seco, R.; Guenther, A.; Karl, T.; Gu, L. Large drought-induced variations in oak leaf volatile organic compound emissions during PINOT NOIR 2012. Chemosphere 2016, 146, 8–21. [Google Scholar] [CrossRef]
- Lavoir, A.; Staudt, M.; Schnitzler, J.; Landais, D.; Massol, F.; Rocheteau, A.; Rodríguez, R.; Zimmer, I.; Rambal, S. Drought reduced monoterpene emissions from Quercus ilex trees: Results from a throughfall displacement experiment within a forest ecosystem. Biogeosci. Discuss. 2009, 6, 863–893. [Google Scholar]
- Staudt, M.; Mir, C.; Joffre, R.; Rambal, S.; Bonin, A.; Landais, D.; Lumaret, R. Isoprenoid emissions of Quercus spp. (Q. suber and Q. ilex) in mixed stands contrasting in interspecific genetic introgression. New Phytol. 2004, 163, 573–584. [Google Scholar] [CrossRef]
- Connell, S.R.; Tracz, D.M.; Nierhaus, K.H.; Taylor, D.E. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 2003, 47, 3675–3681. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Crouzet, J.; Trombik, T.; Fraysse, Å.S.; Boutry, M. Organization and function of the plant pleiotropic drug resistance ABC transporter family. FEBS Lett. 2006, 580, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Kegg Pathway qlo00710. Carbon Fixation by Calvin Cycle—Quercus Lobata (Valley Oak). Available online: https://www.kegg.jp/entry/pathway+qlo00710 (accessed on 10 November 2024).
- Tarkka, M.T.; Herrmann, S.; Wubet, T.; Feldhahn, L.; Recht, S.; Kurth, F.; Mailänder, S.; Bönn, M.; Neef, M.; Angay, O.; et al. OakContig DF 159.1, a reference library for studying differential gene expression in Quercus robur during controlled biotic interactions: Use for quantitative transcriptomic profiling of oak roots in ectomycorrhizal symbiosis. New Phytol. 2013, 199, 529–540. [Google Scholar] [CrossRef]
- Escandón, M.; Castillejo, M.Á.; Jorrín-Novo, J.V.; Rey, M.-D. Molecular Research on Stress Responses in Quercus spp.: From Classical Biochemistry to Systems Biology through Omics Analysis. Forests 2021, 12, 364. [Google Scholar] [CrossRef]
- Zirkle, R.; Ligon, J.M.; MolnáR, I. Cloning, sequence analysis and disruption of the mglA gene involved in swarming motility of Sorangium cellulosum So ce26, a producer of the antifungal polyketide antibiotic soraphen A. J. Biosci. Bioeng. 2004, 97, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A. Isocoumarins, miraculous natural products blessed with diverse pharmacological activities. Eur. J. Med. Chem. 2016, 116, 290–317. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 2019, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Morais, M.C.; Mucha, Â.; Ferreira, H.; Gonçalves, B.; Bacelar, E.; Marques, G. Comparative study of plant growth-promoting bacteria on the physiology, growth and fruit quality of strawberry. J. Sci. Food Agric. 2019, 99, 5341–5349. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef]
- Shah, A.; Smith, D.L. Flavonoids in Agriculture: Chemistry and Roles in, Biotic and Abiotic Stress Responses, and Microbial Associations. Agronomy 2020, 10, 1209. [Google Scholar] [CrossRef]
- Grouzdev, D.S.; Babich, T.L.; Tourova, T.P.; Sokolova, D.S.; Abdullin, R.R.; Poltaraus, A.B.; Schevchenko, M.A.; Toshchakov, S.V.; Nazina, T.N. Draft Genome Sequence of Roseomonas aestuarii Strain JR1/69-1-13 Isolated from Nitrate- and Radionuclide-Contaminated Groundwater in Russia. Genome Announc. 2018, 6, e00583-18. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.Z.; Li, J.L.; Fang, B.Z.; Liu, Z.T.; Wang, P.; Dong, L.; Duan, L.; Luo, X.Q.; Li, S.H.; Li, W.J. Roseomonas ponticola sp. nov., a novel bacterium isolated from Pearl River estuary. Int. J. Syst. Evol. Microbiol. 2021, 71, 004994. [Google Scholar] [CrossRef]
- Shiratori-Takano, H.; Takano, H.; Ueda, K. Whole-Genome Sequence of Filimonas lacunae, a Bacterium of the Family Chitinophagaceae Characterized by Marked Colony Growth under a High-CO2 Atmosphere. Genome Announc. 2016, 4, e00667-16. [Google Scholar] [CrossRef] [PubMed]
- Whitman, T.; Singh, B.P.; Zimmerman, A.R. Priming effects in biochar-amended soils: Implications of biochar-soil organic matter interactions for carbon storage. In Biochar for Environmental Management; Lehmann, L., Joseph, S., Eds.; Routledge: New York, NY, USA, 2015; pp. 455–487. [Google Scholar]
- United States Department of Agriculture, Agriculture Research Service, Eastern Regional Research Center. What Is Pyrolysis? Biomass Pyrolysis Research. 2025. Available online: www.ars.usda.gov/northeast-area/wyndmoor-pa/eastern-regional-research-center/docs/biomass-pyrolysis-research-1/what-is-pyrolysis/#:~:text=Pyrolysis%20is%20the%20heating%20of,strong%20bio-polymers%20mentioned%20above (accessed on 10 January 2025).
- Santín, C.; Doerr, S.H.; Preston, C.M.; González-Rodríguez, G. Pyrogenic organic matter production from wildfires: A missing sink in the global carbon cycle. Glob. Chang. Biol. 2015, 21, 1621–1633. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Energy. Fungal Recyclers: Fungi Reuse Fire-Altered Organic Matter. In Biological and Environmental Research; United States Department of Energy: Washington, DC, USA, 2022. Available online: www.energy.gov/science/ber/articles/fungal-recyclers-fungi-reuse-fire-altered-organic-matter#:~:text=Wildfires%20can%20cause%20significant%20changes,organisms%20to%20use%20as%20food (accessed on 2 November 2024).
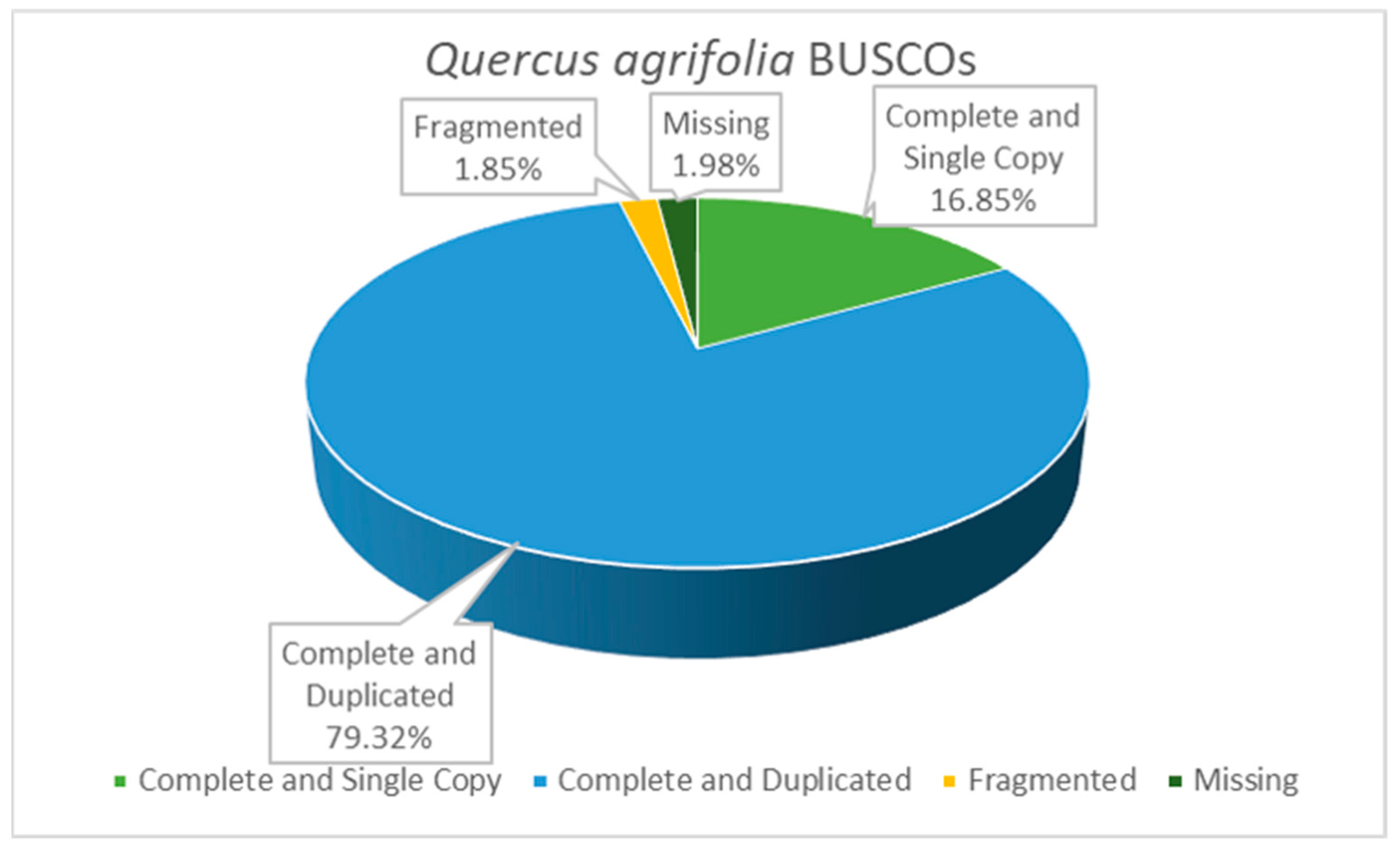
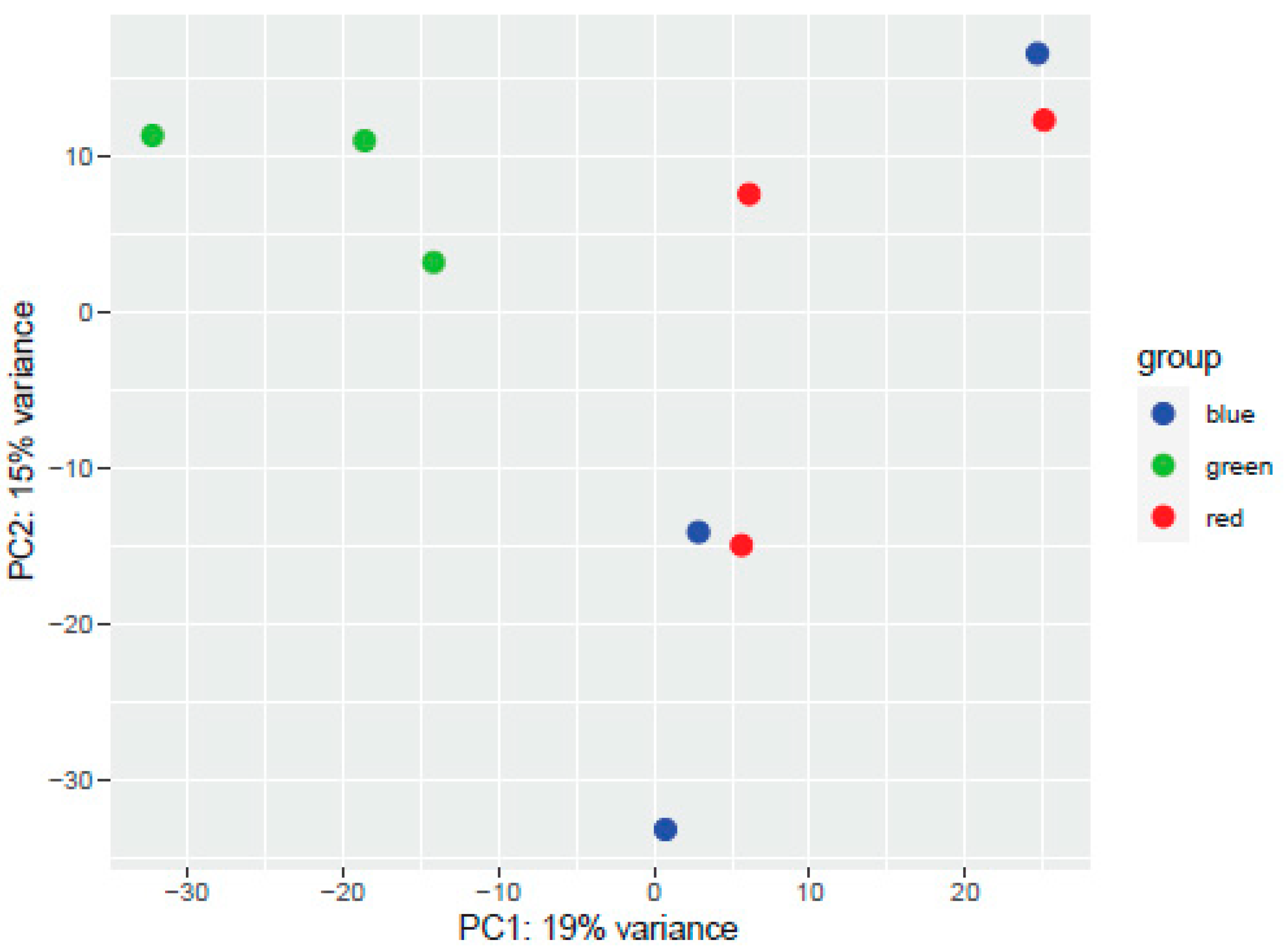
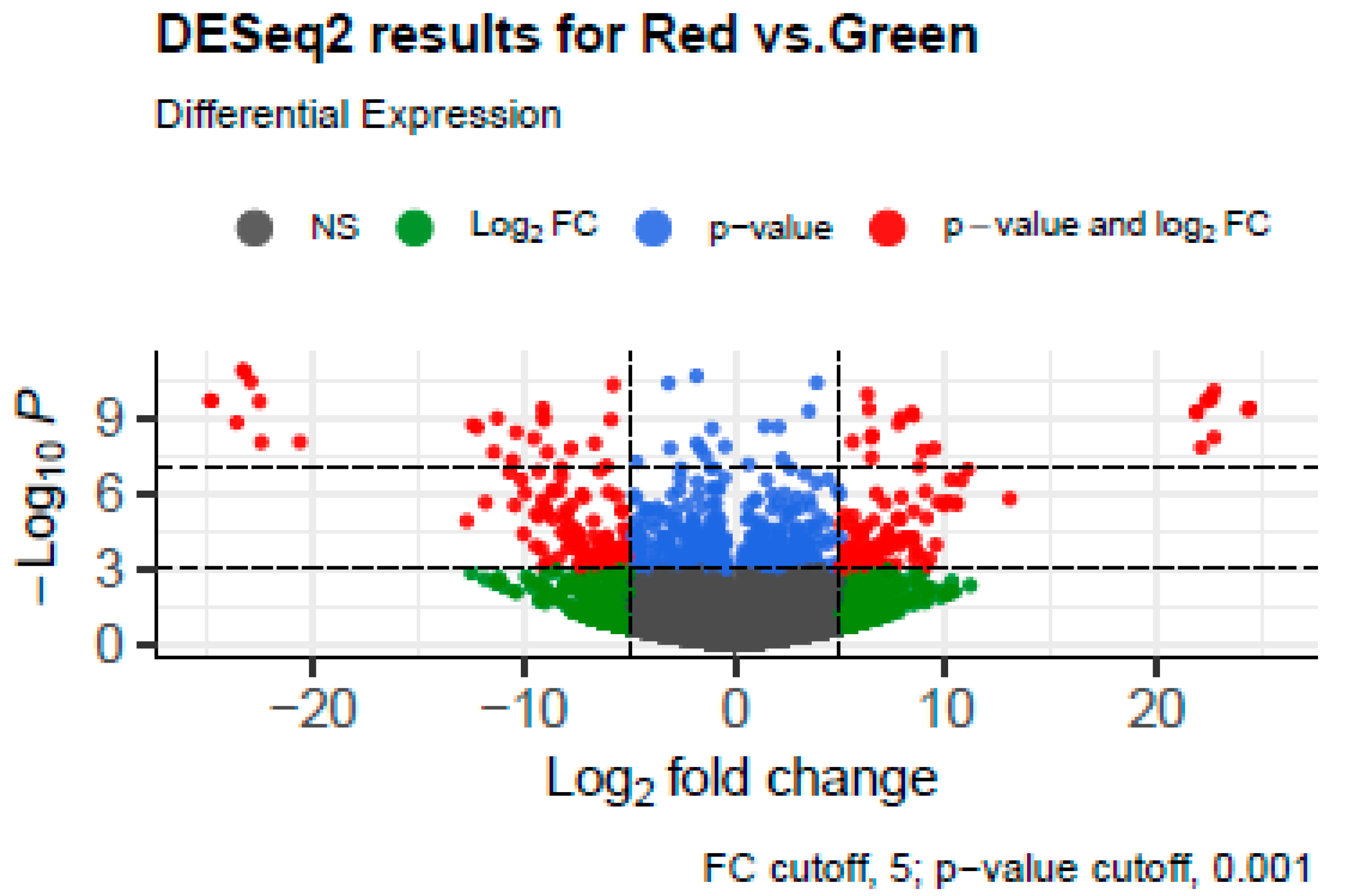


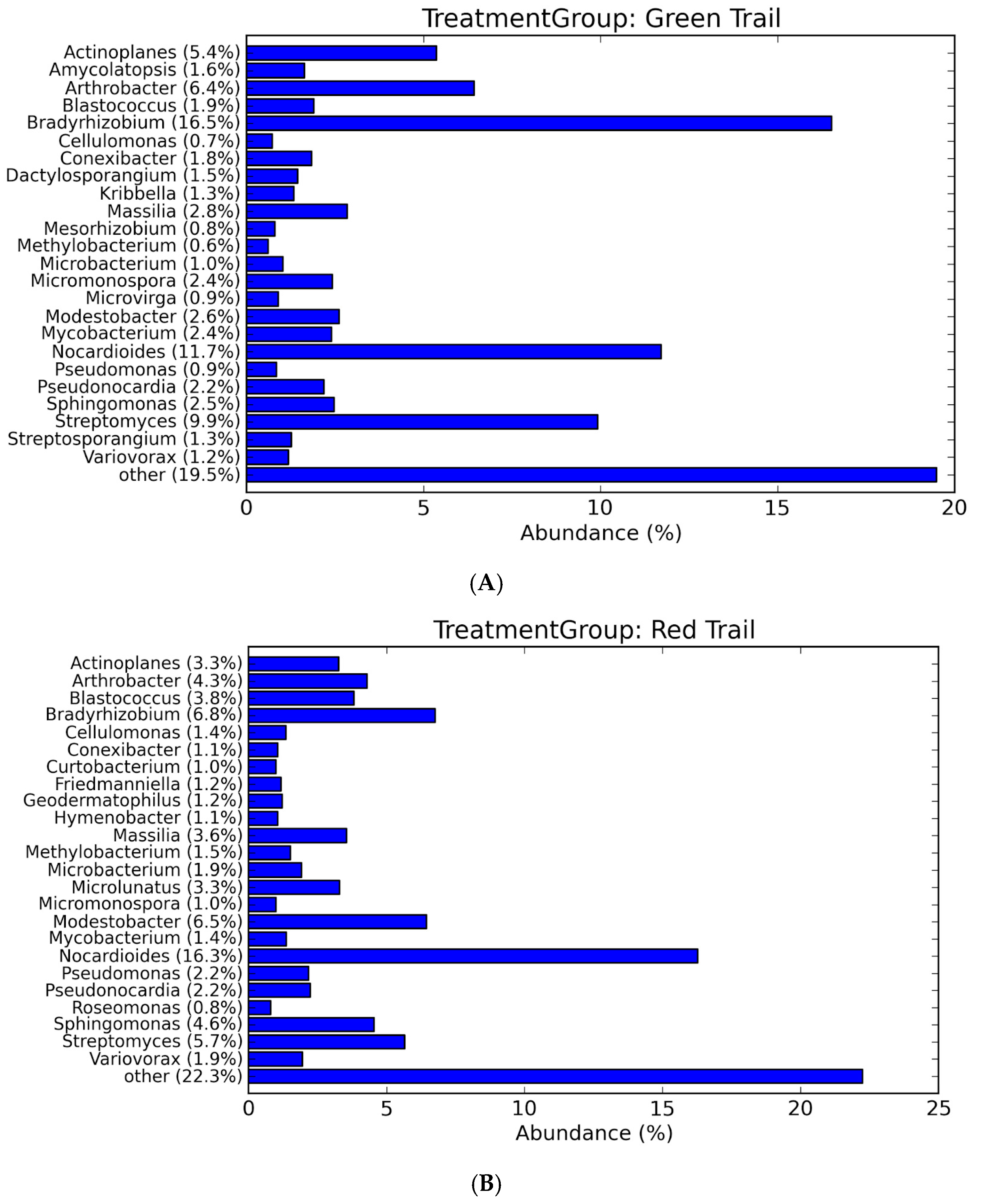
| Number of Sequences | Total Length | Average Length | Min Length | Max Length | L50 | N50 | N80 |
|---|---|---|---|---|---|---|---|
| 521,817 | 4.2 × 108 | 805.2073 | 174 | 15,958 | 87,206 | 1294 | 470 |
| Gene_ID | Base Mean | log2Fold Change | padj | Sprot_Top_BLASTX_Hit |
|---|---|---|---|---|
| TRINITY_DN5071_c0_g1 | 28.23026136 | −8.890413158 | 9.02324 × 10−6 | Putative disease resistance protein At1g50180 |
| TRINITY_DN2917_c2_g1 | 13.61452133 | −6.690330655 | 0.001847702 | 11-beta-hydroxysteroid dehydrogenase-like 6 |
| TRINITY_DN15594_c0_g1 | 31.84983276 | −6.08516735 | 2.9024 × 10−5 | Disease resistance protein Roq1 |
| TRINITY_DN20992_c0_g1 | 30.84011525 | −5.365549041 | 0.000863898 | Disease resistance protein RPS4 |
| TRINITY_DN6739_c0_g2 | 117.1948209 | −2.80631131 | 0.001647927 | Geraniol 8-hydroxylase |
| TRINITY_DN2023_c0_g1 | 148.4293476 | −2.580218229 | 7.44233 × 10−5 | Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase |
| TRINITY_DN35565_c0_g1 | 76.15119378 | −2.474840328 | 0.004799669 | Class V chitinase |
| TRINITY_DN2666_c0_g1 | 108.2599035 | 1.428933703 | 1.22801 × 10−6 | Putative disease resistance protein At3g14460 |
| TRINITY_DN79417_c0_g1 | 85.51571069 | 2.011613378 | 0.00986672 | Pleiotropic drug resistance protein 3 |
| TRINITY_DN3071_c1_g1 | 117.8021701 | 2.116191269 | 0.003537541 | Phosphoenolpyruvate carboxylase 4 |
| TRINITY_DN12296_c0_g1 | 32.40419296 | 4.367923583 | 0.001988292 | Myrcene synthase, chloroplastic |
| TRINITY_DN798_c0_g1 | 86.31811503 | 7.015111442 | 1.00934 × 10−8 | 1-aminocyclopropane-1-carboxylate oxidase homolog 4 |
| TRINITY_DN1798_c0_g2 | 90.52379689 | 2.802793528 | 0.000880927 | Putative disease resistance RPP13-like protein 1 |
| Gene_Id | Quercus sp. NT Hit | %ID | GenBank Accession | Q. agrifolia WGS Contig | %ID | GenBank Accession |
|---|---|---|---|---|---|---|
| TRINITY_DN5071_c0_g1 | Q. robur | 90.63% | XM_050406478.1 | Scaffold6 | 99.62% | JARQAE010000006.1 |
| TRINITY_DN2917_c2_g1 | Q. robur | 98.22% | XM_050400348.1 | Scaffold6 | 100% | JARQAE010000006.1 |
| TRINITY_DN15594_c0_g1 | Q. lobata | 87.19% | XM_031105504.1 | Scaffold5 | 86.77% | JARQAD010000005.1 |
| TRINITY_DN20992_c0_g1 | Q robur | 91.67% | XR_007653105.1 | Scaffold4 | 94.54% | JARQAE010000004.1 |
| TRINITY_DN6739_c0_g2 | Q. robur | 97.41% | XM_050387445.1 | Scaffold11 | 100% | JARQAE010000011.1 |
| TRINITY_DN2023_c0_g1 | Q. lobata | 97.12% | XM_031090969.1 | Scaffold12 | 98.39% | JARQAE010000012.1 |
| TRINITY_DN35565_c0_g1 | Q. suber | 97.05% | XM_024056073.2 | Scaffold1 | 99.92% | JARQAE010000001.1 |
| TRINITY_DN2666_c0_g1 | Q. robur | 95.26% | XM_050423093.1 | Scaffold3 | 95.39% | JARQAD010000003.1 |
| TRINITY_DN79417_c0_g1 | Q. robur | 97.99% | XM_050383676.1 | Scaffold9 | 99.78% | JARQAE010000009.1 |
| TRINITY_DN3071_c1_g1 | Q. lobata | 97.57% | XM_031104334.1 | Scaffold5 | 99.69% | JARQAD010000005.1 |
| TRINITY_DN12296_c0_g1 | Q. lobata | 97.34% | XR_004090747.1 | Scaffold8 | 100% | JARQAE010000008.1 |
| TRINITY_DN798_c0_g1 | Q. robur | 96.03% | XM_050419551.1 | Scaffold4 | 98.29% | JARQAE010000004.1 |
| TRINITY_DN1798_c0_g2 | Q. robur | 86.92% | XM_050427734.1 | Scaffold3 | 85.70% | JARQAD010000003.1 |
| Sample ID | Input Reads | Filtered Reads | % Passed Filter Reads | Feature Count |
|---|---|---|---|---|
| Green Trail 1 | 41,970 | 35,708 | 85.1 | 24,252 |
| Green Trail 2 | 52,137 | 45,766 | 87.8 | 31,251 |
| Green Trail 3 | 36,580 | 32,848 | 89.8 | 23,318 |
| Blue Trail 1 | 52,161 | 44,657 | 85.6 | 30,309 |
| Blue Trail 2 | 38,030 | 33,756 | 88.8 | 23,720 |
| Blue Trail 3 | 34,088 | 29,899 | 87.7 | 19,880 |
| Red Trail 1 | 30,017 | 26,463 | 88.2 | 20,646 |
| Red Trail 2 | 35,076 | 31,725 | 90.5 | 25,632 |
| Red Trail 3 | 34,038 | 30,552 | 89.8 | 24,720 |
| Community standard 1 | 44,898 | 39,605 | 88.2 | 28,068 |
| Community standard 2 | 26,787 | 21,845 | 81.6 | 16,272 |
| Total | 425,782 | 372,824 | 87.6 | 244,348 |
| Genus | FC | Log2(FC) | p Value | Higher Abundance |
|---|---|---|---|---|
| Microlunatus | 0.10638 | −3.2328 | 0.003384 | Red Trail |
| Roseomonas | 0.25615 | −1.965 | 0.000421 | Red Trail |
| Filimonas | 0.29897 | −1.7419 | 0.004025 | Red Trail |
| Pedobacter | 0.32759 | −1.6101 | 0.00882 | Red Trail |
| Sorangium | 3.0202 | 1.5947 | 0.003868 | Green Trail |
| Kibdelosporangium | 3.9514 | 1.9824 | 0.000904 | Green Trail |
| Cohnella | 6.0769 | 2.6033 | 0.000123 | Green Trail |
| Planococcus | 10.571 | 3.4021 | 0.005461 | Green Trail |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senn, S.; Enke, R.A.; Carrell, S.J.; Nations, B.; Best, M.; Kostoglou, M.; Smith, K.; Yan, J.; Ford, J.M.; Vion, L.; et al. De Novo Leaf Transcriptome Assembly and Metagenomic Studies of Coast Live Oak (Quercus agrifolia). Appl. Microbiol. 2025, 5, 24. https://doi.org/10.3390/applmicrobiol5010024
Senn S, Enke RA, Carrell SJ, Nations B, Best M, Kostoglou M, Smith K, Yan J, Ford JM, Vion L, et al. De Novo Leaf Transcriptome Assembly and Metagenomic Studies of Coast Live Oak (Quercus agrifolia). Applied Microbiology. 2025; 5(1):24. https://doi.org/10.3390/applmicrobiol5010024
Chicago/Turabian StyleSenn, Savanah, Ray A. Enke, Steven J. Carrell, Bradley Nations, Meika Best, Mathew Kostoglou, Karu Smith, Jieyao Yan, Jillian M. Ford, Les Vion, and et al. 2025. "De Novo Leaf Transcriptome Assembly and Metagenomic Studies of Coast Live Oak (Quercus agrifolia)" Applied Microbiology 5, no. 1: 24. https://doi.org/10.3390/applmicrobiol5010024
APA StyleSenn, S., Enke, R. A., Carrell, S. J., Nations, B., Best, M., Kostoglou, M., Smith, K., Yan, J., Ford, J. M., Vion, L., & Presley, G. (2025). De Novo Leaf Transcriptome Assembly and Metagenomic Studies of Coast Live Oak (Quercus agrifolia). Applied Microbiology, 5(1), 24. https://doi.org/10.3390/applmicrobiol5010024







