Abstract
The Baltic Sea blue mussel (Mytilus trossulus) plays a crucial role in this brackish water ecosystem, filtering water and accumulating pollutants. This study investigated how exposure to crude oil and dispersants affects the microbiome of M. trossulus at two salinities (5.6 and 15) over 21 days. Results showed that dispersant use significantly increased the accumulation of polycyclic aromatic hydrocarbons (PAHs) in mussel tissues, particularly at lower salinity. The microbial communities in gills and digestive glands were notably affected, with shifts towards hydrocarbon-degrading bacteria like Shewanella and Acinetobacter in samples exposed to chemically enhanced water accommodated fraction of crude oil (CEWAF). Salinity was a key factor in determining both PAH accumulation and microbial diversity, with lower salinity leading to reduced bacterial diversity in dispersant treatments. This study highlights the need for a cautious use of dispersants in sensitive environments like the Baltic Sea, emphasizing the ecological implications of altered microbial communities.
1. Introduction
Petroleum hydrocarbon contamination caused by various types of oil spills poses a constant threat to marine ecosystems. The Baltic Sea is particularly vulnerable to such contamination due to its unique features and the heavy traffic of tankers and other vessels in the area. Oil spill response in the Baltic Sea primarily focuses on mechanical methods, utilizing oil response vessels. However, the use of chemical dispersants can also play a role in oil spill management, preventing oil from polluting shorelines and reducing the smothering of larger animals, such as seabirds. Despite this, recent studies have indicated that the addition of dispersants may not significantly enhance the biodegradation of petroleum hydrocarbons in Baltic Sea water, although they do impact microbial communities [1]. Moreover, the effects of dispersed oil on benthic fauna remain largely unknown.
The Baltic Sea blue mussel, Mytilus trossulus, is a key species in this brackish water ecosystem and is widely used as a bioindicator for environmental pollution, including polycyclic aromatic hydrocarbons (PAHs) and crude oil contamination [2,3]. Mytilus species are particularly suited for such studies because they filter large amounts of water, leading to the accumulation of contaminants, especially PAHs, in their tissues at high concentrations.
It is well-established that the microbiome plays a crucial role in the health of its host [4,5,6]. However, disturbances such as environmental pollution or elevated temperatures [7] can disrupt this relationship. While research on mussel-associated bacteria often focuses on food safety, such as the presence of fecal bacteria or antibiotic resistance genes [8], or the impact of mussel biofiltration on microbial community structure in aquaculture systems [9,10,11], there is limited information on the symbiotic bacteria in mussels, especially in shallow-water species like M. trossulus. Many studies on symbiotic bacteria in mussels have been conducted in deep-sea environments, where endosymbiotic bacteria, such as those involved in sulfide and methane oxidation, are common [12,13,14].
Recent research has shown that microbial communities in Mytilus galloprovincialis differ from the surrounding seawater, and that microbial communities in different tissues of the mussels vary as well [15]. Similarly, Santibáñez et al. [16] characterized the gut microbiome of Mytilus chilensis, revealing the existence of host-specific microbiota. They found that bacterial diversity was higher in mussels from natural habitats compared to those from mussel farms, where anthropogenic impacts were likely affecting the bacterial communities. These findings underscore the importance of understanding marine eukaryotic host-associated microbiomes as a means to predict anthropogenic impacts at both species and ecosystem levels [17].
The effects of crude oil on bacterial communities associated with Mytilus galloprovincialis gills were investigated by Cappello et al. [18], who found that both the quantity and quality of bacteria in the gills were affected. Furthermore, the capacity to degrade xenobiotics has been associated with the microbiome of bivalves like Brachidontes [19]. Polycyclic aromatic hydrocarbon (PAH) degradation capacity in microorganisms has been widely studied using the PAH ring-hydroxylating dioxygenases (PAH-RHDα) gene as a biomarker for polluted sites [20]. Both Gram-negative and Gram-positive bacteria possess this gene, which encodes enzymes involved in PAH degradation. Analyzing both types of bacteria is crucial for evaluating the total PAH-degradation capacity of bacterial communities [21,22].
The primary objective of this study was to evaluate the impact of exposure to water accommodated fraction (WAF) of crude oil and chemically enhanced water accommodated fraction (CEWAF) of crude oil on the microbiome of mussels, specifically focusing on the abundance of hydrocarbon degradation-related genes in the bacterial community. This is the first study to describe the microbiome of the brackish water Baltic Sea blue mussel, Mytilus trossulus, using culture-independent methods and next-generation sequencing (NGS). The experiments were conducted at two different salinity levels, corresponding to those found in the southern and northern Baltic Sea. The results of this study will contribute to a better understanding of how dispersed oil affects mussels and other benthic fauna, with a focus on their microbiomes, which play a vital role in their health. Additionally, these findings will help address the gaps in knowledge regarding the microbiomes of marine species.
2. Materials and Methods
2.1. Collection of the Mussels
Baltic Sea blue mussels (Mytilus trossulus) were collected by scuba diving on October 26, 2016, close to the Tvärminne Zoological Station of the University of Helsinki (southern Finland, northern Baltic Sea) from the depth of 5–6 m. They were transported in cooled thermo-insulated boxes with ambient seawater to the laboratory of the Finnish Environment Institute (Helsinki, Finland), placed into a 10 °C room (ambient temperature at the collection site), and equipped with aeration. After cleaning and measuring the shell length (2–2.5 cm), 200 mussels were placed in each 30 L experimental tank with 20 L of ambient seawater. Two strengths of artificial sea water (ASW) were prepared for the experiment: 5.6, the same as in the collection area, and 15, corresponding to the higher-end salinities present in the southwestern Baltic Sea. The ASW media was prepared by adding Instant Ocean® salt mix to local tap water and leaving it overnight under efficient aeration to remove any chlorine before the use. During a two-week acclimatization period of the mussels in ASW, the water temperature was gradually reduced in each tank to reach 5 °C, corresponding to winter conditions, and in the higher salinity treatment tanks the salinity was gradually increased to 15. The mussels were fed daily with algal cells (Instant Algae®, 3 mL of stock solution diluted 1:2).
2.2. Experimental Set-Up
Mussels were exposed to the water accommodated fraction (WAF) and chemically enhanced water accommodated fraction (CEWAF) of Naphthenic North Atlantic (NNA) crude oil in two different salinities (5.6 and 15) for 21 days. A detailed description of the experimental set-up is provided in Turja et al. [23]. Briefly, both WAF and CEWAF were prepared in 10 L Marionette glass bottles equipped with a tap. The WAF was prepared by adding 20 g of NNA crude oil to 4 L of ASW (5.6 and 15 psu) and the CEWAF was prepared by first mixing dispersant Finasol 51 (Total Fluides, France) into the NNA crude oil with 1:100 ratio. After that, 20 g of this mixture was added to 4 L of ASW (5.6 and 15 psu). All bottles were then gently shaken (200 rpm to avoid vortex) at 10 °C for 48 h. After that, the shaking was stopped and solutions were left let to settle for 2 h before carefully draining the resulting WAFs and CEWAFS into new glass bottles. Fresh WAF and CEWAF were prepared for full renewals of the experimental media every 48 h.
Applying a semi-static system, mussels were exposed to the 5% WAF and 5% CEWAF solutions in the two different salinities for 21 days at 5 °C. The 200 experimental mussels (i.e., 400 per treatment) were placed in 19 L of ASW and one liter of WAF or CEWAF was added on top. Two replicate 30 L tanks containing 200 mussels each in 20 L of experimental medium were set up for each treatment group, including the controls (in ASW). Temperature was kept constant during the experiment by using thermostats (Lauda) placed in the large tap water tank surrounding the experimental tanks. Most of the mussels were sampled for the biological effect measurements studied in Turja et al. [23].
Samples for microbiological analysis were taken after the two-week acclimation period before the experiment started (“in situ”) and after 21 days of exposure, when the experiment was ended. On a sampling day, the gills and digestive gland were (DG) dissected using sterile scalpels and tweezers. Samples were pooled to obtain enough material for DNA extraction (gills or digestive gland of three mussels of same treatment per tube, three of these replicates for each sample). Samples were immediately frozen in sterile tubes and kept in −20 °C until further analysis. Chemical analysis of PAH and oil concentrations in water and PAHs in mussel tissue were performed in SYKE laboratory, as described in Turja et al. [23].
2.3. DNA Extraction and Quantification of PAH-Degradation Genes by qPCR
DNA was extracted from mussel tissues using a Fast DNA Spin Kit (Qbiogene/MPBiomedicals; Carlsbad, CA, USA), according to the manufacturer’s instructions. The samples were homogenized in a FastPrep instrument on setting 4.5 for 30 s. The concentration of DNA was measured with a Qubit fluorometer (Invitrogen Molecular Probes; Carlsbad, CA, USA). qPCR reactions for the PAH ring hydroxylating deoxygenase genes (PAH–RHDα) (Gram-positive and Gram-negative bacteria separately) were carried out in triplicate, as previously described in Reunamo et al. [24], using the following primers: Gram-positive (GP) forward 5′-CGG CGC CGA CAA YTT YGT NGG-3′ and reverse 5′-GGG GAA CAC GGT GCC RTG DAT RAA-3′ or Gram-negative (GN) forward 5′-GAG ATG CAT ACC ACG TKG GTT GGA-3′ and reverse 5′-AGC TGT TGT TCG GGA AGA YWG TGC MGT T-3′ (Cebron et al., 2008). The amplification efficiency of the qPCR reactions, estimated by using the standard curve, was 100% ± 10%. The average gene copy numbers of 16S rDNA and PAH–RHDα genes of Gram-positive and Gram-negative bacteria were calculated per one mussel (gill or digestive gland (DG)).
The high throughput sequencing of bacterial V3-V4 region was performed using an Illumina MiSeq sequencing system at the Institute of Biotechnology of University of Helsinki. The sequences were deposited in the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/, assessed on 1 January 2020) under accession number PRJEB47096. Raw sequences were analyzed with Mothur pipeline version 1.47.0 [25], and bacterial taxonomic identities were determined using the SILVA v138 database [26]. Diversity indices were calculated using Mothur. Additional sequencing and bioinformatics of the commercial algae mix Shellfish Diet 1800 (Reed Mariculture Inc, Campbell USA) was performed by Novogene Company Ltd, (Cambridge UK). The Shellfish Diet 1800 was treated the same way as other samples before sequencing (freezing at −20 °C, DNA extraction).
2.4. PAH Analysis from Mussel Tissues
PAH concentrations in mussels were measured in the SYKE laboratory with the standardized method described in Turja et al. [23]. A total of 10 + 10 mussels were pooled from two replicate aquaria per treatment. Measurements were conducted at the beginning of the experiment, and on day 1, 7, and 21. Altogether, 21 different PAHs were used in the standard mix. These were the 16 EPA PAHs and 1-methylnaphthalene, 2-methylnaphthalene, benzo(e)pyrene, perylene, and triphenylene.
2.5. Statistical Analysis
Ordination of samples, based on the composition of the bacterial community, was obtained using principal coordinate analysis with a Bray–Curtis dissimilarity matrix. To evaluate the differences in bacterial community composition between groups, we employed a Permutational Multivariate Analysis of Variance (PERMANOVA) with 999 permutations. This analysis was conducted using the Vegan package in R [27]. We used the Wilcoxon rank sum test, along with a false discovery rate correction, to identify differentially abundant taxa. To assess variations in diversity values among treatments, we performed a one-way Analysis of Variance (ANOVA) followed by Tukey’s post hoc test.
3. Results
3.1. Concentration of PAHs in Mussel Tissue
After just one day of exposure to WAF and CEWAF, PAHs accumulated in mussel tissues. CEWAF exposure, in particular, led to significantly higher PAH accumulation (Figure S1, Table S1). Although PAH concentrations generally decreased over time, they remained elevated in the 5.6 psu CEWAF treatments compared to other conditions. Additionally, there was an increase in the relative abundance of HMW and methyl PAHs in the mussel tissue.
3.2. Abundance of Bacterial 16S rRNA and PAH Degradation-Related Genes
The 16S rRNA gene count remained steady at approximately 108 copies per gill or digestive gland sample throughout the experiment, except in one 15 psu CEWAF-exposed digestive gland replicate, where it dropped to 104 copies (Table 1). Initially, GN PAH-RHDα genes were undetectable in unexposed mussels’ microbiomes. However, their abundance increased significantly in CEWAF-exposed mussels. GP PAH-RHDα genes were present in various treatments, including controls and in situ digestive gland samples at 5.6 psu, with the highest observed count in the 15 psu CEWAF-treated digestive gland sample.

Table 1.
Copy numbers of the 16S rRNA, PAH-RHDαGP, and PAH-RHDαGN genes calculated per one mussel gill or digestive gland samples in situ and after 21-day incubation.
3.3. Bacterial Community Structure
3.3.1. Bacterial Communities of Mussel Tissues and Impact of Salinity
A total of 32 different bacterial phyla were found in mussel tissues. Bacterial communities differed on a phylum level between low and high salinities (Figure 1). Dominant phyla in lower salinity were Proteobacteria (37.2%), Campylobacterota (33.9%), and Bacteroidota (20.5%). In the in situ samples, Planctomycetota (1.4%) was among the five most abundant phyla, but their abundance decreased in all treatments. In the in situ digestive gland community, the most abundant phyla were Proteobacteria, Planctomycetota, and Actinobacteriota. In higher salinity, Proteobacteria (71.7%) was distinctly the largest phylum, followed by Bacteroidota (20.3%). The amount of Campylobacterota was up to 14.7% in the in situ DG samples. However, the amount of this phylum decreased notably during the experiment.
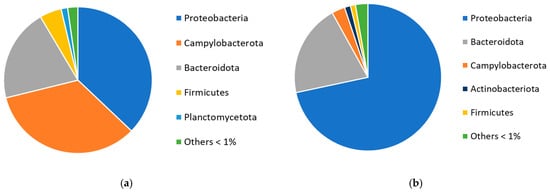
Figure 1.
Most abundant bacterial phyla in mussel tissue in (a) 5.6 psu salinity and (b) 15 psu salinity.
According to PERMANOVA results, bacterial community structure differed significantly in genus level between high and low salinity levels as well as between body locations (Figure 2a,b). At the same time, bacterial community Shannon diversity index values were not different between body locations or salinity levels. In the case of body locations, the following genera were differentially abundant—Flavobacterium (higher in GL) and Pseudomonas (higher in DG). Two salinity levels had an effect on the relative abundance of Colwellia and Flavobacterium (more abundant in low salinity) and Halomonas, Cellulophaga, and Polaribacter (more abundant in high salinity).
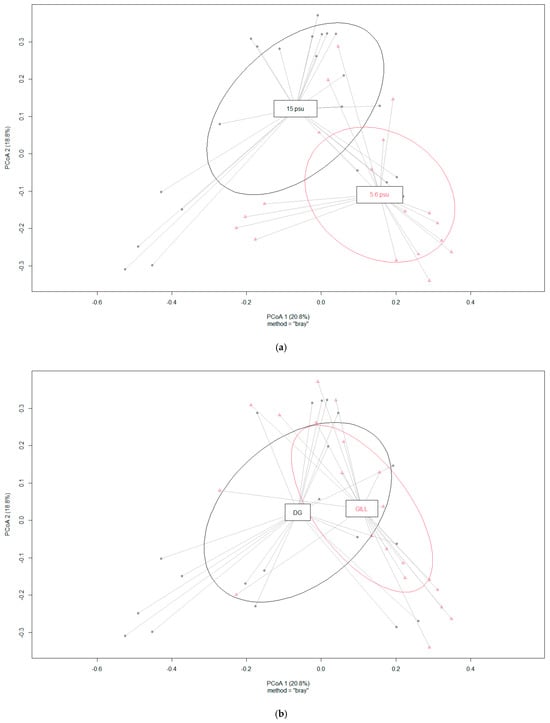
Figure 2.
Ordination of samples based on principal coordinate analysis (PCoA) using Bray–Curtis dissimilarity matrices. (a) Grouping of samples based on two salinity levels (PERMANOVA R2 = 0.104, p = 0.001. (b) Grouping of samples based on the body location (PERMANOVA R2 = 0.083, p = 0.002).
The most abundant genera in the Shellfish Diet controls were the following: Haloferula, Lactobacillus, Roseinatronobacter, Atopostipes, Alkalibacterium, Pelagibacterium, Marivita, Halomonas, Seochaeicola, Paracoccus, Roseovarius, Candidatus Microthrix, Ilumatobacter, and strain ML602J-51 (Microbacteriaceae). Out of these genera, Halomonas (3.7% of total reads) was present in the studied gill and DG samples. Other genera observed in smaller amounts in the gill and DG samples were Lactobacillus (0.02%), Pelagibacterium (0.0005%), Marivita (0.004%), and Paracoccus (0.01%).
3.3.2. Bacterial Community Structure at Low Salinity (5.6 psu)
At 5.6 psu, the in situ bacterial communities in both gills and digestive glands were distinct from those in exposed samples, with a large portion of the community composed of genera with less than 0.5% abundance. In CEWAF-exposed gill samples, the abundance of unclassified Arcobacteraceae and Flavobacterium notably increased, indicating a strong impact of the treatment on community structure.
During the experiment Pseudomonas and Flavobacterium increased, especially in control and WAF incubations in DG samples. In CEWAF-exposed DG samples, unclassified Arcobacteraceae and unclassified Peptostreptococcales-Tissierellales increased, as did CEWAF gill samples (Figure 3). In CEWAF-exposed gill samples, the abundance of unclassified Arcobacteraceae and Flavobacterium notably increased, indicating a strong impact of the treatment on community structure. According to PERMANOVA results, the CEWAF treatment had a strong impact on the bacterial community structure of both gill and DG samples (p < 0.05). Unclassified Peptostreptococcales-Tissierellales (Firmicutes) increased notably in CEWAF treatments. In addition, Shewanella and unclassified Pseudomonadales increased (Figure 3).
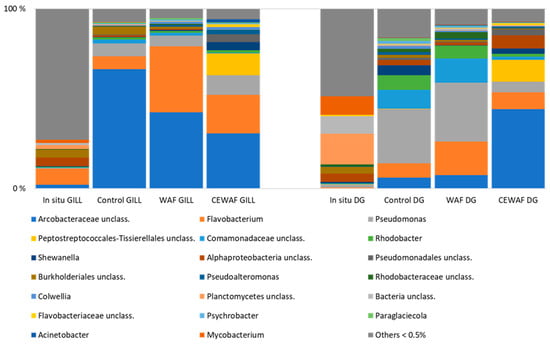
Figure 3.
Relative abundance of bacterial genera representing ≥0.5% of the sequences in the low (5.6) salinity experiment.
3.3.3. Bacterial Community Structure at High Salinity (15 psu)
In the 15 psu salinity gill samples, Flavobacterium dominated in situ, but its abundance decreased in control and WAF treatments over time. In contrast, Pseudomonas increased across all treatments, particularly in CEWAF-exposed samples, indicating a pronounced effect of the CEWAF treatment on bacterial community structure (Figure 4). In the 15 psu salinity in situ DG samples Pseudomonas, Flavobacterium, Shewanella, unclassified Arcobacteraceae, and unclassified Comamonadaceae were the most abundant genera. Pseudomonas increased in all treatments, especially in CEWAF treatments (Figure 4).
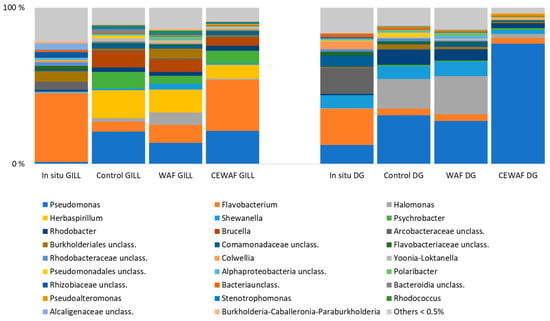
Figure 4.
Relative abundance of bacterial genera representing ≥0.5% of the sequences in the high (15 psu) salinity experiment.
Venn diagrams were constructed to visualize the number of unique bacterial genera in different treatments. Figures show that the proportion of unique bacterial genera in different treatments was relatively small when compared to in situ samples (Figure 5). The number of bacterial OTUs decreased in most of the treatments during the experiment (Table 2). Supplementary Table S2 highlights a variety of bacterial groups that are unique to specific treatments and conditions. This suggests that exposure to different treatments (WAF, CEWAF) and salinity levels has a significant impact on the composition of the mussel microbiome. Five unique genera to dispersed oil (CEWAF) treatments were in low-salinity gills Peptostreptococcales-Tissierellales unclass., Pseudomonadales unclass., Flavobacteriaceae unclass., Acinetobacter, and Rheinheimera. In the digestive gland, the unique genera were Peptostreptococcales-Tissierellales unclass., Acidaminobacter, Proteobacteria unclass., and Aeromonas. In higher salinity, there were no unique genera present in the DG samples. In CEWAF-exposed mussels, the unique genera in gills were Sphingomonadaceae unclass., Pseudoalteromonas, Sulfitobacter, and Sphingorhabdus. (Table S2).
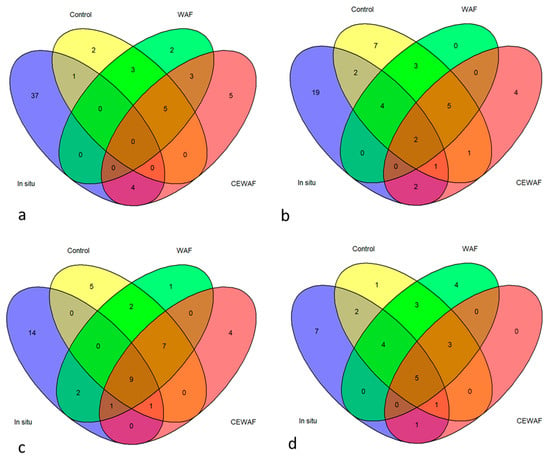
Figure 5.
Venn diagrams showing the number of shared and unique genera between different treatments in low-salinity (a) gills and (b) digestive glands and high-salinity (c) gills and (d) digestive glands. Bacterial genera with a relative abundance greater than or equal to 0.5% were included in the analysis. Numbers in the overlapping regions indicate the number of shared taxa between samples. Blue: In situ, yellow: control, green: WAF and red: CEWAF.

Table 2.
Mean and standard deviation values for number of (processed) sequences, OTUs, and diversity indices (Chao, Shannon, and inverse Simpson’s index).
The Inverse Simpson’s index indicated that community diversity was significantly higher in the digestive gland control samples with a salinity of 5.6 psu compared to the samples treated with CEWAF (p < 0.005, one-way ANOVA, Tukey’s pairwise comparison) (Table 2). It was also higher in the digestive gland samples with an in situ salinity of 15 psu compared to the CEWAF treatments (Table 2). The number of OTUs was lower in the 5.6 psu salinity in situ gill sample; however, sequencing was only possible from one replicate due to the limited DNA amount.
4. Discussion
Ecologically, the Baltic mussel (Baltic Mytilus trossulus) is a key species in the low-diversity brackish-water ecosystem of the Baltic Sea. Salinity is a critical environmental factor since, in the northern Baltic Sea, blue mussels live at the extreme limit of their low-salinity tolerance. In the present study, the salinity effect was studied for the first time on the Baltic Sea blue mussel microbiome. On the genus level, the initial bacterial communities were different in mussels collected from the northern (5.6 psu) and southern (15 psu) Baltic Sea. Overall, salinity was the most important factor affecting the microbiome structure, and also, the communities in the gills and digestive gland differed from each other. Salinity had an effect on the most abundant bacterial groups on the phylum level, Proteobacteria being the most abundant phyla in both salinities: in lower salinity at 37.2% and higher salinity up to 71.7% of the phyla.
The biological effects of different types of oil and PAHs on marine biota have been investigated considerably during recent decades by using various types of biomarkers, both in the laboratory and in the field, e.g., [28,29,30,31,32,33]. Parallel to the present study, a battery of biomarkers was measured in mussels from this same experiment [23], showing increased biological effects in WAF- and CEWAF-exposed mussels, with the highest responses detected in low-salinity (5.6) CEWAF treatments after 7 and 21 days of exposure. Crude oil is a complex mixture of various chemicals influenced by environmental conditions, resulting in intricate response patterns regarding the observed biological effects. The experimental conditions, such as low temperature (5 °C), different salinities, and algal food supply, had an impact on the mussels’ reproductive status. This may have obscured some of the effects of contaminants. However, in the CEWAF treatments, particularly in lower salinity, there was high bioaccumulation of polycyclic aromatic hydrocarbons in mussels, along with increased biochemical, cellular, and tissue-level biomarker responses [23]. Changes in the structure and function of the microbiome observed in the present study might also affect the accumulation and biological effects of PAH compounds in mussels. The observed effects of dispersed oil (CEWAF) related to salinity on the mussel microbiome might give an important insight to the adaptation and survival of this species. Overall, salinity emerged as a critical factor influencing both PAH accumulation and microbial community composition. The stronger effects observed at low salinity highlight the vulnerability of mussels in the northern Baltic Sea to oil spills, particularly when dispersants are used. This finding suggests that salinity should be a key consideration in oil spill response strategies in brackish water environments.
In the present experiment, the use of a dispersant increased the concentrations of petroleum hydrocarbons and PAHs in water and led to a high accumulation of PAHs in mussel tissues. After 21 days, especially high concentrations of PAHs were observed in 5.6 psu salinity CEWAF treatments, where significantly lower bacterial diversity was also observed compared to the corresponding control treatments. By using artificial seawater before the experiment for the acclimatization of mussels for two weeks and during the experiment, the results of our study support the Mytilus-origin of the studied bacterial communities. In previous studies, the solubility and bioavailability of PAHs has been shown to increase with decreasing salinity [34,35] and increased sensitivity to PAHs in mollusks and fish has been observed at lower salinities [36,37]. The higher PAH accumulation in CEWAF treatments, particularly at low salinity, suggests a more significant impact on mussel tissues, which could translate into more severe biological effects. This highlights the potential risks associated with dispersant use in oil spill responses in low-salinity environments like the Baltic Sea.
Bacterial communities underwent changes in all treatments during the experiment, and the main bacterial groups differed between the control and CEWAF samples. In lower-salinity samples, CEWAF-treatment changed the community most visibly: Peptostreptococcales-Tissierellales increased both in gill and DG samples. This order belongs to the class Clostridia (phylum Firmicutes) and has been found, e.g., in the lesions caused by stony coral tissue loss disease [38]. Order Peptostreptococcales-Tissierellales is also known for its capability to produce acetate anaerobically by degrading organic matter [39]. In addition, Shewanella and Acinetobacter, known oil degraders, increased in gills, and unclassified Arcobacteraceae became abundant in CEWAF-exposed digestive glands in low salinity. Genus Arcobacter is commonly found in the marine environment and also as a host associated with shellfish [40]. It is an oil-degrading bacteria adapted to cold conditions [41] and has recently been found to increase due to exposure to oil compounds and dispersants [42]. Cappello et al. [18] showed that known hydrocarbon-degrading bacteria increased in the gills of M. galloprovincialis due to hydrocarbon exposure in a mesocosm experiment. That was the first study to investigate the oil-degrading bacteria in mussels exposed to pollutants. By using selective agar plates with crude oil as a sole source of carbon, they detected bacterial strains originating from M. galloprovincialis gills belonging to Gammaproteobacteria, closely related to the genus Alcanivorax and Marinobacter hydrocarbonoclasticus, known oil-degrading bacteria in the oceans. These genera were not observed to increase in our study, although Gammaproteobacteria was the most abundant class of bacteria in most of the samples. The observed increase in oil-degrading bacteria such as Shewanella and Acinetobacter in CEWAF-treated samples indicates a shift in microbial communities towards those capable of hydrocarbon degradation. This suggests that dispersant use may selectively enrich bacteria with oil-degrading capabilities, potentially altering the natural microbial balance in mussels and affecting their overall health. The significant reduction in bacterial diversity observed in CEWAF treatments may have far-reaching ecological consequences. Reduced diversity can lead to the loss of beneficial microbial functions, potentially compromising the health of mussels and their ability to cope with environmental stressors. Previous research has shown that the mussel microbiome plays a crucial role in processes such as pollution degradation and resistance, enabling the mussel to withstand and ultimately adapt to combined xenobiotic exposure (6).
Observed changes in the bacterial communities were also supported by the results of PAH-degradation gene quantification. The number of GN PAH-RHDα genes was under the detection limit at the beginning of the experiment. GN PAH-RHDα gene abundance increased significantly in CEWAF treatments, particularly in the 5.6 psu gill samples, which also had the highest PAH accumulation. This increase was associated with a rise in GN PAH-RHDα gene-harboring bacteria, such as Pseudomonas and Shewanella. In contrast, GP PAH-RHDα genes were observed more broadly across treatments, with the highest counts detected in 15 psu CEWAF-exposed digestive glands. The Gram-positive phyla Actinobacteria and Firmicutes were among the most abundant phyla in both salinities and GP PAH-RHDα genes can be assumed to originate from bacteria belonging to these groups. The number of bacteria remained constant during the experiment, except in the WAF and CEWAF treatment digestive gland samples in higher salinity. This could indicate that the oil compounds were toxic to some members of microbiota. The increased abundance of GN PAH-RHDα genes in CEWAF-exposed mussels, particularly in low-salinity environments, underscores the role of these genes in hydrocarbon degradation. This increase may indicate an adaptive response of the mussel microbiome to the presence of oil pollutants, potentially aiding in the breakdown of harmful compounds, albeit with unknown long-term effects on mussel health. The study of mussel microbiome is important to understand contaminant-induced effects in these key marine invertebrate species.
5. Conclusions
This study provides critical insights into the impact of crude oil and dispersant exposure on the microbiome of the Baltic Sea blue mussel, Mytilus trossulus. Our findings demonstrate that the use of dispersants, particularly at low salinities, leads to a significant increase in the accumulation of polycyclic aromatic hydrocarbons (PAHs) in mussel tissues. The altered microbial community structure and reduced bacterial diversity observed in CEWAF-treated mussels highlight the potential ecological risks associated with dispersant use in oil spill responses.
Salinity emerged as a key factor influencing both the accumulation of PAHs and the composition of bacterial communities in mussels. The differences observed between low and high salinity environments underscore the vulnerability of mussels in brackish water ecosystems like the Baltic Sea, particularly in areas with lower salinity. Moreover, the increase in hydrocarbon-degrading bacteria and related genes in response to dispersant exposure suggests that the mussel microbiome may adapt to the presence of oil pollutants. However, the long-term consequences of these changes, including potential impacts on mussel health and the stability of the ecosystem, remain uncertain. Overall, this study emphasizes the need for careful consideration of dispersant use in oil spill mitigation strategies, particularly in sensitive environments such as the Baltic Sea. Future research should focus on understanding the long-term effects of such exposures on mussel populations and the broader marine ecosystem.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol5010023/s1. Figure S1: PAH concentrations in mussel tissue.; Table S1: Concentrations of different oil fractions and total oil during the exposure experiments at the salinities of 5.6 and 15.0.; Table S2: Unique bacterial groups in different treatments (In situ, Control, WAF, CEWAF) in different salinities and mussel body parts.
Author Contributions
Conceptualization, A.R. and R.T.; methodology, A.R. and J.T.; validation, A.R.; formal analysis, A.R. and J.T.; investigation, A.R.; resources, K.S.J.; writing—original draft preparation, A.R.; writing—review and editing, A.R., R.T., J.T. and K.S.J.; visualization, A.R.; supervision, K.S.J.; project administration, K.S.J.; funding acquisition, K.S.J. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the Academy of Finland, grant number 289238, and the European Union’s Horizon 2020 research and innovation program under grant agreement No. 679266. Jaak Truu was supported by the Estonian Research Council grant PRG2778.
Data Availability Statement
The sequences were deposited in the European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ (accessed on 1 January 2020)) under accession number PRJEB47096.
Acknowledgments
We want to thank Aino Ahvo for contributing the experimental work and Kari Lehtonen for valuable comments for the text.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| ASW | Artificial seawater |
| WAF | Water accommodated fraction of crude oil |
| CEWAF | Chemically enhanced water accommodated fraction of crude oil |
| NNA | Naphthenic North Atlantic crude oil |
| DG | Digestive gland |
| PAH-RHDα | PAH ring hydroxylating deoxygenase |
References
- Tonteri, O.; Reunamo, A.; Nousiainen, A.; Koskinen, L.; Nuutinen, J.; Truu, J.; Jørgensen, K.S. Effects of Dispersant on the Petroleum Hydrocarbon Biodegradation and Microbial Communities in Seawater from the Baltic Sea and Norwegian Sea. Microorganisms 2023, 11, 882. [Google Scholar] [CrossRef] [PubMed]
- Turja, R.; Soirinsuo, A.; Budzinski, H.; Devier, M.H.; Lehtonen, K.K. Biomarker responses and accumulation of hazardous substances in mussels (Mytilus trossulus) transplanted along a pollution gradient close to an oil terminal in the Gulf of Finland (Baltic Sea). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2013, 157, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Lehtonen, K.K.; D'Errico, G.; Korpinen, S.; Regoli, F.; Ahkola, H.; Kinnunen, T.; Lastumäki, A. Mussel Caging and the Weight of Evidence Approach in the Assessment of Chemical Contamination in Coastal Waters of Finland (Baltic Sea). Front. Mar. Sci. 2019, 6, 688. [Google Scholar] [CrossRef]
- McFall-Ngai, M.; Hadfield, M.G.; Bosch, T.C.G.; Carey, H.V.; Domazet-Lošo, T.; Douglas, A.E.; Dubilier, N.; Eberl, G.; Fukami, T.; Gilbert, S.F.; et al. Animals in a bacterial world, a new imperative for the life sciences. Proc. Natl. Acad. Sci. USA 2013, 110, 3229–3236. [Google Scholar] [CrossRef]
- Apprill, A. Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Front. Mar. Sci. 2017, 4, 222. [Google Scholar] [CrossRef]
- Palladino, G.; Rampelli, S.; Scicchitano, D.; Nanetti, E.; Iuffrida, L.; Wathsala, R.H.G.R.; Interino, N.; Marini, M.; Porru, E.; Turroni, S.; et al. Seasonal dynamics of the microbiome-host response to pharmaceuticals and pesticides in Mytilus galloprovincialis farmed in the Northwestern Adriatic Sea. Sci. Total. Environ. 2023, 887, 163948. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.F.; Xu, J.K.; Chen, Y.W.; Ding, W.Y.; Shao, A.Q.; Liang, X.; Zhu, Y.T.; Yang, J.L. Characterization of Gut Microbiome in the Mussel Mytilus galloprovincialis in Response to Thermal Stress. Front. Physiol. 2019, 10, 1086. [Google Scholar] [CrossRef]
- Bighiu, M.A.; Haldén, A.N.; Goedkoop, W.; Ottoson, J. Assessing microbial contamination and antibiotic resistant bacteria using zebra mussels (Dreissena polymorpha). Sci. Total Environ. 2019, 650, 2141–2149. [Google Scholar] [CrossRef]
- Stabili, L.; Acquaviva, M.I.; Cavallo, R.A. Mytilus galloprovincialis filter feeding on the bacterial community in a Mediterranean coastal area (Northern Ionian Sea, Italy). Water Res. 2005, 39, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Voudanta, E.; Kormas, K.A.; Monchy, S.; Delegrange, A.; Vincent, D.; Genitsaris, S.; Christaki, U. Mussel biofiltration effects on attached bacteria and unicellular eukaryotes in fish-rearing seawater. PeerJ 2016, 4, e1829. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bozcal, E.; Dagdeviren, M. Bacterial metagenome analysis of Mytilus galloprovincialis collected from Istanbul and Izmir coastal stations of Turkey. Environ. Monit. Assess. 2020, 192, 186. [Google Scholar] [CrossRef] [PubMed]
- Kochevar, R.E.; Childress, J.J.; Fisher, C.R.; Minnich, E. The methane mussel: Roles of symbiont and host in the metabolic utilization of methane. Mar. Biol. 1992, 112, 389–401. [Google Scholar] [CrossRef]
- Duperron, S.; Nadalig, T.; Caprais, J.C.; Sibuet, M.; Fiala-Médioni, A.; Amann, R.; Dubilier, N. Dual symbiosis in a Bathymodiolus sp. mussel from a methane seep on the Gabon continental margin (Southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills. Appl. Environ. Microbiol. 2005, 71, 1694–1700. [Google Scholar] [CrossRef] [PubMed]
- Sayavedra, L.; Kleiner, M.; Ponnudurai, R.; Wetzel, S.; Pelletier, E.; Barbe, V.; Satoh, N.; Shoguchi, E.; Fink, D.; Breusing, C.; et al. Abundant toxin-related genes in the genomes of beneficial symbionts from deep-sea hydrothermal vent mussels. eLife 2015, 4, e07966. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Musella, M.; Wathsala, R.; Tavella, T.; Rampelli, S.; Barone, M.; Palladino, G.; Biagi, E.; Brigidi, P.; Turroni, S.; Franzellitti, S.; et al. Tissue-scale microbiota of the Mediterranean mussel (Mytilus galloprovincialis) and its relationship with the environment. Sci. Total. Environ. 2020, 717, 137209. [Google Scholar] [CrossRef]
- Santibáñez, P.; Romalde, J.; Maldonado, J.; Fuentes, D.; Figueroa, J. First characterization of the gut microbiome associated with Mytilus chilensis collected at a mussel farm and from a natural environment in Chile. Aquaculture 2021, 548, 737644. [Google Scholar] [CrossRef]
- Wilkins, L.G.E.; Leray, M.; O’dea, A.; Yuen, B.; Peixoto, R.S.; Pereira, T.J.; Bik, H.M.; Coil, D.A.; Duffy, J.E.; Herre, E.A.; et al. Host-associated microbiomes drive structure and function of marine ecosystems. PLOS Biol. 2019, 17, e3000533. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cappello, S.; Russo, D.; Santisi, S.; Calogero, R.; Gertler, C.; Crisafi, F.; De Domenico, M.; Yakimov, M.M. Presence of hydrocarbon-degrading bacteria in the gills of mussel Mytilus galloprovincialis in a contaminated environment: A mesoscale simulation study. Chem. Ecol. 2012, 28, 239–252. [Google Scholar] [CrossRef]
- Cleary, D.F.R.; Becking, L.E.; Polónia, A.R.M.; Freitas, R.M.; Gomes, N.C.M. Composition and predicted functional ecology of mussel-associated bacteria in Indonesian marine lakes. Antonie Van Leeuwenhoek 2015, 107, 821–834. [Google Scholar] [CrossRef]
- Liang, C.; Huang, Y.; Wang, Y.; Ye, Q.; Zhang, Z.; Wang, H. Distribution of bacterial polycyclic aromatic hydrocarbon (PAH) ring-hydroxylating dioxygenases genes in oilfield soils and mangrove sediments explored by gene-targeted metagenomics. Appl. Microbiol. Biotechnol. 2019, 103, 2427–2440. [Google Scholar] [CrossRef]
- Habe, H.; Omori, T. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 2003, 67, 225–243. [Google Scholar] [CrossRef] [PubMed]
- Cébron, A.; Norini, M.-P.; Beguiristain, T.; Leyval, C. Real-Time PCR quantification of PAH-ring hydroxylating dioxygenase (PAH-RHDα) genes from Gram positive and Gram negative bacteria in soil and sediment samples. J. Microbiol. Methods 2008, 73, 148–159. [Google Scholar] [PubMed]
- Turja, R.; Benito, D.; Ahvo, A.; Izagirre, U.; Lekube, X.; Stankevičiūtė, M.; Butrimavičienė, L.; Soto, M.; Lehtonen, K.K. Biomarker responses in mussels (Mytilus trossulus) from the Baltic Sea exposed to water-accommodated fraction of crude oil and a dispersant at different salinities. Mar. Pollut. Bull. 2023, 192, 115100. [Google Scholar] [CrossRef] [PubMed]
- Reunamo, A.; Yli-Hemminki, P.; Nuutinen, J.; Jörgensen, K.S.; Lehtoranta, J. Degradation of crude oil and PAHs by microbial communities of iron-manganese concretions and sediment in the northern Baltic Sea. Geomicrobiol. J. 2016, 34, 385–399. [Google Scholar] [CrossRef]
- Schloss, P.D. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS ONE 2009, 4, e8230. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Sólymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package, 2012, Software. Available online: http://CRAN.R-project.org/package=vegan (accessed on 10 February 2025).
- van der Oost, R.; Beyer, J.; Vermeulen, N.P. Fish bioaccumulation and biomarkers in environmental risk assessment: A review. Environ. Toxicol. Pharmacol. 2003, 13, 57–149. [Google Scholar] [CrossRef] [PubMed]
- Garmendia, L.; Soto, M.; Vicario, U.; Kim, Y.; Cajaraville, M.P.; Marigómez, I. Application of a battery of biomarkers in mussel digestive gland to assess long-term effects of the Prestige oil spill in Galicia and Bay of Biscay: Tissue-level biomarkers and histopathology. J. Environ. Monit. 2011, 13, 915–932. [Google Scholar] [CrossRef] [PubMed]
- Vethaak, A.D.; Davies, I.M.; Thain, J.E.; Gubbins, M.J.; Martínez-Gómez, C.; Robinson, C.D.; Moffat, C.F.; Burgeot, T.; Maes, T.; Wosniok, W.; et al. Integrated indicator framework and methodology for monitoring and assessment of hazardous substances and their effects in the marine environment. Mar. Environ. Res. 2017, 124, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Sanni, S.; Björkblom, C.; Jonsson, H.; Godal, B.F.; Liewenborg, B.; Lyng, E.; Pampanin, D.M. I: Biomarker quantification in fish exposed to crude oil as input to species sensitivity distributions and threshold values for environmental monitoring. Mar. Environ. Res. 2017, 125, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Beyer, J.; Green, N.W.; Brooks, S.; Allan, I.J.; Ruus, A.; Gomes, T.; Bråte, I.L.N.; Schøyen, M. Blue mussels (Mytilus edulis spp.) as sentinel organisms in coastal pollution monitoring: A review. Mar. Environ. Res. 2017, 130, 338–365. [Google Scholar] [CrossRef] [PubMed]
- Turja, R.; Sanni, S.; Stankevičiūtė, M.; Butrimavičienė, L.; Devier, M.H.; Budzinski, H.; Lehtonen, K.K. Biomarker responses and accumulation of polycyclic aromatic hydrocarbons in Mytilus trossulus and Gammarus oceanicus during exposure to crude oil. Environ. Sci. Pollut. Res. 2020, 27, 15498–15514. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shukla, P.; Gopalani, M.; Ramteke, D.S.; Wate, S.R. Influence of salinity on PAH Uptake from water soluble fraction of crude oil in Tilapia mossambica. Bull. Environ. Contam. Toxicol. 2007, 79, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Gan, N.; Martin, L.; Xu, W. Impact of Polycyclic Aromatic Hydrocarbon Accumulation on Oyster Health. FFront. Physiol. 2021, 12, 734463. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ramachandran, S.D.; Sweezey, M.J.; Hodson, P.V.; Boudreau, M.; Courtenay, S.C.; Lee, K.; King, T.; Dixon, J.A. Influence of salinity and fish species on PAH uptake from dispersed crude oil. Mar. Pollut. Bull. 2006, 52, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- DeLorenzo, M.E.; Key, P.B.; Chung, K.W.; Aaby, K.; Hausman, D.; Jean, C.; Pennington, P.L.; Pisarski, E.C.; Wirth, E.F. Multi-stressor Effects of Ultraviolet Light, Temperature, and Salinity on Louisiana Sweet Crude Oil Toxicity in Larval Estuarine Organisms. Arch. Environ. Contam. Toxicol. 2021, 80, 461–473. [Google Scholar] [CrossRef] [PubMed]
- Rosales, S.M.; Huebner, L.K.; Evans, J.S.; Apprill, A.; Baker, A.C.; Becker, C.C.; Bellantuono, A.J.; E Brandt, M.; Clark, A.S.; del Campo, J.; et al. A meta-analysis of the stony coral tissue loss disease microbiome finds key bacteria in unaffected and lesion tissue in diseased colonies. ISME Commun. 2023, 3, 19. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Owusu-Agyeman, I.; Plaza, E.; Cetecioglu, Z. Long-term alkaline volatile fatty acids production from waste streams: Impact of pH and dominance of Dysgonomonadaceae. Bioresour. Technol. 2022, 346, 126621. [Google Scholar] [CrossRef]
- Rahman, F.U.; Andree, K.B.; Salas-Massó, N.; Fernandez-Tejedor, M.; Sanjuan, A.; Figueras, M.J.; Furones, M.D. Improved culture enrichment broth for isolation of Arcobacter-like species from the marine environment. Sci. Rep. 2020, 10, 14547. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bagi, A.; Pampanin, D.M.; Lanzén, A.; Bilstad, T.; Kommedal, R. Naphthalene biodegradation in temperate and arctic marine microcosms. Biodegradation 2014, 25, 111–125. [Google Scholar] [CrossRef]
- Thomas, G.E.; Brant, J.L.; Campo, P.; Clark, D.R.; Coulon, F.; Gregson, B.H.; McGenity, T.J.; McKew, B.A. Effects of dispersants and biosurfactants on crude-oil biodegradation and bacterial community succession. Microorganisms 2021, 9, 1200. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).