Evaluation of qPCR for the Selective Detection of Enteric Adenovirus Followed by Sequence-Based Genetic Characterization of F Strains Circulating in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Stool Collection and Ethics Statement
2.2. Viral DNA Extraction
2.3. HAdV-F Detection and Quantification
2.4. HAdV-F Genetic Diversity and Nucleotide Sequencing
2.5. Phylogenetic and Mutation Analysis of F40/41
2.6. Statistical Analysis
3. Results
3.1. Detection of HAdV-F40/41 Using a Specific qPCR Assay
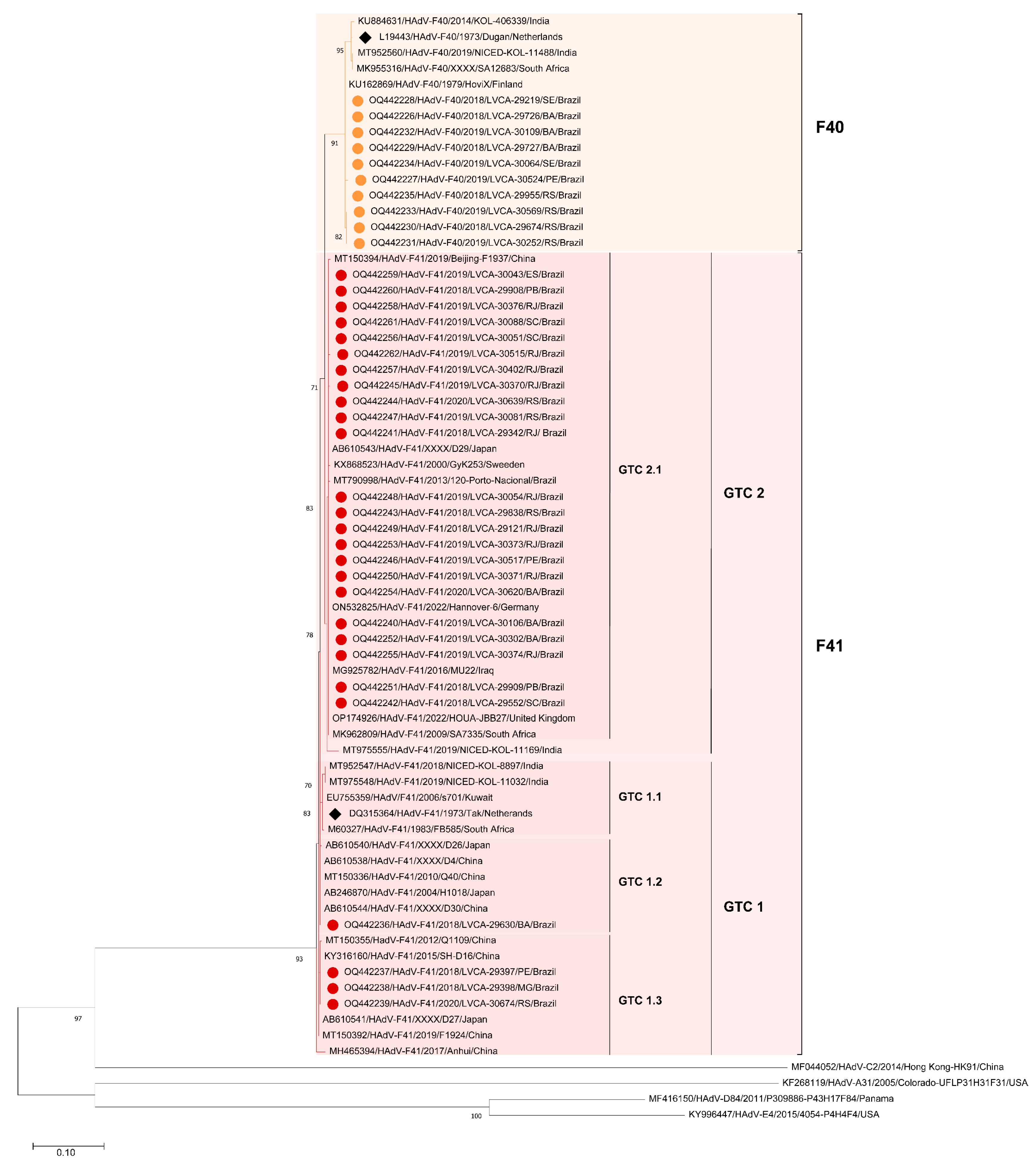
3.2. Phylogenetic Analysis of HAdV-F Hexon and Fiber Genes
3.2.1. Hexon Gene
3.2.2. Fiber Gene
3.3. Mutation Analysis of the Hexon and Fiber Protein of HAdV-F Strains Circulating in Brazil
3.3.1. Hexon Gene
3.3.2. Fiber Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2016 Diarrhoeal Disease Collaborators. Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Diarrhoea in 195 Countries: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1211–1228. [Google Scholar] [CrossRef] [PubMed]
- Wardlaw, T.; Salama, P.; Brocklehurst, C.; Chopra, M.; Mason, E. Diarrhoea: Why Children Are Still Dying and What Can Be Done. Lancet 2010, 375, 870–872. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.L.; Platts-Mills, J.A.; Nakamura, T.; Operario, D.J.; Antoni, S.; Mwenda, J.M.; Weldegebriel, G.; Rey-Benito, G.; de Oliveira, L.H.; Ortiz, C.; et al. Aetiology and Incidence of Diarrhoea Requiring Hospitalisation in Children under 5 Years of Age in 28 Low-Income and Middle-Income Countries: Findings from the Global Pediatric Diarrhea Surveillance network. BMJ Glob. Health 2022, 7, e009548. [Google Scholar] [CrossRef] [PubMed]
- Kotloff, K.L.; Nataro, J.P.; Blackwelder, W.C.; Nasrin, D.; Farag, T.H.; Panchalingam, S.; Wu, Y.; Sow, S.O.; Sur, D.; Breiman, R.F.; et al. Burden and Aetiology of Diarrhoeal Disease in Infants and Young Children in Developing Countries (The Global Enteric Multicenter Study, GEMS): A Prospective, Case-Control Study. Lancet 2013, 382, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Operario, D.J.; Platts-Mills, J.A.; Nadan, S.; Page, N.; Seheri, M.; Mphahlele, J.; Praharaj, I.; Kang, G.; Araujo, I.T.; Leite, J.P.G.; et al. Etiology of Severe Acute Watery Diarrhea in Children in the Global Rotavirus Surveillance Network Using Quantitative Polymerase Chain Reaction. J. Infect. Dis. 2017, 216, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Platts-Mills, J.A.; Liu, J.; Rogawski, E.T.; Kabir, F.; Lertsethtakarn, P.; Siguas, M.; Khan, S.S.; Praharaj, I.; Murei, A.; Nshama, R.; et al. Use of Quantitative Molecular Diagnostic Methods to Assess the Aetiology, Burden, and Clinical Characteristics of Diarrhoea in Children in Low-Resource Settings: A Reanalysis of the MAL-ED Cohort Study. Lancet Glob. Health 2018, 6, e1309–e1318. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Benkő, M.; Harrach, B. Genetic Content and Evolution of Adenoviruses. J. Gen. Virol. 2003, 84, 2895–2908. [Google Scholar] [CrossRef] [PubMed]
- Seto, D.; Chodosh, J.; Brister, J.R.; Jones, M.S.; Members of the Adenovirus Research Community. Using the Whole-Genome Sequence to Characterize and Name Human Adenoviruses. J. Virol. 2011, 85, 5701–5702. [Google Scholar] [CrossRef]
- Lion, T. Adenovirus Infections in Immunocompetent and Immunocompromised Patients. Clin. Microbiol. Rev. 2014, 27, 441–462. [Google Scholar] [CrossRef]
- Lynch, J.P.; Kajon, A.E. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention. Semin. Respir. Crit. Care Med. 2016, 37, 586–602. [Google Scholar] [CrossRef]
- Shieh, W.-J. Human Adenovirus Infections in Pediatric Population—An Update on Clinico-Pathologic Correlation. Biomed. J. 2021, 45, 38–49. [Google Scholar] [CrossRef]
- Lee, B.; Damon, C.F.; Platts-Mills, J.A. Pediatric Acute Gastroenteritis Associated with Adenovirus 40/41 in Low-Income and Middle-Income Countries. Curr. Opin. Infect. Dis. 2020, 33, 398–403. [Google Scholar] [CrossRef]
- Afrad, M.H.; Avzun, T.; Haque, J.; Haque, W.; Hossain, M.E.; Rahman, A.R.; Ahmed, S.; Faruque, A.S.G.; Rahman, M.Z.; Rahman, M. Detection of Enteric- and Non-Enteric Adenoviruses in Gastroenteritis Patients, Bangladesh, 2012–2015. J. Med. Virol. 2018, 90, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Kumthip, K.; Khamrin, P.; Ushijima, H.; Maneekarn, N. Enteric and Non-Enteric Adenoviruses Associated with Acute Gastroenteritis in Pediatric Patients in Thailand, 2011 to 2017. PLoS ONE 2019, 14, e0220263. [Google Scholar] [CrossRef]
- Nascimento, L.G.; Fialho, A.M.; Andrade, J.d.S.R.; Assis, R.M.S.; Fumian, T.M. Human Enteric Adenovirus F40/41 as a Major Cause of Acute Gastroenteritis in Children in Brazil, 2018 to 2020. Sci. Rep. 2022, 12, 11220. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-L.; Bessey, T.; Wang, S.-M.; Mo, Z.-J.; Barclay, L.; Wang, J.-X.; Zhang, C.-J.; Ma, J.-C.; Qiu, C.; Zhao, G.; et al. Burden and Etiology of Moderate and Severe Diarrhea in Children Less than 5 Years of Age Living in North and South of China: Prospective, Population-Based Surveillance. Gut Pathog. 2021, 13, 33. [Google Scholar] [CrossRef]
- Marsh, K.; Tayler, R.; Pollock, L.; Roy, K.; Lakha, F.; Ho, A.; Henderson, D.; Divala, T.; Currie, S.; Yirrell, D.; et al. Investigation into Cases of Hepatitis of Unknown Aetiology among Young Children, Scotland, 1 January 2022 to 12 April 2022. Euro Surveill. 2022, 27, 2200318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.-Y.; Huang, L.-S.; Yue, Y.-H.; Fawaz, R.; Lim, J.K.; Fan, J.-G. Acute Hepatitis of Unknown Origin in Children: Early Observations from the 2022 Outbreak. J. Clin. Transl. Hepatol. 2022, 10, 522–530. [Google Scholar] [CrossRef]
- Grand, R.J. Pathogenicity and Virulence of Human Adenovirus F41: Possible Links to Severe Hepatitis in Children. Virulence 2023, 14, 2242544. [Google Scholar] [CrossRef]
- Liu, J.; Gratz, J.; Amour, C.; Nshama, R.; Walongo, T.; Maro, A.; Mduma, E.; Platts-Mills, J.; Boisen, N.; Nataro, J.; et al. Optimization of Quantitative PCR Methods for Enteropathogen Detection. PLoS ONE 2016, 11, e0158199. [Google Scholar] [CrossRef]
- Hernroth, B.E.; Conden-Hansson, A.-C.; Rehnstam-Holm, A.-S.; Girones, R.; Allard, A.K. Environmental Factors Influencing human Viral Pathogens and Their Potential Indicator Organisms in the Blue Mussel, Mytilus Edulis: The First Scandinavian Report. Appl. Environ. Microbiol. 2002, 68, 4523–4533. [Google Scholar] [CrossRef]
- Allard, A.; Albinsson, B.; Wadell, G. Rapid Typing of Human Adenoviruses by a General PCR Combined with Restriction Endonuclease Analysis. J. Clin. Microbiol. 2001, 39. [Google Scholar] [CrossRef]
- Li, L.; Shimizu, H.; Doan, L.T.P.; Tung, P.G.; Okitsu, S.; Nishio, O.; Suzuki, E.; Seo, J.K.; Kim, K.S.; Müller, W.E.G.; et al. Characterizations of Adenovirus Type 41 Isolates from Children with Acute Gastroenteritis in Japan, Vietnam, and Korea. J. Clin. Microbiol. 2004, 42, 4032–4039. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; McDonough, M.C.; Erdman, D.D. Species-Specific Identification of Human Adenoviruses by a Multiplex PCR Assay. J. Clin. Microbiol. 2000, 38, 4114–4120. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Platts-Mills, J.A.; Juma, J.; Kabir, F.; Nkeze, J.; Okoi, C.; Operario, D.J.; Uddin, J.; Ahmed, S.; Alonso, P.L.; et al. Use of Quantitative Molecular Diagnostic Methods to Identify Causes of Diarrhoea in Children: A Reanalysis of the GEMS Case-Control Study. Lancet 2016, 388, 1291–1301. [Google Scholar] [CrossRef]
- Sarmento, S.K.; de Andrade, J.d.S.R.; Miagostovich, M.P.; Fumian, T.M. Virological and Epidemiological Features of Norovirus Infections in Brazil, 2017–2018. Viruses 2021, 13, 1724. [Google Scholar] [CrossRef]
- Gutierrez, M.B.; Fialho, A.M.; Maranhão, A.G.; Malta, F.C.; Andrade, J.d.S.R.d.; Assis, R.M.S.d.; Mouta, S.d.S.e.; Miagostovich, M.P.; Leite, J.P.G.; Machado Fumian, T. Rotavirus A in Brazil: Molecular Epidemiology and Surveillance during 2018–2019. Pathogens 2020, 9, 515. [Google Scholar] [CrossRef]
- Gelaw, A.; Pietsch, C.; Liebert, U.G. Genetic Diversity of Human Adenovirus and Human Astrovirus in Children with Acute Gastroenteritis in Northwest Ethiopia. Arch. Virol. 2019, 164, 2985–2993. [Google Scholar] [CrossRef] [PubMed]
- Portal, T.M.; Reymão, T.K.A.; Quinderé Neto, G.A.; Fiuza, M.K.D.C.; Teixeira, D.M.; Lima, I.C.G.; Sousa Júnior, E.C.; Bandeira, R.D.S.; De Deus, D.R.; Justino, M.C.A.; et al. Detection and Genotyping of Enteric Viruses in Hospitalized Children with Acute Gastroenteritis in Belém, Brazil: Occurrence of Adenovirus Viremia by Species F, Types 40/41. J. Med. Virol. 2019, 91, 378–384. [Google Scholar] [CrossRef]
- Primo, D.; Pacheco, G.T.; Timenetsky, M.d.C.S.T.; Luchs, A. Surveillance and Molecular Characterization of Human Adenovirus in Patients with Acute Gastroenteritis in the Era of Rotavirus Vaccine, Brazil, 2012–2017. J. Clin. Virol. 2018, 109, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Portes, S.A.R.; Volotão, E.d.M.; Rocha, M.S.; Rebelo, M.C.; Xavier, M.d.P.T.P.; Assis, R.M.d.; Rose, T.L.; Miagostovich, M.P.; Leite, J.P.G.; Carvalho-Costa, F.A. A Non-Enteric Adenovirus A12 Gastroenteritis Outbreak in Rio de Janeiro, Brazil. Mem. Inst. Oswaldo Cruz 2016, 111, 403–406. [Google Scholar] [CrossRef] [PubMed]
- Qiu, F.-Z.; Shen, X.-X.; Li, G.-X.; Zhao, L.; Chen, C.; Duan, S.-X.; Guo, J.-Y.; Zhao, M.-C.; Yan, T.-F.; Qi, J.-J.; et al. Adenovirus Associated with Acute Diarrhea: A Case-Control Study. BMC Infect. Dis. 2018, 18, 450. [Google Scholar] [CrossRef] [PubMed]
- De Francesco, M.A.; Lorenzin, G.; Meini, A.; Schumacher, R.F.; Caruso, A. Nonenteric Adenoviruses Associated with Gastroenteritis in Hospitalized Children. Microbiol. Spectr. 2021, 9, e0030021. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, A.; Hage, E.; Ganzenmueller, T.; Böttcher, S.; Hofmann, J.; Hamprecht, K.; Obermeier, P.; Rath, B.; Hausmann, F.; Dobner, T.; et al. Molecular Evolution of Human Adenovirus (HAdV) Species C. Sci. Rep. 2019, 9, 1039. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A.N.; Ivanova, O.E.; Eremeeva, T.P.; Iggo, R.D. Evidence of Frequent Recombination among Human Adenoviruses. J. Gen. Virol. 2008, 89, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Robinson, C.M.; Singh, G.; Lee, J.Y.; Dehghan, S.; Rajaiya, J.; Liu, E.B.; Yousuf, M.A.; Betensky, R.A.; Jones, M.S.; Dyer, D.W.; et al. Molecular Evolution of Human Adenoviruses. Sci. Rep. 2013, 3, 1812. [Google Scholar] [CrossRef] [PubMed]
- Götting, J.; Cordes, A.K.; Steinbrück, L.; Heim, A. Molecular Phylogeny of Human Adenovirus Type 41 Lineages. Virus Evol. 2022, 8, veac098. [Google Scholar] [CrossRef]
- Maes, M.; Khokhar, F.; Wilkinson, S.A.J.; Smith, A.D.; Kovalenko, G.; Dougan, G.; Quick, J.; Loman, N.J.; Baker, S.; Curran, M.D.; et al. Multiplex MinION Sequencing Suggests Enteric Adenovirus F41 Genetic Diversity Comparable to Pre-COVID-19 Era. Microb. Genomics 2023, 9, mgen000920. [Google Scholar] [CrossRef] [PubMed]
- Lambisia, A.W.; Makori, T.O.; Mutunga, M.; Cheruiyot, R.; Murunga, N.; Quick, J.; Githinji, G.; Nokes, D.J.; Houldcroft, C.J.; Agoti, C.N. Genomic Epidemiology of Human Adenovirus F40 and F41 in Coastal Kenya: A Retrospective Hospital-Based Surveillance Study (2013–2022). Virus Evol. 2023, 9, vead023. [Google Scholar] [CrossRef] [PubMed]
- Chandra, P.; Lo, M.; Mitra, S.; Banerjee, A.; Saha, P.; Okamoto, K.; Deb, A.K.; Ghosh, S.K.; Manna, A.; Dutta, S.; et al. Genetic Characterization and Phylogenetic Variations of Human Adenovirus-F Strains Circulating in Eastern India during 2017–2020. J. Med. Virol. 2021, 93, 6180–6190. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, L.H.G.; Shiau, H.; Baker, J.M.; Saaybi, S.; Buchfellner, M.; Britt, W.; Sanchez, V.; Potter, J.L.; Ingram, L.A.; Kelly, D.; et al. A Case Series of Children with Acute Hepatitis and Human Adenovirus Infection. N. Engl. J. Med. 2022, 387, 620–630. [Google Scholar] [CrossRef] [PubMed]
- Kelgeri, C.; Couper, M.; Gupte, G.L.; Brant, A.; Patel, M.; Johansen, L.; Valamparampil, J.; Ong, E.; Hartog, H.; Perera, M.T.P.R.; et al. Clinical Spectrum of Children with Acute Hepatitis of Unknown Cause. N. Engl. J. Med. 2022, 387, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Morfopoulou, S.; Buddle, S.; Torres Montaguth, O.E.; Atkinson, L.; Guerra-Assunção, J.A.; Moradi Marjaneh, M.; Zennezini Chiozzi, R.; Storey, N.; Campos, L.; Hutchinson, J.C.; et al. Genomic Investigations of Unexplained Acute Hepatitis in Children. Nature 2023, 617, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.; Eslick, G.D.; Elliott, E.J. Demystifying the Global Outbreak of Severe Acute Hepatitis of Unknown Aetiology in Children: A Systematic Review and Meta-Analysis. J. Infect. 2024, 88, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.A.; Gonzalez, G.; Reynolds, L.J.; Bennett, C.; Campbell, C.; Nolan, T.M.; Byrne, A.; Fennema, S.; Holohan, N.; Kuntamukkula, S.R.; et al. Adeno-Associated Virus 2 and Human Adenovirus F41 in Wastewater during Outbreak of Severe Acute Hepatitis in Children, Ireland. Emerg. Infect. Dis. 2023, 29, 751–760. [Google Scholar] [CrossRef]
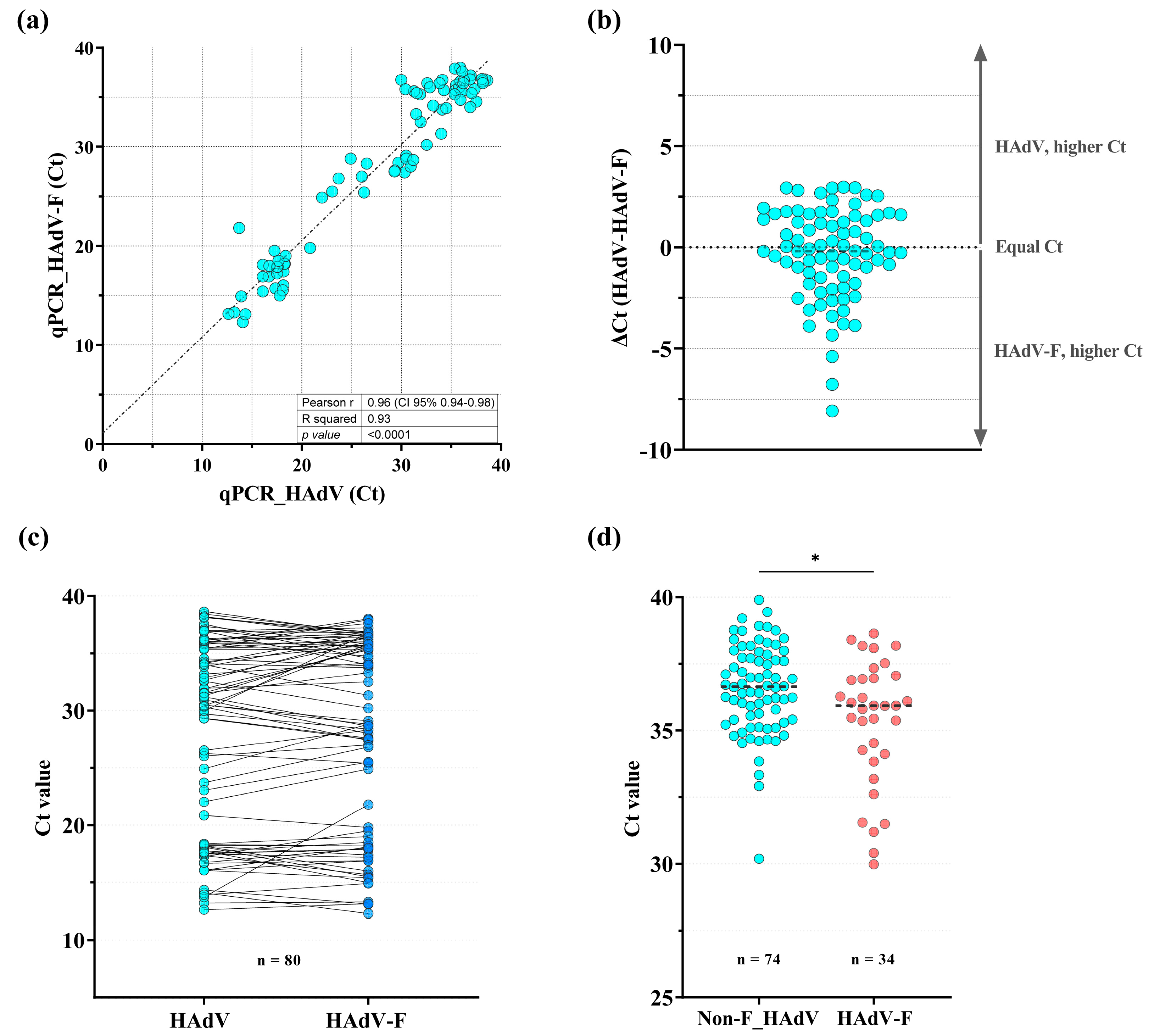


| HAdV Classification | HAdV Degenerated qPCR 1 | HAdV-F qPCR | Positivity (%) | |
|---|---|---|---|---|
| N° of HAdV-Positive Tested Samples | Positive | Negative | ||
| Enteric types | 46 | 43 | 3 | 93.5 |
| Non-enteric types | 66 | 3 | 63 | 4.5 |
| Non-sequenced HAdV | 108 | 34 | 74 | 31.5 |
| Total | 220 | 80 | 140 | 36.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nascimento, L.G.d.; Sarmento, S.K.; Röpke Junior, R.; Fumian, T.M. Evaluation of qPCR for the Selective Detection of Enteric Adenovirus Followed by Sequence-Based Genetic Characterization of F Strains Circulating in Brazil. Appl. Microbiol. 2024, 4, 1016-1029. https://doi.org/10.3390/applmicrobiol4030069
Nascimento LGd, Sarmento SK, Röpke Junior R, Fumian TM. Evaluation of qPCR for the Selective Detection of Enteric Adenovirus Followed by Sequence-Based Genetic Characterization of F Strains Circulating in Brazil. Applied Microbiology. 2024; 4(3):1016-1029. https://doi.org/10.3390/applmicrobiol4030069
Chicago/Turabian StyleNascimento, Lilian Gonçalves do, Sylvia Kahwage Sarmento, Reinaldo Röpke Junior, and Tulio Machado Fumian. 2024. "Evaluation of qPCR for the Selective Detection of Enteric Adenovirus Followed by Sequence-Based Genetic Characterization of F Strains Circulating in Brazil" Applied Microbiology 4, no. 3: 1016-1029. https://doi.org/10.3390/applmicrobiol4030069
APA StyleNascimento, L. G. d., Sarmento, S. K., Röpke Junior, R., & Fumian, T. M. (2024). Evaluation of qPCR for the Selective Detection of Enteric Adenovirus Followed by Sequence-Based Genetic Characterization of F Strains Circulating in Brazil. Applied Microbiology, 4(3), 1016-1029. https://doi.org/10.3390/applmicrobiol4030069







