First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolation and Morphological Identification
2.2. Morphological and Cultural Characterization
2.3. DNA Extraction, PCR Amplifications and Sequencing
2.4. Phylogenetic Analyses
3. Results
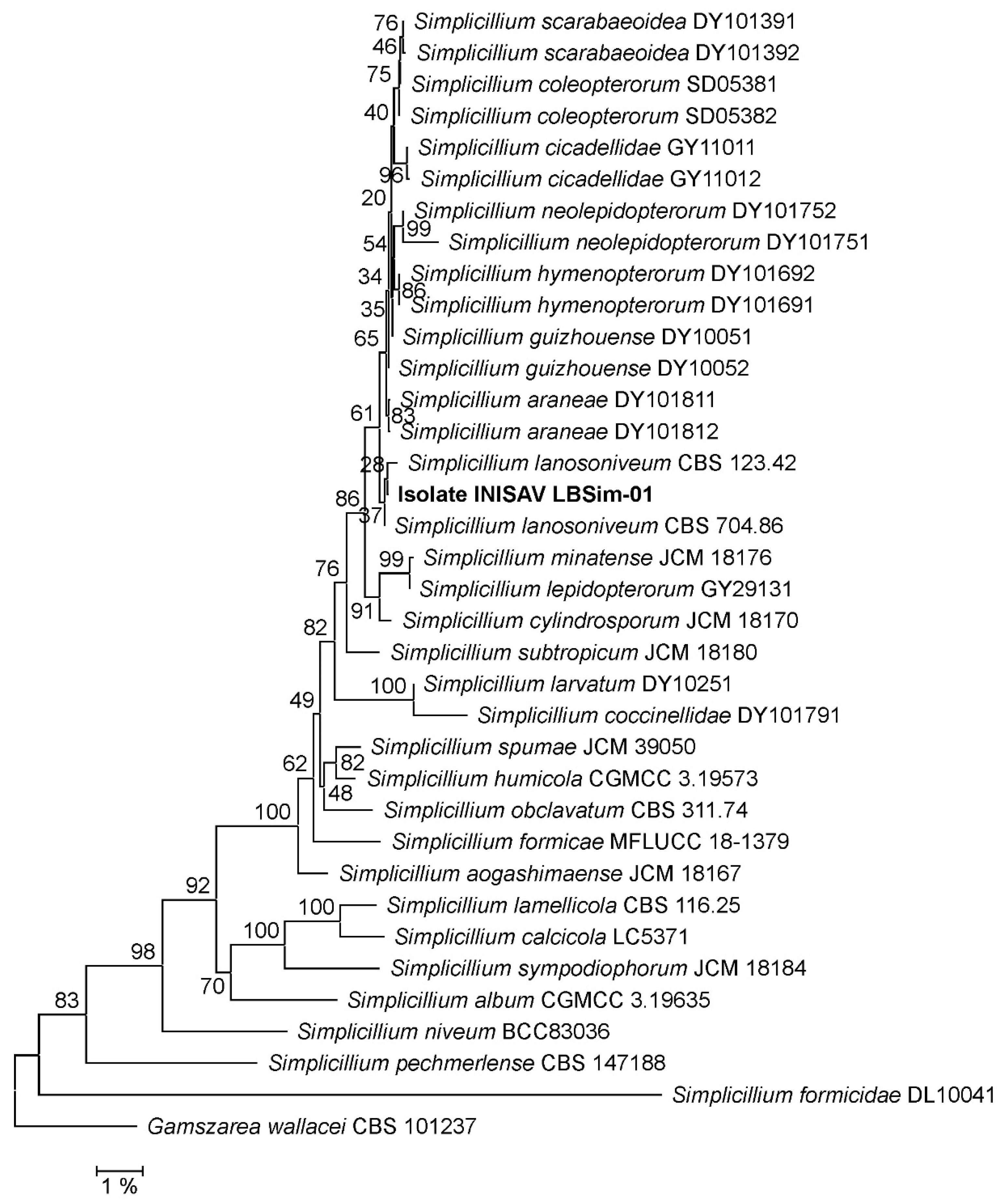
3.1. Phylogeny
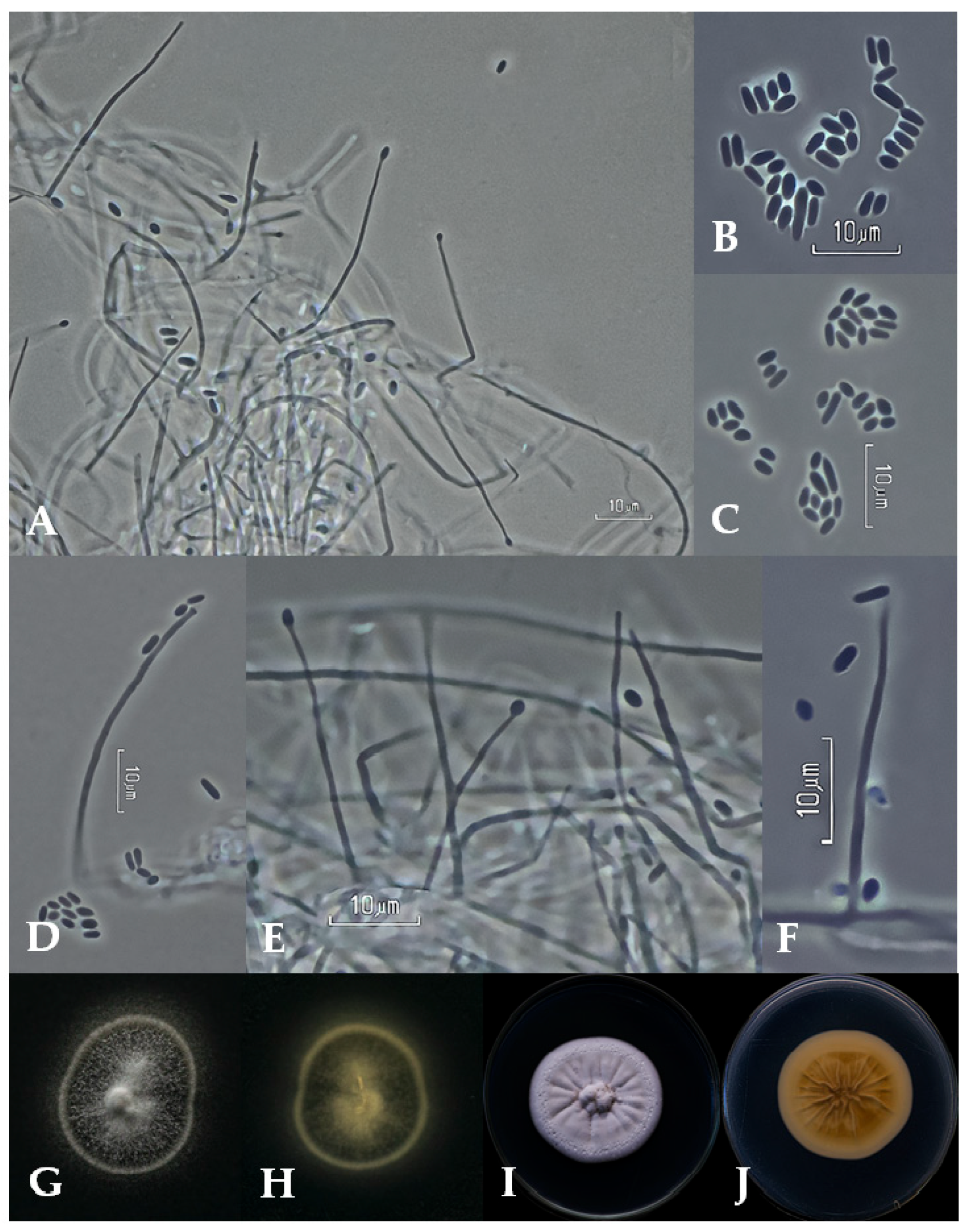
3.2. Taxonomy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Markets and Trade: Coffee. 2022. Available online: https://www.fao.org/markets-and-trade/commodities/coffee/en/ (accessed on 22 March 2023).
- Bermudez, S.; Voora, V.; Larrea, C. Global Market Report: Coffee Prices and Sustainability (Sustainable Commodities Marketplace Series); International Institute for Sustainable Development: Winnipeg, MB, Canada, 2022; Available online: https://www.iisd.org/sites/default/files/publications/ssi-global-market-report-coffee.pdf (accessed on 31 May 2023).
- Pérez, L. La roya del cafeto en Cuba. Evolución al manejo alternativo de la enfermedad. In Memorias del Seminario Científico Internacional “Manejo Agroecológico de la Roya del Café”; FAO: Rome, Italy, 2015. [Google Scholar]
- Subit Lamí, D.; Ricabal, P.M.S.; Pérez-Vicente, L. Efectividad biológica del plaguicida Verdadero GD 600 para el control de la Roya del Café (Hemileia vastatrix Berkeley & Broome) en la Provincia Cienfuegos. Rev. Cient. Agroecosist. 2020, 7, 163–168. [Google Scholar]
- Setiawati, R.; Widiastuti, A.; Wibowo, A.; Priyatmojo, A. Variability of Lecanicillium spp. Mycoparasite of Coffee Leaf Rust Pathogen (Hemileia vastatrix) in Indonesia. Pak. J. Biol. Sci. 2021, 24, 588–598. [Google Scholar] [CrossRef]
- Manfrino, R.; Gutierrez, A.; Diez del Valle, F.; Schuster, C.; Ben Gharsa, H.; López Lastra, C.; Leclerque, A. First Description of Akanthomyces uredinophilus comb. nov. from Hemipteran Insects in America. Diversity 2022, 14, 1118. [Google Scholar] [CrossRef]
- García, N.G. Evaluación de la Calidad y Métodos de Producción de Bioplaguicidas para el Manejo de Hemileia vastatrix en Plantaciones de Café. Ph.D. Thesis, Centro Agronómico Tropical de Investigación y Enseñanza, Turrialba, Costa Rica, 2018. Available online: http://repositorio.bibliotecaorton.catie.ac.cr/bitstream/handle/11554/8823/Evaluacion_de_la_calidad.pdf?sequence=1&isAllowed=y (accessed on 22 March 2023).
- Ward, N.A.; Robertson, C.L.; Chanda, A.K.; Schneider, R.W. Effects of Simplicillium lanosoniveum on Phakopsora pachyrhizi, the soybean rust pathogen, and its use as a biological control agent. Phytopathology 2012, 102, 749–760. [Google Scholar] [CrossRef]
- Wang, N.; Fan, X.; Zhang, S.; Liu, B.; He, M.; Chen, X.; Tang, C.; Kang, Z.; Wang, X. Identification of a hyperparasitic Simplicillium obclavatum strain affecting the infection dynamics of Puccinia striiformis f. sp. tritici on wheat. Front. Microbiol. 2020, 11, 1277. [Google Scholar] [CrossRef]
- Lim, S.Y.; Lee, S.; Kong, H.G.; Lee, J. Entomopathogenicity of Simplicillium lanosoniveum isolated in Korea. Mycobiology 2014, 42, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Zare, R.; Gams, W.; Culham, A.A. Revision of Verticillium sect. Prostrata I. Phylogenetic studies using ITS sequences. Nova Hedwig. 2000, 71, 465–480. [Google Scholar] [CrossRef]
- Zare, R.; Gams, W. A revision of Verticillium section Prostrata. IV. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwig. 2001, 73, 1–50. [Google Scholar] [CrossRef]
- Sung, G.; Spatafora, J.W.; Zare, R.; Hodge, K.T.; Gams, W. A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedwig. 2001, 72, 311–328. [Google Scholar] [CrossRef]
- Sung, G.; Hywel-Jones, N.; Sung, J.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Lumbsch, H.T.; Liu, J.K.; Maharachchikumbura, S.S.N.; Ekanayaka, A.H.; Tian, Q.; Phookamsak, R. Outline of Ascomycota: 2017. Fungal Divers. 2018, 88, 167–263. [Google Scholar] [CrossRef]
- Chen, W.; Liang, J.; Ren, X.; Zhao, J.; Han, Y.; Liang, Z. Multigene phylogeny, phylogenetic network, and morphological characterizations reveal four new arthropod- associated Simplicillium species and their evolutional relationship. Front. Microbiol. 2022, 13, 950773. [Google Scholar] [CrossRef]
- Yan, Q.H.; Ni, Q.R.; Gu, W.J.; Liu, H.W.; Yuan, X.Y.; Sun, J.Z. Simplicillium sinense sp. nov., a novel potential pathogen of tinea faciei. Front. Microbiol. 2023, 14, 1156027. [Google Scholar] [CrossRef] [PubMed]
- Rayner, R.W.A. Mycological Colour Chart; Commonwealth Mycological Institute: London, UK, 1970. [Google Scholar]
- Humber, R.A. Fungi: Identification. In Manual of Techniques in Insect Pathology; Lacey, L., Ed.; Academic Press: San Diego, CA, USA, 1997; pp. 153–189. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 2nd ed.; Innis, M.A., Gelfand, D.H., Snisky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Baró, Y.; Schuster, C.; Gato, Y.; Márquez, M.E.; Leclerque, A. Characterization, identification and virulence of Metarhizium species from Cuba to control the sweet potato weevil, Cylas formicarius Fabricius (Coleoptera: Brentidae). J. Appl. Microbiol. 2022, 132, 3705–3716. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D. MAFFT multiple sequence alignment software version improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- James, T.; Marino, J.; Perfecto, I.; Vandermeer, J. Identification of putative coffee rust micoparasites via single-molecule DNA sequencing of infected pustules. Appl. Environ. Microbiol. 2016, 83, 631–639. [Google Scholar] [CrossRef]
- Si, B.; Wang, H.; Bai, J.; Zhang, Y.; Cao, Y. Evaluating the Utility of Simplicillium lanosoniveum, a Hyperparasitic Fungus of Puccinia graminis f. sp. tritici, as a Biological Control Agent against Wheat Stem Rust. Pathogens 2022, 12, 22. [Google Scholar] [CrossRef]
- Zhu, M.; Xiao, D.; Pengkun, C.; Yong, F.; Zongbo, Q. Deciphering the genome of Simplicillium aogashimaense to understand its mechanisms against the wheat powdery mildew fungus Blumeria graminis f. sp. tritici. Phytopathol. Res. 2022, 4, 16. [Google Scholar] [CrossRef]
- Chen, W.H.; Han, Y.F.; Liang, J.D.; Liang, Z.Q. Taxonomic and phylogenetic characterizations reveal four new species of Simplicillium (Cordycipitaceae, Hypocreales) from Guizhou, China. Sci. Rep. 2021, 11, 15300. [Google Scholar] [CrossRef] [PubMed]
- Gams, W. A contribution to the knowledge of nematophagous species of Verticillium. Neth. J. Plant Pathol. 1988, 94, 123–148. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, B.; Li, L.Y.; Zhu, X.F.; Wang, Y.Y.; Wang, J.Q.; Duan, Y.X.; Chen, L.J. Simplicillium chinense: A biological control agent against plant parasitic nematodes. Biocontrol Sci. Technol. 2013, 23, 980–986. [Google Scholar] [CrossRef]
- Luyen, V.T. Identification of the entomopathogenic fungi sample DL0069 by combination of morphological and molecular phylogenetic analyses. Vietnam J. Sci. Technol. 2017, 55, 117–123. [Google Scholar] [CrossRef]
- Ward, N.; Schneider, R.W.; Aime, M.C. Colonization of soybean rust sori by Simplicillium lanosoniveum. Fungal Ecol. 2011, 4, 303–308. [Google Scholar] [CrossRef]
- Gauthier, N.W.; Maruthachalam, K.; Subbarao, K.V.; Brown, M.; Xiao, Y.; Robertson, C.L.; Schneider, R.W. Mycoparasitism of Phakopsora pachyrhizi, the soybean rust pathogen, by Simplicillium lanosoniveum. Biol. Control 2014, 76, 87–94. [Google Scholar] [CrossRef]
- Chen, R.; Huang, C.; Li, J.; Tsay, J. Evaluation of characteristics of Simplicillium lanosoniveum on pathogenicity to aphids and in vitro antifungal potency against plant pathogenic fungi. Int. J. Environ. Agric. Res. 2017, 3, 55–61. Available online: https://ijoear.com/Paper-January-2017/IJOEAR-JAN-2017-7.pdf (accessed on 7 June 2023).

| Primer Designation | Primer Sequence (5′→3′) | Annealing Temperature (°C) | Elongation Time (sec) | Reference |
|---|---|---|---|---|
| LR0R | GTACCCGCTGAACTTAAGC | 58 | 90 | [23] |
| LR5 | ATCCTGAGGGAAACTTC | [23] | ||
| ITS4 | TCCTCCGCTTATTGATATGC | 52 | 60 | [24] |
| ITS5 | GGAAGTAAAAGTCGTAACAAGG | [24] | ||
| EF1A-983F | GCYCCYGGHCAYCGTGAYTTYAT | 52 | 90 | [25] |
| EF1A-2318R | ATGACACCRACRGCRACRGTYTG | [25] | ||
| EF1A-1567R | ACHGTRCCRATACCACCSATCTT | sequencing primer | [25] | |
| EF1A-1577F | CARGAYGTBTACAAGATYGGTGG | sequencing primer | [26] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baró Robaina, Y.; González Marrero, I.; Lorenzo Nicao, M.E.; Castañeda Ruiz, R.F.; Li, D.-W.; Ponce de la Cal, A.; Ben Gharsa, H.; Manfrino, R.G.; Schuster, C.; Leclerque, A. First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba. Appl. Microbiol. 2024, 4, 275-283. https://doi.org/10.3390/applmicrobiol4010018
Baró Robaina Y, González Marrero I, Lorenzo Nicao ME, Castañeda Ruiz RF, Li D-W, Ponce de la Cal A, Ben Gharsa H, Manfrino RG, Schuster C, Leclerque A. First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba. Applied Microbiology. 2024; 4(1):275-283. https://doi.org/10.3390/applmicrobiol4010018
Chicago/Turabian StyleBaró Robaina, Yamilé, Isel González Marrero, María Elena Lorenzo Nicao, Rafael F. Castañeda Ruiz, De-Wei Li, Amaia Ponce de la Cal, Haifa Ben Gharsa, Romina G. Manfrino, Christina Schuster, and Andreas Leclerque. 2024. "First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba" Applied Microbiology 4, no. 1: 275-283. https://doi.org/10.3390/applmicrobiol4010018
APA StyleBaró Robaina, Y., González Marrero, I., Lorenzo Nicao, M. E., Castañeda Ruiz, R. F., Li, D.-W., Ponce de la Cal, A., Ben Gharsa, H., Manfrino, R. G., Schuster, C., & Leclerque, A. (2024). First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba. Applied Microbiology, 4(1), 275-283. https://doi.org/10.3390/applmicrobiol4010018








