Toward Effects of Hydrophobicity on Biosurfactant Production by Bacillus subtilis Isolates from Crude-Oil-Exposed Environments
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Screening of Crude Oil Degrading and Biosurfactant-Producing Bacterial Isolates
2.3. Measurement of Surface Tension
2.4. 16S rRNA Sequence Analysis
2.5. Quantitative Lipopeptide Analysis
2.6. Quantification of Glucose Consumption during Cultivation
2.7. Cultivation Conditions for Lipopeptide Production
2.8. Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)
2.9. Extraction of Hexadecane and Calculation of Biodegradation Efficiency
2.10. Calculation of Yield and Productivity
2.11. Sample Preparation for Mass Spectrometric Proteome Determination
2.12. LC-MS/MS Analysis
2.13. MS Data Analysis and Protein Quantification
3. Results
3.1. Characterization of the Crude-Oil-Degrading and Biosurfactant-Producing Strains Isolated from Crude-Oil-Exposed Environments
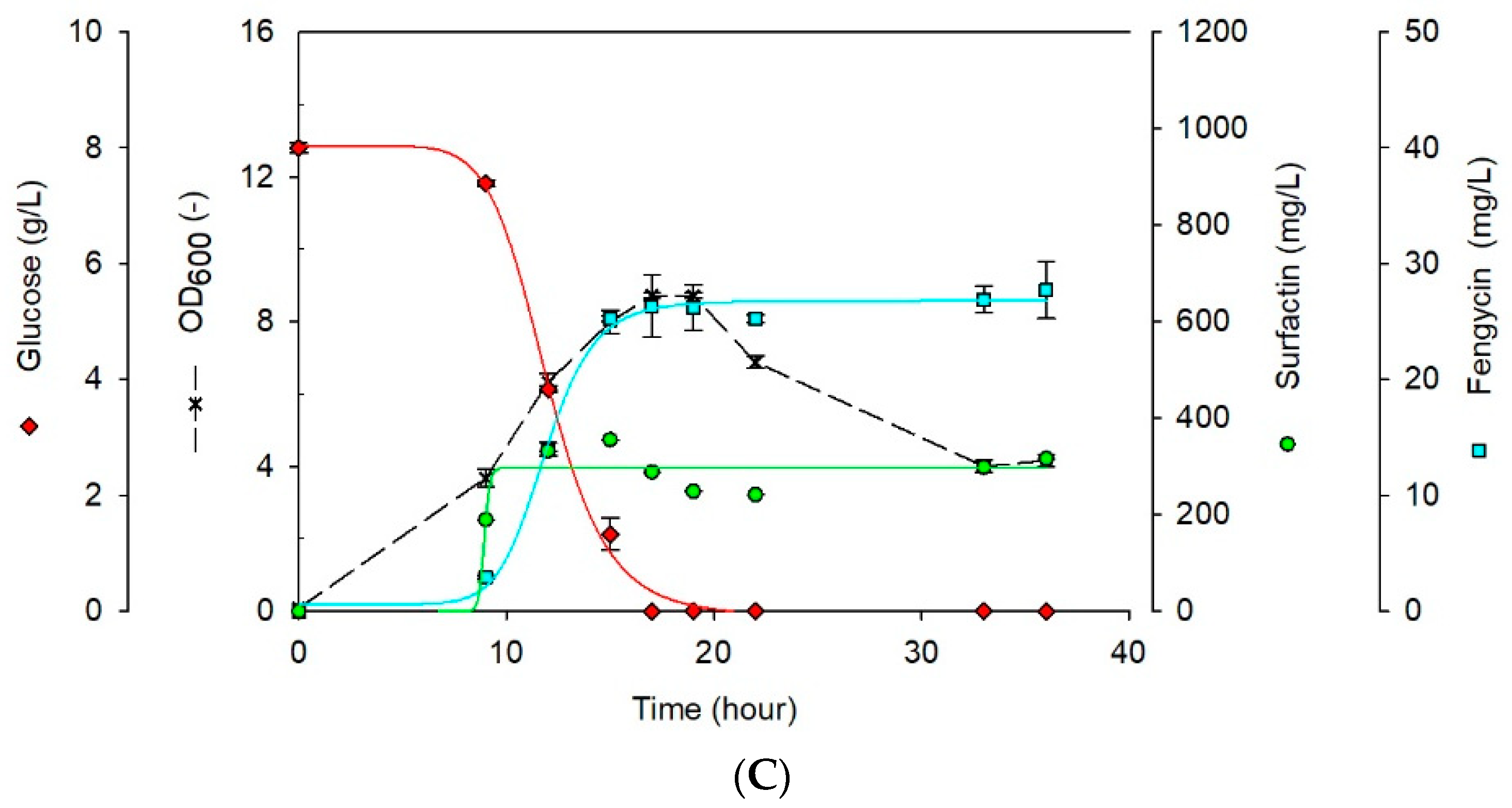
3.2. Lipopeptide Production by B. subtilis Strains ZH1 and P7
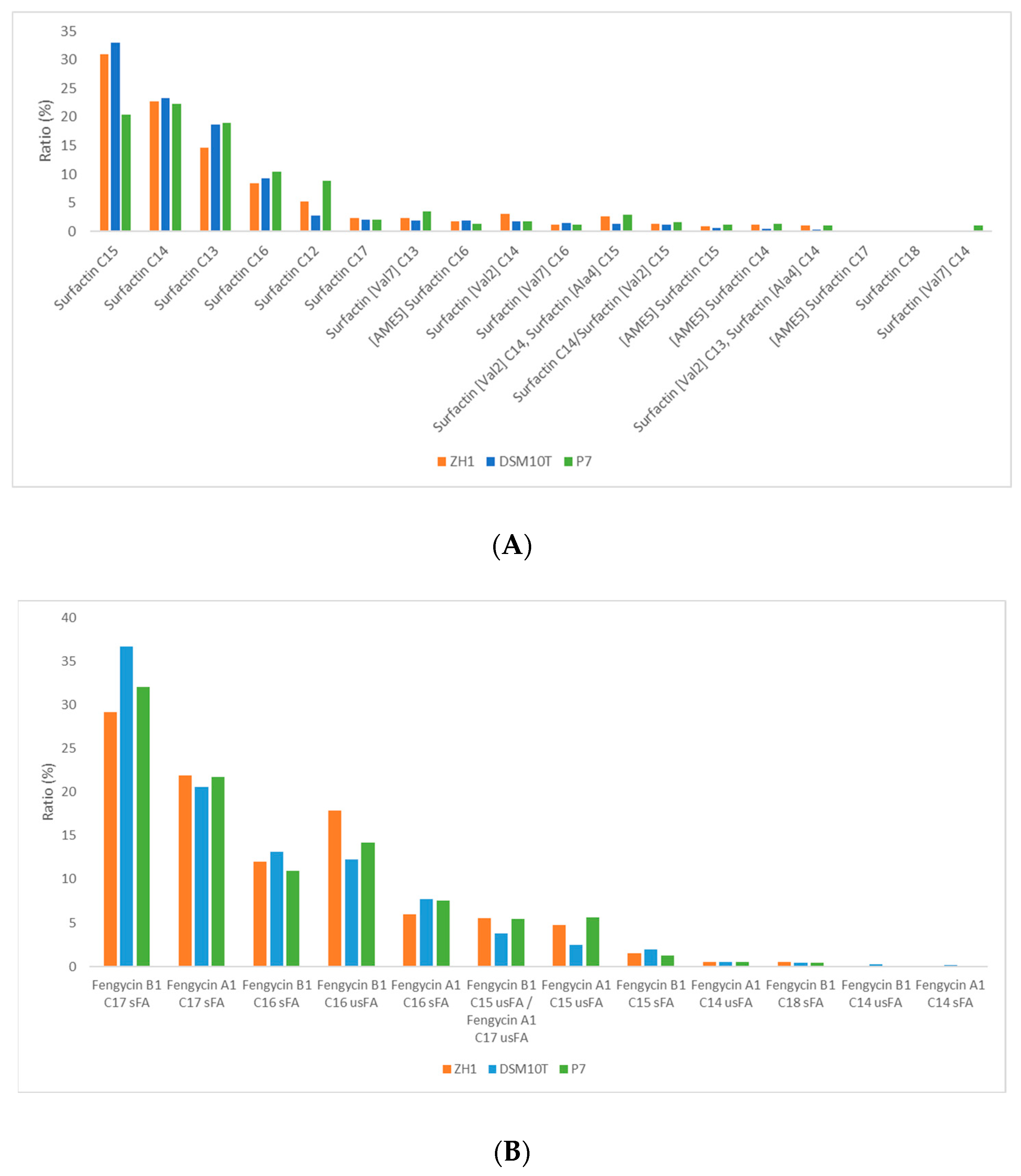
3.3. Comparative Mass Spectrometric Analyses of Lipopeptide Variants Produced by B. subtilis Isolates ZH1 and P7
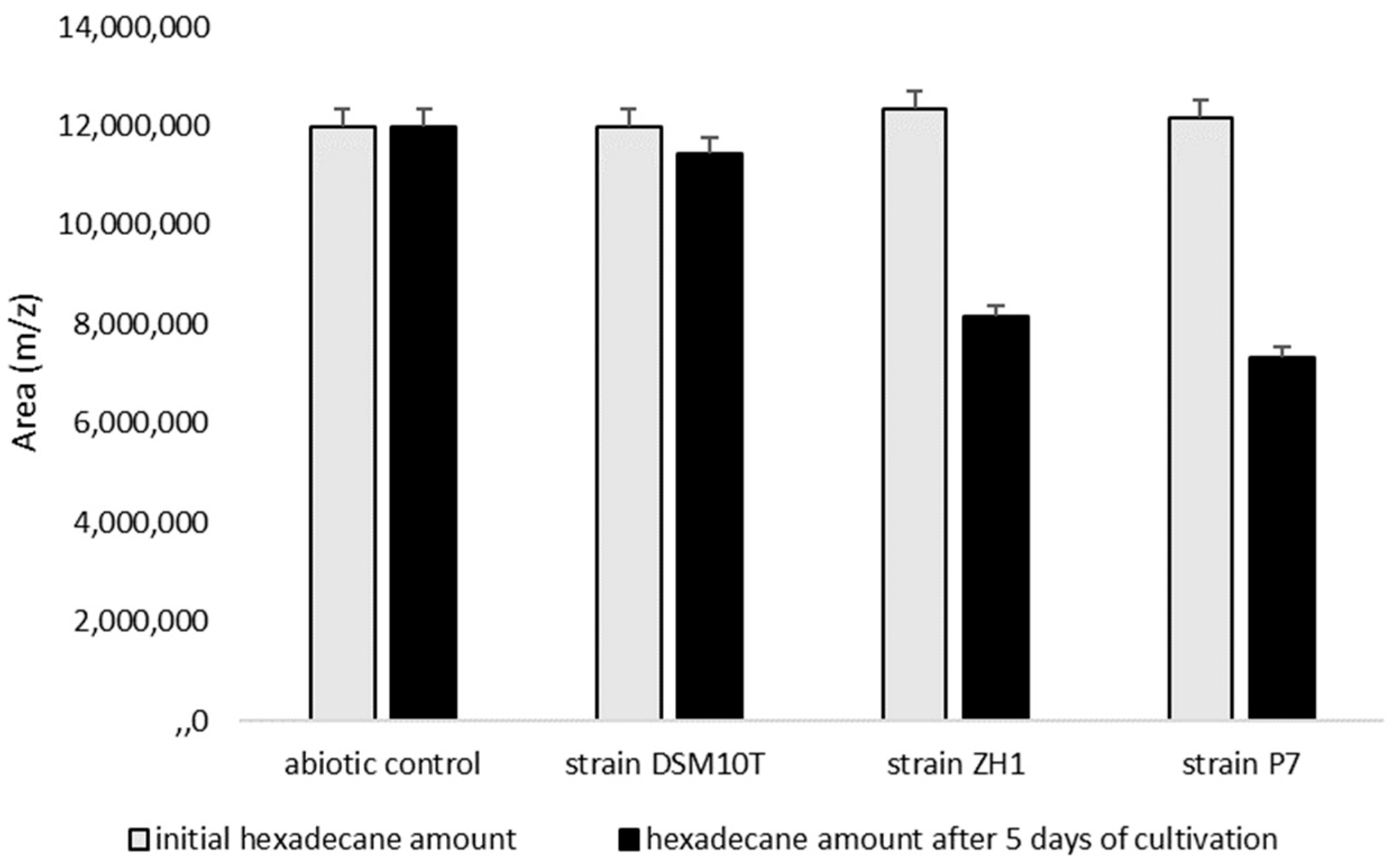
3.4. Degradation of Hexadecane as a Hydrophobic Substrate by B. subtilis Strains ZH1 and P7
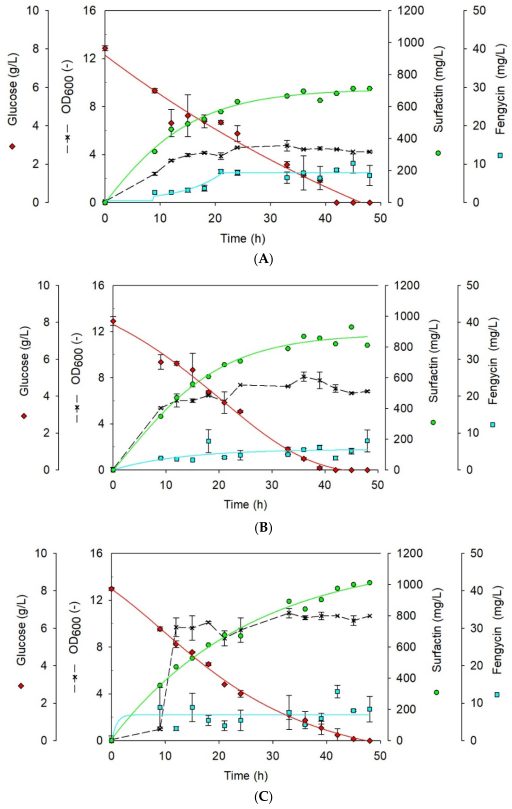
3.5. Effect of Hydrophobic Environment on Surfactin Production
3.6. Proteomic Adaptation to Polystyrene-Bead-Mediated Hydrophobic Environment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Araji, L.; Noor, R.; Raja, Z.; Rahman, A.; Basri, M.; Salleh, A.B. Minireview Microbial Surfactant. Asia Pac. J. Mol. Biol. Biotechnol. 2007, 15, 99–105. [Google Scholar]
- Geetha, S.J.; Banat, I.M.; Joshi, S.J. Biosurfactants: Production and potential applications in microbial enhanced oil recovery (MEOR). Biocatal. Agric. Biotechnol. 2018, 14, 23–32. [Google Scholar] [CrossRef]
- Sarubbo, L.A.; Silva, M.D.G.C.; Durval, I.J.B.; Bezerra, K.G.O.; Ribeiro, B.G.; Silva, I.A.; Twigg, M.S.; Banat, I.M. Biosurfactants: Production, properties, applications, trends, and general perspectives. Biochem. Eng. J. 2022, 181, 108377. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour. Technol. 2017, 232, 389–397. [Google Scholar] [CrossRef]
- Henkel, M.; Müller, M.M.; Kügler, J.H.; Lovaglio, R.B.; Contiero, J.; Syldatk, C.; Hausmann, R. Rhamnolipids as biosurfactants from renewable resources: Concepts for next-generation rhamnolipid production. Process. Biochem. 2012, 47, 1207–1219. [Google Scholar] [CrossRef]
- Farias, C.B.B.; Almeida, F.C.; Silva, I.A.; Souza, T.C.; Meira, H.M.; Soares da Silva, R.D.C.F.; Luna, J.M.; Santos, V.A.; Converti, A.; Banat, I.M.; et al. Production of green surfactants: Market prospects. Electron. J. Biotechnol. 2021, 51, 28–39. [Google Scholar] [CrossRef]
- Tunsagool, P.; Leelasuphakul, W.; Jaresitthikunchai, J.; Phaonakrop, N.; Roytrakul, S.; Jutidamrongphan, W. Targeted transcriptional and proteomic studies explicate specific roles of Bacillus subtilis iturin A, fengycin, and surfactin on elicitation of defensive systems in mandarin fruit during stress. PLoS ONE 2019, 14, e0217202. [Google Scholar] [CrossRef]
- Vahidinasab, M.; Lilge, L.; Reinfurt, A.; Pfannstiel, J.; Henkel, M.; Morabbi Heravi, K.; Hausmann, R. Construction and description of a constitutive plipastatin mono-producing Bacillus subtilis. Microb. Cell. Fact. 2020, 19, 205. [Google Scholar] [CrossRef]
- Lilge, L.; Ersig, N.; Hubel, P.; Aschern, M.; Pillai, E.; Klausmann, P.; Pfannstiel, J.; Henkel, M.; Morabbi Heravi, K.; Hausmann, R. Surfactin Shows Relatively Low Antimicrobial Activity against Bacillus subtilis and Other Bacterial Model Organisms in the Absence of Synergistic Metabolites. Microorganisms 2022, 10, 779. [Google Scholar] [CrossRef]
- Vollenbroich, D.; Vater, J.; Maria Kamp, R.; Pauli, G. Mechanism of Inactivation of Enveloped Viruses by the Biosurfactant Surfactin from Bacillus subtilis. Biologicals 1997, 25, 289–297. [Google Scholar] [CrossRef]
- Haddaji, N.; Ncib, K.; Bahia, W.; Ghorbel, M.; Leban, N.; Bouali, N.; Bechambi, O.; Mzoughi, R.; Mahdhi, A. Control of Multidrug-Resistant Pathogenic Staphylococci Associated with Vaginal Infection Using Biosurfactants Derived from Potential Probiotic Bacillus Strain. Fermentation 2022, 8, 19. [Google Scholar] [CrossRef]
- Teixeira Souza, K.S.; Gudiña, E.J.; Schwan, R.F.; Rodrigues, L.R.; Dias, D.R.; Teixeira, J.A. Improvement of biosurfactant production by Wickerhamomyces anomalus CCMA 0358 and its potential application in bioremediation. J. Hazard. Mater 2018, 346, 152–158. [Google Scholar] [CrossRef]
- Dubey, K.V.; Charde, P.N.; Meshram, S.U.; Shendre, L.P.; Dubey, V.S.; Juwarkar, A.A. Surface-active potential of biosurfactants produced in curd whey by Pseudomonas aeruginosa strain-PP2 and Kocuria turfanesis strain-J at extreme environmental conditions. Bioresour. Technol. 2012, 126, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Nitschke, M.; Costa, S.G.V.A.O. Biosurfactants in food industry. Trends. Food. Sci. Technol. 2007, 18, 252–259. [Google Scholar] [CrossRef]
- Drakontis, C.E.; Amin, S. Biosurfactants: Formulations, properties, and applications. Curr. Opin. Colloid Interface Sci. 2020, 48, 77–90. [Google Scholar] [CrossRef]
- Makkar, R.S.; Cameotra, S.S.; Banat, I.M. Advances in utilization of renewable substrates for biosurfactant production. AMB Express 2011, 1, 5. Available online: http://www.amb-express.com/content/1/1/5 (accessed on 23 December 2023). [CrossRef]
- Treinen, C.; Magosch, O.; Hoffmann, M.; Klausmann, P.; Würtz, B.; Pfannstiel, J.; Morabbi Heravi, K.; Lilge, L.; Hausmann, R.; Henkel, M. Modeling the time cource of ComX: Towards molecular process control for Bacillus wild-type cultivations. AMB Express 2021, 11, 144. [Google Scholar] [CrossRef]
- Fenibo, E.O.; Douglas, S.I.; Stanley, H.O. A Review on Microbial Surfactants: Production, Classifications, Properties and Characterization. J. Adv. Microbiol. 2019, 18, 1–22. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, H.; Zeng, F.; Jiang, L.; Atakpa, E.O.; Chen, G.; Zhang, C.; Xie, Q. A biosurfactant-producing yeast Rhodotorula sp.CC01 utilizing landfill leachate as nitrogen source and its broad degradation spectra of petroleum hydrocarbons. World. J. Microbiol. Biotechnol. 2022, 38, 68. [Google Scholar] [CrossRef]
- Souza, E.C.; Vessoni-Penna, T.C.; De Souza Oliveira, R.P. Biosurfactant-enhanced hydrocarbon bioremediation: An overview. Int. Biodeterior. Biodegradation 2014, 89, 88–94. [Google Scholar] [CrossRef]
- Lai, C.C.; Huang, Y.C.; Wie, Y.H.; Chang, J.S. Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J. Hazard. Mater. 2009, 167, 609–614. [Google Scholar] [CrossRef]
- Soberón-Chávez, G. Biosurfactants: Research and Development. Available online: https://www.elsevier.com/books-and-journals/book-series/books-series- (accessed on 23 December 2023).
- Singh, A.; Van Hamme, J.D.; Ward, O.P. Surfactants in microbiology and biotechnology: Part 2. Application aspects. Biotechnol. Adv. 2007, 25, 99–121. [Google Scholar] [CrossRef] [PubMed]
- Althalb, H.A.; Elmusrati, I.M.; Banat, I.M. Correction: Althalb et al. A Novel Approach to Enhance Crude Oil Recovery Ratio Using Selected Bacterial Species. Appl. Sci. 2021, 11, 10492. [Google Scholar] [CrossRef]
- de Oliveira Schmidt, V.K.; de Souza Carvalho, J.; de Oliveira, D.; de Andrade, C.J. Biosurfactant inducers for enhanced production of surfactin and rhamnolipids: An overview. World. J. Microbiol. Biotechnol. 2021, 37, 21. [Google Scholar] [CrossRef]
- de Oliveira Schmidt, V.K.; Moraes, P.A.D.; Cesca, K.; Pereira, L.P.S.; de Andrade, L.M.; Mendes, M.A.; de Oliverira, D.; de Andrade, C.J. Enhanced production of surfactin using cassava wastewater and hydrophobic inducers: A prospection on new homologues. World. J. Microbiol. Biotechnol. 2023, 39, 82. [Google Scholar] [CrossRef]
- Meneses, D.P.; Gudiña, E.J.; Fernandes, F.; Gonçalves, L.R.B.; Rodrigues, L.R.; Rodrigues, S. The yeast-like fungus Aureobasidium thailandense LB01 produces a new biosurfactant using olive oil mill wastewater as an inducer. Microbiol. Res. 2017, 204, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Nurfarahin, A.H.; Mohamed, M.S.; Phang, L.Y. Culture medium development for microbial-derived surfactants production—An overview. Molecules 2018, 23, 1049. [Google Scholar] [CrossRef]
- Salam, J.A.; Das, N. Induced Biosurfactant Production and Degradation of Lindane by Soil Basidiomycetes Yeast, Rhodotorula sp. VITJzN03. Res. J.Pharm. Biol. Chem. Sci. 2013, 4, 664–670. [Google Scholar]
- Gudiña, E.J.; Rodrigues, A.I.; de Freitas, V.; Azevedo, Z.; Teixeira, J.A.; Rodrigues, L.R. Valorization of agro-industrial wastes towards the production of rhamnolipids. Biores. Technol. 2016, 212, 144–150. [Google Scholar] [CrossRef]
- Zhang, L.; Pemberton, J.E.; Maier, R.M. Effect of fatty acid substrate chain length on Pseudomonas aeruginosa ATCC 9027 monorhamnolipid yield and congener distribution. Process Biochem 2014, 49, 989–995. [Google Scholar] [CrossRef]
- Hashemi, S.Z.; Fooladi, J.; Ebrahimipour, G.; Khodayari, S. Isolation and identification of crude oil degrading and biosurfactant producing bacteria from the oil-contaminated soils of Gachsaran. Appl. Food Biotechnol. 2016, 3, 83–89. [Google Scholar] [CrossRef]
- Amani, H.; Mehrnia, M.R.; Sarrafzadeh, M.H.; Haghighi, M.; Soudi, M.R. Scale up and application of biosurfactant from bacillus subtilis in enhanced oil recovery. Appl. Biochem. Biotechnol. 2010, 162, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Najmi, Z.; Ebrahimipour, G.; Franzetti, A. Investigation of Physicho-chemical Properties and Characterization of Produced Biosurfactant by Selected Indigenous Oil-degrading Bacterium. Iran. J. Public. Health 2018, 47, 1151–1159. [Google Scholar] [PubMed]
- Satpute, S.K.; Banpurkar, A.G.; Dhakephalkar, P.K.; Banat, I.M.; Chopade, B.A. Methods for investigating biosurfactants and bioemulsifiers: A review. Crit. Rev. Biotechnol. 2010, 30, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Walter, V.; Syldatk, C.; Hausmann, R. Screening concepts for the isolation of biosurfactant producing microorganisms. Adv. Exp. Med. Biol. 2010, 672, 1–13. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20545270 (accessed on 23 December 2023). [PubMed]
- Babazadeh, F.; Gharavi, S.; Soudi, M.R.; Zarrabi, M.; Talebpour, Z. Potential for Polyethylene Terephthalate (PET) Degradation Revealed by Metabarcoding and Bacterial Isolates from Soil Around a Bitumen Source in Southwestern Iran. J. Polym. Environ. 2023, 31, 1279–1291. [Google Scholar] [CrossRef]
- Geissler, M.; Oellig, C.; Moss, K.; Schwack, W.; Henkel, M.; Hausmann, R. High-performance thin-layer chromatography (HPTLC) for the simultaneous quantification of the cyclic lipopeptides Surfactin, Iturin A and Fengycin in culture samples of Bacillus species. J. Chromatogr. B 2017, 1044–1045, 214–224. [Google Scholar] [CrossRef]
- Geissler, M.; Heravi, K.M.; Henkel, M.; Hausmann, R. Lipopeptide Biosurfactants From Bacillus Species. In Biobased Surfactants: Synthesis, Properties, and Applications; Elsevier: Amsterdam, The Netherlands, 2019; pp. 205–240. [Google Scholar] [CrossRef]
- Willenbacher, J.; Zwick, M.; Mohr, T.; Schmid, F.; Syldatk, C.; Hausmann, R. Evaluation of different Bacillus strains in respect of their ability to produce Surfactin in a model fermentation process with integrated foam fractionation. Appl. Microbiol. Biotechnol. 2014, 98, 9623–9632. [Google Scholar] [CrossRef]
- Bóka, B.; Manczinger, L.; Kecskeméti, A.; Chandrasekaran, M.; Kadaikunnan, S.; Alharbi, N.S.; Vágvölgyi, C.; Szekeres, A. Ion trap mass spectrometry of surfactins produced by Bacillus subtilis SZMC6179J reveals novel fragmentation features of cyclic lipopeptides. Rapid Commun. Mass Spectrom. 2016, 30, 1581–1590. [Google Scholar] [CrossRef]
- Kecskeméti, A.; Bartal, A.; Bóka, B.; Kredics, L.; Manczinger, L.; Shine, K.; Alharby, N.S.; Khaled, J.M.; Varga, M.; Vágvölgyi, C.; et al. High-frequency occurrence of surfactin monomethyl isoforms in the ferment broth of a bacillus subtilis strain revealed by ion trap mass spectrometry. Molecules 2018, 23, 2224. [Google Scholar] [CrossRef]
- Lin, L.Z.; Zheng, Q.W.; Wei, T.; Zhang, Z.Q.; Zhao, C.F.; Zhong, H.; Xu, Q.Y.; Lin, J.F.; Guo, L.O. Isolation and Characterization of Fengycins Produced by Bacillus amyloliquefaciens JFL21 and Its Broad-Spectrum Antimicrobial Potential Against Multidrug-Resistant Foodborne Pathogens. Front. Microbiol. 2020, 11, 579621. [Google Scholar] [CrossRef]
- Pathak, K.V.; Keharia, H.; Gupta, K.; Thakur, S.S.; Balaram, P. Lipopeptides from the banyan endophyte, Bacillus subtilis K1: Mass spectrometric characterization of a library of fengycins. J. Am. Soc. Mass Spectrom. 2012, 23, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Rughöft, S.; Jehmlich, N.; Gutierrez, T.; Kleindienst, S. Comparative proteomics of Marinobacter sp. Tt1 reveals corexit impacts on hydrocarbon metabolism, chemotactic motility, and biofilm formation. Microorganisms 2021, 9, 3. [Google Scholar] [CrossRef]
- Wang, X.B.; Chi, C.Q.; Nie, Y.; Tang, Y.Q.; Tan, Y.; Wu, G.; Wu, X.L. Degradation of petroleum hydrocarbons (C6-C40) and crude oil by a novel Dietzia strain. Biores. Technol. 2011, 102, 7755–7761. [Google Scholar] [CrossRef] [PubMed]
- Klausmann, P.; Hennemann, K.; Hoffmann, M.; Treinen, C.; Aschern, M.; Lilge, L.; Morabbi Heravi, K.; Hausmann, R. Bacillus subtilis High Cell Density Fermentation Using a Sporulation-Deficient Strain for the Production of Surfactin. Appl. Microbiol. Biotechnol. 2021, 105, 4141–4151. [Google Scholar] [CrossRef] [PubMed]
- Hughes, C.S.; Foehr, S.; Garfield, D.A.; Furlong, E.E.; Steinmetz, L.M.; Krijgsveld, J. Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 2014, 10, 757. [Google Scholar] [CrossRef]
- Olsen, J.V.; de Godoy, L.M.F.; Li, G.; Macek, B.; Mortensen, P.; Pesch, R.; Makarov, A.; Lange, O.; Horning, S.; Mann, M. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteom. 2005, 4, 2010–2021. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. Perseus platform for proteomics data The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef]
- Lilge, L.; Hertel, R.; Morabbi Heravi, K.; Henkel, M.; Commichau, F.M.; Hausmann, R. Draft Genome Sequence of the Type Strain Bacillus subtilis subsp. subtilis DSM10. Microbiol. Resourc. Announc. 2021, 10, e00158-21. [Google Scholar] [CrossRef]
- Lilge, L.; Vahidinasab, M.; Adiek, I.; Becker, P.; Kuppusamy Nesamani, C.; Treinen, C.; Hoffmann, M.; Morabbi Heravi, K.; Henkel, M.; Hausmann, R. Expression of degQ gene and its effect on lipopeptide production as well as formation of secretory proteases in Bacillus subtilis strains. MicrobiologyOpen 2021, 10, e1241. [Google Scholar] [CrossRef]
- Zhu, B.; Stülke, J. SubtiWiki in 2018: From genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res. 2018, 46, D743–D748. [Google Scholar] [CrossRef] [PubMed]
- Ismail, W.; Al-Rowaihi, I.S.; Al-Humam, A.A.; Hamza, R.Y.; El Nayal, A.M.; Bououdina, M. Characterization of a lipopeptide biosurfactant produced by a crude-oil-emulsifying Bacillus sp. I-15. Int. Biodeterior. Biodegradation 2013, 84, 168–178. [Google Scholar] [CrossRef]
- Calvo, C.; Toledo, F.L.; González-López, J. Surfactant activity of a naphthalene degrading Bacillus pumilus strain isolated from oil sludge. J. Biotech. 2004, 109, 255–262. [Google Scholar] [CrossRef]
- Gentili, A.R.; Cubitto, M.A.; Ferrero, M.; Rodriguéz, M.S. Bioremediation of crude oil polluted seawater by a hydrocarbon-degrading bacterial strain immobilized on chitin and chitosan flakes. Int. Biodeterior. Biodegradation 2006, 57, 222–228. [Google Scholar] [CrossRef]
- Bachmann, D.; Pal, U.; Bockwoldt, J.A.; Schaffert, L.; Roentgen, R.; Büchs, J.; Kalinowski, J.; Blank, L.M.; Tiso, T. C-, N-, S-, and P-Substrate Spectra in and the Impact of Abiotic Factors on Assessing the Biotechnological Potential of Paracoccus pantotrophus. Appl. Microbiol. 2023, 3, 175–198. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by Bacillus subtilis CN2. Biochem. Eng. J. 2015, 101, 168–178. [Google Scholar] [CrossRef]
- Li, S.W.; Liu, M.Y.; Yang, R.Q. Comparative Genome Characterization of a Petroleum-Degrading Bacillus subtilis Strain DM2. Int. J. Genomics 2019, 2019, 7410823. [Google Scholar] [CrossRef] [PubMed]
- Parthipan, P.; Preetham, E.; Machuca, L.L.; Rahman, P.K.S.M.; Murugan, K.; Rajasekar, A. Biosurfactant and degradative enzymes mediated crude oil degradation by bacterium Bacillus subtilis A1. Front. Microbiol. 2017, 8, 193. [Google Scholar] [CrossRef]
- Rahimi, T.; Niazi, A.; Deihimi, T.; Taghavi, S.M.; Ayatollahi, S.; Ebrahimie, E. Genome annotation and comparative genomic analysis of Bacillus subtilis MJ01, a new bio-degradation strain isolated from oil-contaminated soil. Funct. Integr. Genomics 2018, 18, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Willenbacher, J.; Yeremchuk, W.; Mohr, T.; Syldatk, C.; Hausmann, R. Enhancement of Surfactin yield by improving the medium composition and fermentation process. AMB Express 2015, 5, 145. [Google Scholar] [CrossRef]
- Cameotra, S.S.; Singh, P. Synthesis of rhamnolipid biosurfactant and mode of hexadecane uptake by Pseudomonas species. Microb. Cell Factories 2009, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Solyanikova, I.P.; Golovleva, L.A. Hexadecane and Hexadecane-Degrading Bacteria: Mechanisms of Interaction. Microbiology 2019, 88, 15–26. [Google Scholar] [CrossRef]
- Cooper, D.G.; Macdonald, C.R.; Duff, S.J.B.; Kosaric, N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 1981, 42, 408–412. [Google Scholar] [CrossRef]
- Kim, H.-S.; Yoon, B.-D.; Lee, C.-H.; Suh, H.-H.; Oh, H.-M.; Katsuragi, T.; Tani, Y. Production and Properties of a Lipopeptide Biosurfactant from Bacillus subtilis C9. J. Ferment. Bioeng. 1997, 84, 41–46. [Google Scholar] [CrossRef]
- Deshpande, M.; Daniels, L. Evaluation of sophorolipid biosurfactant production by Candida bombicola using animal fat. Bioresour. Technol. 1995, 54, 143–150. [Google Scholar] [CrossRef]
- Bartolini, M.; Cogliati, S.; Vileta, D.; Bauman, C.; Ramirez, W.; Grau, R. Stress-Responsive Alternative Sigma Factor SigB Plays a Positive Role in the Antifungal Proficiency of Bacillus subtilis. Appl. Environ. Microbiol. 2019, 85, e00178-19. [Google Scholar] [CrossRef]
- Brück, H.L.; Delvigne, F.; Dhulster, P.; Jacques, P.; Coutte, F. Molecular strategies for adapting Bacillus subtilis 168 biosurfactant production to biofilm cultivation mode. Bioresour. Technol. 2019, 293, 122090. [Google Scholar] [CrossRef]
- Akintayo, S.O.; Treinen, C.; Vahidinasab, M.; Pfannstiel, J.; Bertsche, U.; Fadahunsi, I.; Oellig, C.; Granvogl, M.; Henkel, M.; Lilge, L.; et al. Exploaration of surfactin production by newly isolated Bacillus and Lysinibacillus strains from food-related sources. Lett. Appl. Microbiol. 2022, 75, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Akintayo, S.O.; Hosseini, B.; Vahidinasab, M.; Messmer, M.; Pfannstiel, J.; Bertsche, U.; Hubel, P.; Henkel, M.; Hausmann, R.; Voegele, R.T.; et al. Characterization ofantifungal properties of lipopeptide-producing Bacillus velezensis strains and their proteome-based response to the phytopathogens, Diaporthe spp. Front. Bioeng. Biotechnol. 2023, 11, 1228386. [Google Scholar] [CrossRef]
- Akintayo, S.O.; Neumann, B.; Fischer, M.; Henkel, M.; Lilge, L.; Hausmann, R. Lysinibacillus irui sp. nov., isolated from Iru, fermented African locust beans. Int. J. Syst. Evol. Microbiol. 2023, 73, 006167. [Google Scholar] [CrossRef]
- Jabiri, S.; Legrifi, I.; Benhammou, M.; Laasli, S.-E.; Mokrini, F.; Bendriss Amraoui, M.; Lahlali, R. Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity. Appl. Microbiol. 2023, 3, 948–967. [Google Scholar] [CrossRef]
- Takishita, Y.; Souleimanov, A.; Bourguet, C.; Ohlund, L.B.; Arnold, A.A.; Sleno, L.; Smith, D.L. Pseudomonas entomophila 23S Produces a Novel Antagonistic Compound against Clavibacter michiganensis subsp. michiganensis, a Pathogen of Tomato Bacterial Canker. Appl. Microbiol. 2021, 1, 60–73. [Google Scholar] [CrossRef]
- Bodoura, A.A.; Miller-Maier, R.M. Application of a modified drop-collapse technique for surfactant quantitation and screening of biosurfactant-producing mi-croorganisms. J. Microbiol. Methods 1998, 32, 273–280. [Google Scholar] [CrossRef]
- Tugrul, T.; Cansunar, E. Detecting surfactant-producing microorganisms by the drop-collapse test. World J. Microbiol. Biotechnol. 2005, 21, 851–853. [Google Scholar] [CrossRef]
- Siegmund, I.; Wagner, F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]


| Control | Hexadecane Supplementation | ||||||
|---|---|---|---|---|---|---|---|
| Surfactin | Strains | ZH1 | P7 | DSM10T | ZH1 | P7 | DSM10T |
| Max. concentration (g/L) | 1.05 ± 0.02 | 0.81 ± 0.13 | 0.36 ± 0.00 | 0.89 ± 0.02 | 0.68 ± 0.01 | 0.33 ± 0.01 | |
| time [h] | 15 | 15 | 15 | 15 | 17 | 12 | |
| Biomass (CDW) (g) | 1.95 ± 0.03 | 2.19 ± 0.33 | 2.4 ± 0.01 | 2.46 ± 0.18 | 2.91 ± 0.15 | 2.44 ± 0.15 | |
| Specific productivity (g/gh) | 0.036 | 0.025 | 0.010 | 0.024 | 0.014 | 0.011 | |
| Fengycin | Max. concentration (mg/L) | 15.73 ± 1.26 | 10.55 ± 1.4 | 27.72 ± 2.45 | 21.65 ± 1.43 | 11.41 ± 1.56 | 30 ± 1.62 |
| time (h) | 19 | 22 | 36 | 15 | 17 | 36 | |
| Biomass (CDW) (g) | 2.27 ± 0.13 | 2.19 ± 0.18 | 1.25 ± 0.05 | 2.46 ± 0.18 | 2.91 ± 0.15 | 1.2 ± 0.07 | |
| Specific productivity (mg/gh) | 0.365 | 0.219 | 0.617 | 0.587 | 0.231 | 0.694 | |
| Surfactin | Fengycin | |||||||
|---|---|---|---|---|---|---|---|---|
| Polystyrene Beads (%) | Max. Concentration (g/L) | Biomass (CDW) (g) | Time (h) | Specific Productivity (g/gh) | Max. Concentration (mg/L) | Biomass (CDW) (g) | Time (h) | Specific Productivity (mg/gh) |
| 1 | 0.71 ± 0.00 | 1.27 ± 0.00 | 45 | 0.0125 | 10.18 ± 2.45 | 1.27 ± 0.00 | 45 | 0.1781 |
| 5 | 0.93 ± 0.00 | 2.27 ± 0.08 | 45 | 0.0091 | 7.90 ± 2.97 | 2.05 ± 0.08 | 48 | 0.0802 |
| 10 | 1.01 ± 0.00 | 3.89 ± 0.00 | 48 | 0.0054 | 13.11 ± 1.69 | 3.20 ± 0.00 | 42 | 0.0975 |
| Control | 1.05 ± 0.02 | 1.95 ± 0.03 | 15 | 0.3600 | 15.72 ± 1.26 | 2.25 ± 0.45 | 19 | 0.3677 |
| Gene Name | Log2-fold Change | Regulons | Functions | Categories |
|---|---|---|---|---|
| Positively affected proteins | ||||
| mdxE | 7.91 | MdxR | Maltodextrin utilization | Uptake of carbon sources, utilization of starch/maltodextrin |
| yqjP | 7.66 | - | Similar to metallohydrolase | Proteins of unknown function |
| acoB | 5.92 | CcpA, SigL, AcoR, Fnr | Acetoin utilization | Utilization of acetoin |
| tilS | 5.67 | SigM, SigF, HprT, TilS | tRNA modification, control of ftsH expression | tRNA modification and maturation |
| ribE | 5.44 | FMN-box | Riboflavin biosynthesis | Biosynthesis/acquisition of riboflavin/FAD |
| mtlA | 5.36 | MtlR | Mannitol uptake and phosphorylation, control of MtlR activity | Sugar-specific PTS proteins, utilization of mannitol |
| glvA | 5.28 | CcpA, GlvR | Maltose utilization | Utilization of maltose |
| yflK | 4.93 | - | Similar to hydroxylaminopurine resistance protein | Detoxification reactions |
| kbl | 4.89 | - | Threonine utilization | Utilization of threonine/glycine |
| cotJA | 4.68 | SpoIIID, SigE | Polypeptide composition of the spore coat | Spore coat proteins |
| yjkB | 4.54 | - | Similar to amino acid ABC transporter | - |
| yabR | 4.51 | SigE, SigM, SigW, SigD, SigX | Unknown | - |
| yhjG | 4.48 | - | Similar to monooxygenase | - |
| lcfA | 4.34 | CcpA, FadR | Fatty acid degradation | Utilization of fatty acids |
| mntC | 4.27 | MntR | Manganese uptake | Trace metal homeostasis |
| rghR | 4.16 | RghR | Regulation of sporulation initiation | Sporulation, phosphorelay |
| bbmA | 4.09 | MdxR | Starch and maltodextrin utilization | Utilization of starch/maltodextrin |
| malP | 4.03 | CcpA, GlvR | Maltose uptake and phosphorylation | Sugar-specific PTS proteins, utilization of maltose |
| ispA | 4.01 | CodY | Protein degradation | Utilization of proteins |
| ykvR | 4.00 | LexA | Unknown | - |
| Negatively affected proteins | ||||
| lipA | −7.00 | - | Lipoyl synthase | Synthesis of lipoic acid |
| gsiB | −6.87 | SigB, SigI | Response to water deficits | General stress protein |
| spoVM | −6.61 | SpoIIID, SigE | Initiation of spore coat assembly | Spore coat proteins |
| yqeI | −6.59 | - | Similar to RNA-binding protein, putatively involved in ribosome assembly | Translation |
| ydbP | −6.14 | - | Similar to thioredoxin | - |
| yuxO | −6.01 | - | Similar to thioesterase | - |
| spoIIAA | −5.85 | SigF, Spo0A, AbrB, SigG, SinR, SigH | Control of SigF activity | Sporulation proteins |
| cspD | −5.67 | - | RNA chaperone | Transcription elongation/termination RNA chaperones |
| yurT (glxB) | −5.53 | - | Detoxification of methylglyoxal | Resistance against oxidative and electrophile stress |
| ytpP | −5.44 | Spx | Cleavage of CoA moieties from modified proteins | Protein modification |
| yhfF | −5.15 | - | Unknown | - |
| ykuS | −5.00 | - | Unknown | - |
| nin | −5.00 | ComK | Genetic transformation, DNA uptake | Genetic competence |
| trxA | −5.00 | CtsR, SigB, Spx | Protection of proteins against oxidative damage | Resistance against oxidative and electrophile stress |
| yraH | −4.68 | - | Unknown | - |
| yhaN (sbcE) | −4.67 | LexA | DNA double-strand break repair and competence | DNA repair/recombination |
| flgL | −4.61 | SigD, ComK, DegU, ScoC | Motility and chemotaxis | Flagellar proteins, swarming |
| yolI (bdbA) | −4.59 | AbrB, YvrHb, Rok, DnaA, Abh | Oxidative folding of proteins | Chaperones/protein folding |
| fliH | −4.42 | CodY, SigD, DegU, Spo0A, SwrA | Movement and chemotaxis | Flagellar proteins, swarming |
| crh | −4.33 | CcpA | Control of carbon flux | Control of transcription factor |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hashemi, S.Z.; Fooladi, J.; Vahidinasab, M.; Hubel, P.; Pfannstiel, J.; Pillai, E.; Hrenn, H.; Hausmann, R.; Lilge, L. Toward Effects of Hydrophobicity on Biosurfactant Production by Bacillus subtilis Isolates from Crude-Oil-Exposed Environments. Appl. Microbiol. 2024, 4, 215-236. https://doi.org/10.3390/applmicrobiol4010015
Hashemi SZ, Fooladi J, Vahidinasab M, Hubel P, Pfannstiel J, Pillai E, Hrenn H, Hausmann R, Lilge L. Toward Effects of Hydrophobicity on Biosurfactant Production by Bacillus subtilis Isolates from Crude-Oil-Exposed Environments. Applied Microbiology. 2024; 4(1):215-236. https://doi.org/10.3390/applmicrobiol4010015
Chicago/Turabian StyleHashemi, Seyedeh Zahra, Jamshid Fooladi, Maliheh Vahidinasab, Philipp Hubel, Jens Pfannstiel, Evelina Pillai, Holger Hrenn, Rudolf Hausmann, and Lars Lilge. 2024. "Toward Effects of Hydrophobicity on Biosurfactant Production by Bacillus subtilis Isolates from Crude-Oil-Exposed Environments" Applied Microbiology 4, no. 1: 215-236. https://doi.org/10.3390/applmicrobiol4010015
APA StyleHashemi, S. Z., Fooladi, J., Vahidinasab, M., Hubel, P., Pfannstiel, J., Pillai, E., Hrenn, H., Hausmann, R., & Lilge, L. (2024). Toward Effects of Hydrophobicity on Biosurfactant Production by Bacillus subtilis Isolates from Crude-Oil-Exposed Environments. Applied Microbiology, 4(1), 215-236. https://doi.org/10.3390/applmicrobiol4010015






