Abstract
This research aims to develop a standardised protocol for monitoring the disinfection efficacy of healthcare laundry processes in view of numerous differential methodologies currently being employed within the healthcare laundry sector, including agitation and surface sampling for post-laundering decontamination assessment and swatch and bioindicator testing for in-wash-process efficacy. Enterococcus faecium as an indicator species within industrial wash systems is preferable due to its high thermal and disinfectant tolerance. Methods for measuring laundry disinfection were compared; commercially available E. faecium bioindicators and contaminated cotton swatches (loose, in cloth bags or within nylon membranes) were laundered industrially at ambient temperature and microbial recovery determined. E. faecium was lost from cotton during laundering but retained by the bioindicator membrane, which allows disinfection efficacy to be measured without loss of microorganisms from the test swatch. Commercially available bioindicators were only permeable to disinfectants and detergents at ≥60 °C. Subsequently, polyethersulphone membranes for enclosing contaminated swatches were developed for low-temperature laundering, with permeability to industrial laundry chemistries at below ≤60 °C. This study demonstrates that bioindicators are the recommended methodology for laundry disinfection validation. The use of a universal healthcare laundry disinfection methodology will lead to standardised microbiological testing across the industry and improvements in infection control.
1. Introduction
Reusable healthcare textiles, including bed linen, are typically laundered in offsite industrial laundries using thermal and/or chemical disinfection parameters [1,2]. The aim of healthcare laundering is to clean and disinfect textiles by removing soiling and potential pathogens in order to reduce the risk of infection or cross-contamination between patients [3,4].
Textiles become contaminated during use in healthcare settings, and potential pathogens can persist for several days [5]. The onward transfer of microorganisms from textiles to other surfaces including skin and bioaerosol generation from soiled linen have also been documented [6,7]. The role of reusable healthcare textiles in the transmission of infections has been debated, with the risk currently considered to be low due to a lack of epidemiological evidence [8,9,10]. A small number of hospital outbreaks have been associated with textiles, linked primarily to contamination of textiles with opportunistic environmental pathogens during or after laundering [11]. This highlights the importance of effective laundering procedures to protect susceptible patients [1].
In the EU and UK, the EN14065 (European Committee for Standardization) Risk Analysis and Biocontamination Control (RABC) system and HTM 01-04 require laundries to determine microbiological hazards and implement control measures to ensure decontamination and prevent recontamination of linen [4,12,13]. However, there is no internationally recognised standard method for microbial monitoring of healthcare laundering. In the USA, the CDC does not recommend routine microbiological testing; that said, the Textile Rental Services Association (TRSA) recently introduced certification based on testing of the microbial bioburden on processed articles. Laundered textiles must have <20 CFU/dm2 total aerobic microorganisms, yeast and moulds using RODAC plate sampling, and no pathogens detected using the United States Pharmacopoeia 62 method [14]. No specific validation tests for the disinfection efficacy of the wash cycle are required. European guidelines for healthcare laundering set out processes for monitoring the microbial load of processed textiles, with requirements varying. In Germany, RODAC plates should be used to test for post-laundering microbial load, and the disinfection efficacy of the wash process must be assessed by laundering artificially contaminated textile samples, where no viable microorganisms should be detected after the wash [15]. In England and Wales, a standardised protocol is not prescribed; chemo-thermal processes are validated by laundering sterile swatches, which should remain sterile after the wash cycle. Additional best practice involves validation of the disinfection efficacy of the wash process. This is achieved using bioindicators, which are test strips containing 1 cm2 cotton swatches inoculated with specified loads of the microorganism (typically 3, 4, 5 and 6 log10 CFU Enterococcus faecium) in discrete compartments of a semi-permeable membrane [16,17]. The semi-permeable membrane reportedly prevents microorganisms from being lost in the wash process to dilution and agitation, whilst allowing disinfectants and detergents to pass through and interact with the test microorganism, thereby measuring the biocidal efficacy (≥5 log10 reduction) of the wash process using a semi-quantitative enumeration method. In order to measure true disinfection within a laundry process, the test methodology must assess the destruction of microorganisms and not the overall dilution factor of the wash. The lack of standardisation both between and within countries may lead to variation in the reliability of methods and acceptance criteria, and, in turn, the disinfection standards between laundries [18]. In addition, the industrial laundry industry is looking to move towards low-temperature washing due to the adverse environmental effects of high temperature laundering. Currently, there is no standardised microbiological test methodology for validating an alternative process that will allow for this transition to low-temperature laundering to occur. This is of particular importance due to potential infection transmission from the reduction of thermal disinfection, which poses an increased risk from healthcare laundry.
This study aimed to establish a standard protocol for monitoring both the disinfection efficacy of healthcare laundry processes and the recontamination of healthcare textiles post-disinfection. The research was divided into two phases: the assessment of current practices utilised within the global industrial laundry sector and, secondly, the development of a bioindicator for wash process efficacy testing.
2. Methods
2.1. Chemicals
All chemicals were from Oxoid (Basingstoke, UK) unless otherwise stated.
2.2. Microorganisms
The microorganisms selected were representative of thermotolerant Enterococcus sp. currently used as indicator species within the laundry and food industries. In addition, representative Gram-positive, Gram-negative and spore-bearing microorganisms, which are used as representative species in other disinfection test standards, were utilised to establish recovery from textiles and survival in wash processes.
Escherichia coli NCTC 10538, Enterococcus hirae ATCC 10541, Enterococcus faecium ATCC 6057, Enterococcus faecium NCIMB 2699 and Staphylococcus aureus ATCC 6538 were cultured aerobically at 37 °C for 18–24 h using nutrient agar. Test suspensions were prepared by suspending colonies in phosphate-buffered saline (PBS, BR0014G (Oxoid) pH 7.4, 1 tablet per 100 mL). Bacillus cereus CIP105151 spore suspensions were generated by culturing in 2 l Leighton Doi broth (7 days; 37 °C; 180 rpm shaking), washing thrice with chilled, sterile, distilled water and resuspending to 40 mL. Sporulation efficiency was determined by comparing the viable counts of untreated and heat-shocked (70 °C; 30 min) spore samples. Spore suspensions were stored at 4 °C and diluted as required in PBS.
2.3. Phase 1: To Determine the Efficacy of Current Methods for Measuring Disinfection within Healthcare Laundry Wash Processes
Efficiency of Methods for the Recovery of Microorganisms from Textiles.
Information on the current methodologies used to determine decontamination of industrially laundered healthcare textiles was obtained from 5 commercial laundries across the UK, Europe and the USA. These data informed the choice of the recovery methods to be assessed including suspension, surface sampling and bioindicator methods.
Sample Preparation
Sterile 25 cm2 swatches of 100% cotton were inoculated with 500 μL of either 8, 2 or 1 log10 CFU/mL of E. coli, E. faecium, S. aureus or B. cereus spores and allowed to dry overnight at ~21 °C and 44% ± 1% relative humidity (RH) [19].
2.4. Suspension Methods
2.4.1. Recovery Media
Inoculated cotton samples were immersed in 30 mL PBS or maximum recovery diluent (MRD) ± 2 g/L tween-80 (Fisher Scientific, Loughborough, UK) and vortexed for 1 min. The supernatant was viable-counted using spiral plating (Interscience, Saint Nom la Brétèche, France), spread plating (1 mL) or membrane filtration (0.45 μm; cellulose acetate) onto nutrient agar before incubation (24 h; 37 °C).
2.4.2. Agitation Method
Inoculated cotton samples were immersed in 30 mL PBS-T and vortexed for 1 min [19], shaken by hand 30 times, stomached for 30 secs or 1 min or shaken (100 rpm) for 10 min with 5 g glass beads. Recovered microorganisms were enumerated as described above. Hereafter, microorganisms were recovered from textile samples by immersing in 30 mL PBS-T and shaking by hand 30 times, unless otherwise stated.
2.4.3. Recovery Agar
Microorganisms were recovered from inoculated cotton samples as stated above (Section 2.4.2) and plated on nutrient agar and mannitol egg yolk polymyxin (MYP) agar (B. cereus spores), membrane lactose glucuronide agar (MLGA; E. coli), Enterococcus selective agar (Sigma Aldrich, Gillingham, UK; E. faecium) or Baird-Parker agar (S. aureus). Nutrient agar was incubated at 37 °C for 48 h and selective agars were incubated according to the manufacturers’ instructions prior to enumeration.
2.5. Surface Sampling Methods
Test species were recovered from inoculated cotton samples using tryptone soya agar RODAC contact plates or total count agar dip slides (Scientific Laboratory Supplies, Willford, UK) at 100 g pressure for 10 s. RODAC and dip slides were incubated at 37 °C for 24 h and the number of colonies enumerated. Cotton samples were also swabbed with a moistened cotton swab (Scientific Laboratory Supplies, UK) and vortexed for 30 s in 5 mL PBS-T, and the supernatant was spread, plated or membrane-filtered onto nutrient agar plates.
2.6. Bioindicators
Commercially available bioindicators for assessing microbial kill within laundry processes containing either E. faecium, E. coli (106, 105, 104 and 103 CFU/sample) or Bacillus subtilis spores (106 CFU) (DES controllers) were obtained from CCD, Netherlands.
2.6.1. Recovery of Microorganisms from DES Controller Bioindicators
Each swatch from within the DES controller bioindicators was removed, and microorganisms were recovered in PBS-T and enumerated on nutrient agar as described above (Section 2.4.2). For comparison, 1 cm2 sterile cotton swatches were inoculated with 20 μL of E. faecium (6, 5, 4 and 3 log10 CFU/swatch) and allowed to dry overnight (~21 °C; 44% ± 1% RH) before being recovered and enumerated as described above. DES controller bioindicator swatches were also incubated (37 °C; 48 h) in 10 mL tryptone soya broth (TSB) as a control.
2.6.2. E. faecium DES Controller Bioindicator Retention Efficacy
E. faecium DES controller bioindicator samples were laundered with and without the membrane in an industrial washer–extractor machine (JLA, Ripponden UK) under a 2 kg load using polycotton makeweights without temperature. Washes were conducted with and without 20 g ECE Standard Reference Detergent A (ISO 6330:2012) [20]. For comparison, 1 cm2 cotton swatches inoculated with E. faecium (106 CFU/swatch, prepared as per Section 2.6.1) were also laundered. The swatches were laundered loose or enclosed within a laundry bag (Webknot), reusable autoclave bag (Elis, Leicester, UK), sewn cotton bag (400 cm2) or cotton bag (400 cm2) sealed with iron-on hemming web or heat-sealed sterile nylon membrane filters (0.22 µm; 47 mm, Fisher Scientific).
Following laundering, E. faecium was enumerated as described above. Unlaundered swatches were included as a control.
2.6.3. E. faecium DES Controller Bioindicator Permeability to Disinfectants and Detergents
Sublethal concentrations of peracetic acid (OxoniaTM, Ecolab, Northwich, UK), sodium hypochlorite (HygenilTM, Ecolab, UK), benzalkonium chloride (BAC) (Sigma Aldrich, UK), didecyldimethylammonium chloride (DDAC) (Lonza, Switzerland), hydrogen peroxide (Christeyns, UK), hypochlorous acid (ProchlorTM, Contec™, Fisher Scientific) and sodium dodecyl sulphate (SDS) were determined using an adapted disinfectant suspension test (Table S1). Collectively, anionic and cationic surfactants, alongside chlorine-based, quaternary ammonium-based and peroxide-based disinfectants, represent the majority of the chemical components commonly found in washing detergents.
A neutraliser comprising 30 g/L polysorbate 80 (Fisher Scientific, UK), 8.5 g/L sodium chloride (Sigma Aldrich, UK), 5 g/L sodium thiosulphate (Sigma Aldrich, UK), 3 g/L lecithin (Fisher Scientific, UK) and 1 g/L tryptone (Oxoid, UK) was validated as non-toxic and efficacious according to the BS EN 1040:2005 [21]; for hydrogen peroxide, an increased concentration of sodium thiosulphate was used (20 g/L).
DES controller bioindicators were laundered with 0.64 mL/L peracetic acid, 0.8 mL/L sodium hypochlorite, 320 μM BAC, 414 μM DDAC, 138 mM hydrogen peroxide, 596 μM hypochlorous acid or 2 mM SDS in a 10 min wash without temperature (23.44 ± 0.06 °C) or textile load. Identical unlaundered DES controller bioindicator swatches were placed in a 10 mL volume of the disinfectant or detergent solutions for 13 min. Washes were also conducted at 30 °C, 40 °C and 50 °C (SDS) and 60°C (SDS and BAC). The wash temperature was monitored using an iButton Thermochron data logger (Measurement Systems, Newbury, UK). Surviving microorganisms were neutralized for 5 min by shaking 30 times in 30 mL neutraliser and vortexing for 30 s prior to spread plating onto nutrient agar. E. faecium was enumerated after 48 h incubation at 37 °C. Water-only controls were included, and the log10 reduction of the disinfectant was calculated from the water control as follows:
SDS treatment solutions were also membrane-filtered to confirm that a loss of microorganisms from the swatch was associated with biocidal activity rather than surfactant activity. Further validation was conducted using reference detergent against E. faecium on inoculated cotton swatches (Table S2).
2.7. Comparison of Semi-Quantitative and Quantitative Enumeration Methods
The efficacy of the manufacturer-recommended semi-quantitative DES controller bioindicator enumeration method was compared to that of the quantitative recovery method developed in this study. E. faecium, E. coli and B. subtilis spore DES controller bioindicators were laundered using two industrial wash cycles under a 2 kg load of polycotton makeweights: (1) 35 °C, 3 min pre-wash and 67 °C, 10 min main wash with and without industrial detergent (Cool asepsis, Christeyns, Bradford, UK) and (2) 40 °C pre-wash and 75 °C main wash without detergent. Domestic laundering was also conducted using a standard 60 °C wash cycle (Indesit IWSD61251 Eco) and 2 kg polycotton makeweights, with biological detergent (Persil) and non-biological detergent (ECE standard reference detergent A). Unlaundered DES controller bioindicators, and DES controller bioindicators laundered with water only (60 °C and ambient temperature) were included as controls.
Laundered bioindicators were enumerated quantitatively by neutralising and plating on nutrient agar as described above (Section 2.6.3). Semi-quantitative enumeration was achieved by incubating test swatches in 10 mL TSB (37 °C; 48 h) before subculturing positive vials on selective and nutrient agar. Uninoculated broths were included as a control. Log10 reductions were calculated based on growth on selective agar compared to the manufacturer-stated inoculum load.
2.8. Field Test
E. faecium, E. coli and B. subtilis spore DES controller bioindicators were laundered in a continuous batch washer using a standard 75 °C wash programme with detergent (Cool Asepsis, Christeyns, UK) and survivors enumerated using the quantitative and semi-quantitative methods at an on-site industrial laundry laboratory. Unlaundered DES controller bioindicators were included as a control.
2.9. Phase 2: Development of Alternative E. faecium Strains and a Bioindicator Membrane for Low-Temperature Laundering
Due to the commercially available DES controller bioindicators containing a biosafety level 2 microorganism, alternative Enterococcus sp. were explored.
2.10. Disinfectant, Detergent and Thermal Tolerance on Cotton
Sterile cotton swatches (1 cm2) were inoculated with 20 μL E. faecium ATCC 6057, E. faecium NCIMB 2699 or E. hirae ATCC 10541 (106 CFU/swatch, equivalent to the bioindicator) and dried overnight at room temperature. Samples were immersed in 10 mL distilled water for 13 min at ambient temperature, 60 °C and 70 °C. An untreated control was included. Cotton samples were neutralised and plated as described above (Section 2.6.3).
Chemical susceptibility was assessed using a method adapted from the BS EN 1040:2005 suspension test. Sterile cotton swatches (1 cm2) inoculated with 20 μL E. faecium ATCC 6057, E. faecium NCIMB 2699 or E. hirae were prepared as described above. Samples were immersed in 10 mL of SDS (166.67 mM and 2 mM), sodium hypochlorite (2 mL/L, 0.8 mL/L and 0.2 mL/L) or peracetic acid (1.2 mL/L, 0.64 mL/L and 0.04 mL/L) and incubated for 13 min at room temperature. Surviving microorganisms were recovered in neutraliser and enumerated as above.
2.11. Bioindicator Membrane Assessment for Low-Temperature Laundering
A polyethersulphone (PES) membrane with a pore size of 0.2 µm was selected for assessment as an alternative bioindicator membrane for low-temperature laundering due to its low inherent molecular charge, which should limit charged chemicals being repelled from the membrane surface.
Flow Rate of the Solution through PES
The flow rate of distilled water through PES membranes was compared to 100% cotton, 100% polyester and DES controller bioindicator membrane samples. This was determined by measuring the volume of water that had diffused though membrane samples (1.5 cm diameter) after 30 min, except for 100% cotton, where the time needed for 10 mL of water to pass through the fabric was measured.
2.12. Detergent and Disinfectant Membrane Permeability
The permeabilities of the 100% cotton, 100% polyester, DES controller bioindicator and PES membranes to various detergents and disinfectants were determined by filtering the chemical solutions through each membrane by gravity. The detergents and disinfectants tested were 414 µM DDAC, 321 µM BAC, 2mM SDS (w/v), 1mM Tergitol 15-S-15 (Sigma-Aldrich, UK) (non-ionic surfactant often used in washing processes), 138 mM hydrogen peroxide, 596 µM hypochlorous acid, 0.8 mL/L sodium hypochlorite and 0.64 mL/L peracetic acid. The concentration of the chemical solution before and after filtration was determined by high-resolution 1H NMR analysis (DDAC, BAC, SDS, Tergitol and peracetic acid) or spectrophotometry (sodium hypochlorite, hypochlorous acid and hydrogen peroxide).
2.12.1. Preparation of Samples for 1H NMR Analysis
Aliquots (0.54 mL) of both unfiltered and filtered disinfectants and detergents were added to 0.06 mL of 2H2O containing 5.80 × 10−3 mol./L of t 3-(trimethylsilyl)propionic-2,2,3,3-d4 acid (TSP) sodium salt. In exception, SDS samples were prepared by diluting 0.30 mL in HPLC-grade water to a 0.54 mL volume before addition of 0.06 mL TSP solution in 2H2O. Admixtures were rotamixed before transfer to 5 mm diameter NMR tubes. Samples were either prepared and run immediately or were retained frozen at –20 °C for no longer than 3 days. Samples were thawed at room temperature (approximately 1 h) prior to1H NMR analysis.
2.12.2. Acquisition of 1H NMR Spectra
1H NMR spectra for each test solution were acquired on a Jeol JNM-ECZ600R/S1 spectrometer equipped with a 5 mm broadband gradient auto-tunable probe operating at a frequency of 600.17 Hz for 1H and a probe temperature of 298K. Spectra were acquired with suppression of the very intense H2O/HOD resonance (δ = 4.68 ppm) by use of the Robust-5 pulse sequence. The pulsing conditions were sweep width 8993 Hz; 16,384 datapoints; acquisition time 1.82 s; relaxation delay 1.00 s; and 124 transients. Four pre-scans were acquired.
All signal intensities were electronically integrated using ACD Spectrus Processor software (Version 2021 for Microsoft Windows), and these were normalised with respect to that of the pre-fixed concentration TSP internal standard (δ = 0.00 ppm) for all samples evaluated. In this manner, the extent of membrane passage for each biocide tested was determined from its 1H NMR resonance. The chemical shift regions selected for the integration of these signals were constant for each analyte determined.
For peracetic acid and its acetic acid degradation products, singlet resonances located at δ = 2.13 and 2.05 ppm, respectively, were employed for quantification purposes.
2.12.3. Spectrophotometric Determinations of Hypochlorite, Hypochlorous acid and Hydrogen Peroxide
Spectrophotometric determinations of hypochlorite anion (OCl−) and hypochlorous acid (HOCl) along with H2O2 were performed using an Evolution 201 spectrophotometer applied to diluted samples of these biocides for both the control (pre-membrane passage) and post-membrane passage samples. Aliquots (1.00 mL) of sodium hypochlorite analyte solution were treated with a 10 μL volume of a 1.00 mol./L sodium hydroxide solution in order to ensure the conversion of any residual HOCl to OCl− according to the equation:
HOCl + OH− ↔ H2O + OCl−
Samples were then thoroughly rotamixed, and spectra were acquired at 220–500 nm; OCl− concentrations were determined from the λmax. value at 292 nm. However, HOCl concentrations were determined from absorbance at 300 nm in order to avoid any interferences from absorption bands arising from any other, albeit contaminating, agents present in the product tested [22].
Similarly, H2O2 levels in these control and membrane-passaged samples were monitored at a wavelength of 240 nm [23].
2.12.4. PES Bioindicator Permeability to Disinfectants and Detergents
PES bioindicators were prepared as follows. Cotton swatches were inoculated with E. faecium NCIMB 2699 (106 CFU/swatch) and incubated for 18 h at room temperature (~21 °C, 44% ± 1% RH) before loading with either 20 μL defibrinated sheep blood (EO Labs, UK) as an interfering substance or peptone salt solution as a control. Swatches were dried for 1 h at room temperature prior to heat sealing within a PES membrane (47 mm circular membrane folded in half, 0.22 μm pore size).
The PES bioindicators were laundered with either 1.1 mM sodium hypochlorite or 2 mM SDS in a 10 min wash without heating (23.44 ± 0.06 °C) or a textile load. Identical bioindicator swatches were placed in 10 mL of the disinfectant or detergent solutions for 13 min. Water-only controls were included. Surviving microorganisms were recovered from the swatches in 10 mL neutraliser and viable-counted as described above.
2.12.5. Comparison of PES Bioindicators and Swatch Methodologies within the Wash Process
The PES bioindicators were compared to two different swatch methodologies: BS EN 16616 [24] and the DES controller bioindicator. PES bioindicators were prepared as described above; in addition, PES bioindicators comprising 106 CFU E. faecium ATCC 6057 with 20 μL of tryptone salt were also assessed. For comparison, the wash procedure was performed as described in the BS EN 16616 standard. In brief, ten inoculated (E. faecium ATCC 6057) and six non-inoculated BS EN 16616 swatches, three PES bioindicators of each type and three DES controller bioindicators were placed in polycotton pillowcases. The pillowcases, the ballast load (70% of the maximum capacity) and defibrinated sheep blood (12.5 mL/kg) were placed inside the industrial machine drum (JLA, UK). Swatches were washed at ambient temperature using a 4 min prewash cycle and a 10 min wash cycle without rinsing. Microorganisms were processed according to BS EN 16616 and enumerated by pour plating in nutrient agar. All swatches were washed with and without detergents. The detergents (Cool Asepsis, Cristeyns, Bradford, UK) consisted of 2.5 mL/kg Power Extract and 3 mL/kg Cool Care and, for the wash, 16 mL/kg of Cool Asepsis. The inoculum concentrations were determined by processing three untreated swatches with each method.
2.13. Statistical Analysis
All investigations were conducted to a minimum of n = 4. Significant differences (p ≤ 0.05) in log10 CFU/swatch recoveries were analysed using independent-sample t-tests or one-way analysis of variance (ANOVA) with Tukey’s post-hoc (equal variances) or Games–Howell post-hoc test (unequal variances). Where data were not normally distributed, Mann–Whitney U tests or independent-samples Kruskal–Wallis tests were performed.
Means, standard deviations, standard errors and 95% confidence intervals (CIs) for the percentage of microbicidal agent passaging through all membranes were computed using XLSTAT2020 software options (Addinsoft, Paris, France). For this analysis, these parameters were primarily computed for the entire dataset irrespective of the membranes employed. An analysis-of-variance (ANOVA) model (XLSTAT2020) was also employed to determine CI values and the statistical significance of any differences observed between the % passage of microbicidal agents through each of the materials featured herein.
3. Results
3.1. Phase 1: The Efficacy of Current Methods for Measuring Disinfection within Healthcare Laundry Wash Processes
Methods for the Recovery of Microorganisms from Textiles
The viability of E. faecium and B. cereus spores on cotton remained stable over 18 h air drying (p > 0.05); E. coli and S. aureus showed a significant (p ≤ 0.05) decline in recovery (0.33–1.02 log10 reduction; Figure S1). There were no significant differences (p > 0.05) in recovery from cotton between all recovery media (Table 1). The agitation method did not significantly (p > 0.05) affect the recovery of all test species at 2 log10 CFU or of E. coli or E. faecium at 8 log10 CFU, yet stomaching resulted in inconsistent recovery depending on duration (30 s or 1 min), while shaking by hand recovered significantly (p ≤ 0.05) greater numbers of B. cereus spores compared to vortexing.

Table 1.
Log10 CFU recovery (% recovery) of S. aureus, E. faecium, E. coli and B. cereus spores from 25 cm2 cotton according to recovery medium and recovery method (mean, n = 4 ± SEM).
At 2 log10 CFU, surface-testing methods generally recovered significantly (p ≤ 0.05) lower numbers of the test species compared to shake-out methods (Table 1). At 8 log10 CFU, quantification was not possible for dip slide and RODAC plates due to a lack of discrete colonies. Shaking by hand recovered marginally higher numbers of S. aureus, E. faecium and B. cereus spores at 8 log10 CFU and the greatest quantity of S. aureus at 2 log10 CFU (2.40%), which had the lowest overall recovery of the test species. At 1 log10 CFU, shaking by hand recovered all species except for E. coli (Table 1). Nutrient agar and selective agar showed no significant difference (p > 0.05) in recovery across all species tested.
3.2. Recovery of Microorganisms from DES Controller Bioindicators
Recovery of E. faecium from DES controller bioindicators ranged from 5.59 log10 CFU for the 6 log10 CFU swatch to 4.68 log10 CFU for the 3 log10 CFU swatch (Table 2), with higher-than-specified counts for the 4 log10 CFU and 3 log10 CFU swatches. The recovery of E. coli from bioindicators was, conversely, 0.54–1.93 log10 CFU lower than specified. The recovery of B. subtilis spores was in line with the expected count at 5.96 log10 CFU, including after 30 min at 70 °C, indicating spores were present.

Table 2.
Viable counts (log10 CFU) of E. faecium, E. coli and B. subtilis spore DES controller bioindicators (mean, n = 4 ± SEM).
3.3. E. faecium DES Controller Bioindicator Retention Efficacy
There was a significant reduction (p ≤ 0.05) in E. faecium on loose cotton samples after laundering at ambient temperature with and without detergent (Table 3). E. faecium was reduced to a similar extent (p > 0.05) when laundered within a laundry bag, sewn cotton bag, hemming web-sealed bag or reusable autoclave bag. Nylon membrane filters and commercially available DES controller bioindicators retained significantly (p ≤ 0.05) greater numbers of E. faecium than loose samples, with no significant difference (p > 0.05) between the recovery of E. faecium pre- and post-laundering; all other conditions exhibited a significant (p ≤ 0.05) loss of cells post-laundering with detergent.

Table 3.
Log10 CFU/swatch recovery of E. faecium (6 log10 CFU/swatch) from 1 cm2 cotton swatches pre- and post-laundering at ambient temperature without detergent within different bags and comparison to commercially available bioindicators (n = 4 ± SEM).
3.4. E. faecium DES Controller Bioindicator Permeability to Disinfectants and Detergents
There was no significant difference (p > 0.05) in the recovery of E. faecium from DES controller bioindicators using 30 mL PBS-T or neutraliser (6.48 ± 0.12 versus 6.34 ± 0.03 log10 CFU, respectively). Recovery of E. faecium from cotton (25 and 1 cm2) using neutraliser was comparable to or greater than with PBS-T (Figure S2 and Table S3).
The DES controller bioindicator membrane did not significantly (p ≤ 0.05) affect the antimicrobial activity of peracetic acid, sodium hypochlorite, hydrogen peroxide or hypochlorous acid at ambient temperature (Table 4). A marginal reduction (p > 0.05; 0.94 log10) of E. faecium on loose swatches compared to enclosed swatches by DDAC suggests that permeability was limited. The DES controller bioindicator membrane also limited the reductions of E. faecium by SDS and BAC compared to samples treated without the membrane. No viable E. faecium was recovered from SDS swatch treatment solutions, suggesting a bactericidal effect rather than loss from the textile associated with surfactant action.

Table 4.
Permeability of DES controller bioindicators towards disinfectants and detergents at ambient temperature as determined by the log10 CFU/swatch recovery of E. faecium from DES controller bioindicator swatches after treatment in solution without a membrane compared to an industrial wash without heating and detergent enclosed within a membrane. Log10 reduction of E. faecium following treatment was calculated relative to the pre-treatment microbial load (6.34 log10 CFU/swatch) and water-only control (n = 4 ± SEM).
Washing at 60 °C significantly (p ≤ 0.05) improved the permeability of the DES controller bioindicator membrane to SDS, with a 2.47 log10 CFU reduction compared to samples in solution (Table 4). There was limited permeability at 50 °C and below.
3.5. Comparison of Semi-Quantitative and Quantitative Enumeration Methods for Commercially Available DES Controller Bioindicators
Using a quantitative viable-count method, E. faecium was reduced by 5.25 log10 compared to the water-only control using a 67 °C wash cycle without detergent. E. faecium was completely inhibited (6.03 log10 reduction) by industrial laundering at 67 °C with detergent and 75 °C without detergent, compared to 4.80–6.53 log10 CFU using a 60 °C domestic wash cycle with biological or non-biological detergent (Table 5).

Table 5.
Log10 reduction of E. faecium, E. coli and B. subtilis spores following domestic and industrial laundering processes as determined using both quantitative and semi-quantitative methodologies (mean, n = 4 ± SEM). Log10 reduction calculated from the unlaundered or ambient water-only controls for the quantitative method and from expected inoculum size for the semi-quantitative method.
E. coli was completely reduced by the domestic (60 °C) and industrial wash cycles (67 and 75 °C) with and without detergent (Table 5). B. subtilis spores were reduced by <1 log10 CFU compared to the water-only control by domestic wash cycles with and without detergent and by industrial cycles (67 and 75 °C) without detergent. A 1.68 log10 reduction was achieved by the 67 °C industrial wash cycle with detergent (Table 5).
The semi-quantitative method followed a similar pattern to the quantitative method; discrepancies were observed for the domestic wash cycles with detergent against E. faecium, where only a 4 log10 CFU reduction was indicated compared to 4.8–6.35 log10 reductions calculated using the quantitative method (Table 5). There were also inconsistencies in growth for 60 °C water-only washes against E. faecium (3, 5 and 6 log10 CFU swatches positive for growth, 4 log10 CFU negative) for three out of four repeats, suggesting that the method could lead to difficulties in the interpretation of results. The 1.68 log10 reduction of B. subtilis spores was not detected, with only a 6 log10 CFU swatch included in B. subtilis spore bioindicators. Moreover, the lower count of E. coli than stated by the manufacturer (6 log10 CFU) led to the log10 reduction being calculated as higher than that observed using the quantitative method.
An in-field test conducted using a continuous batch washer (CBW) demonstrated that E. faecium was completely reduced according to both the quantitative and semi-quantitative methods (Table 6). The quantitative method indicated that B. subtilis spores were completely reduced by laundering in two out of four repeats, while viable spores were detected on the remaining samples, leading to an average reduction of ≥4.51 log10 CFU (Table 6). In accordance, growth was present in two out of four repeats for the semi-quantitative method, indicating that a 6 log10 reduction was achieved for these two samples. The reduction of E. coli during laundering was only quantifiable to ≥3.03 log10 CFU (Table 6).

Table 6.
Log10 reduction of E. faecium, E. coli and B. subtilis spores following an in-field test using a CBW (75 °C with industrial detergent) as determined using both quantitative and semi-quantitative methodologies (mean, n = 4 ± SEM). Log10 reduction calculated from the unlaundered control for the quantitative method and from expected inoculum size for the semi-quantitative method.
3.6. Phase 2: Development of Alternative E. faecium Strains and a Bioindicator Membrane for Low-Temperature Laundering
Disinfectant, Detergent and Thermal Tolerance on Cotton
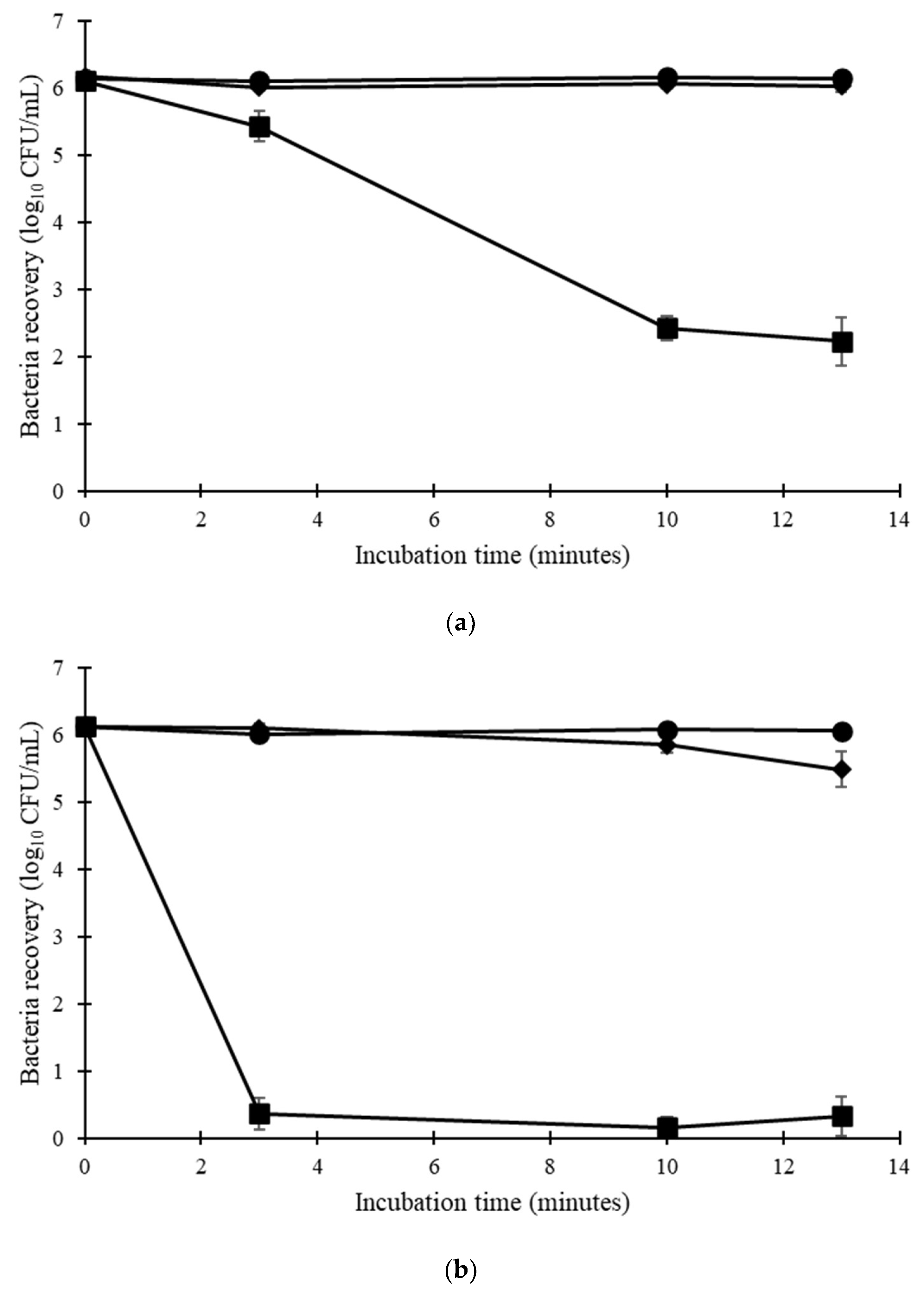
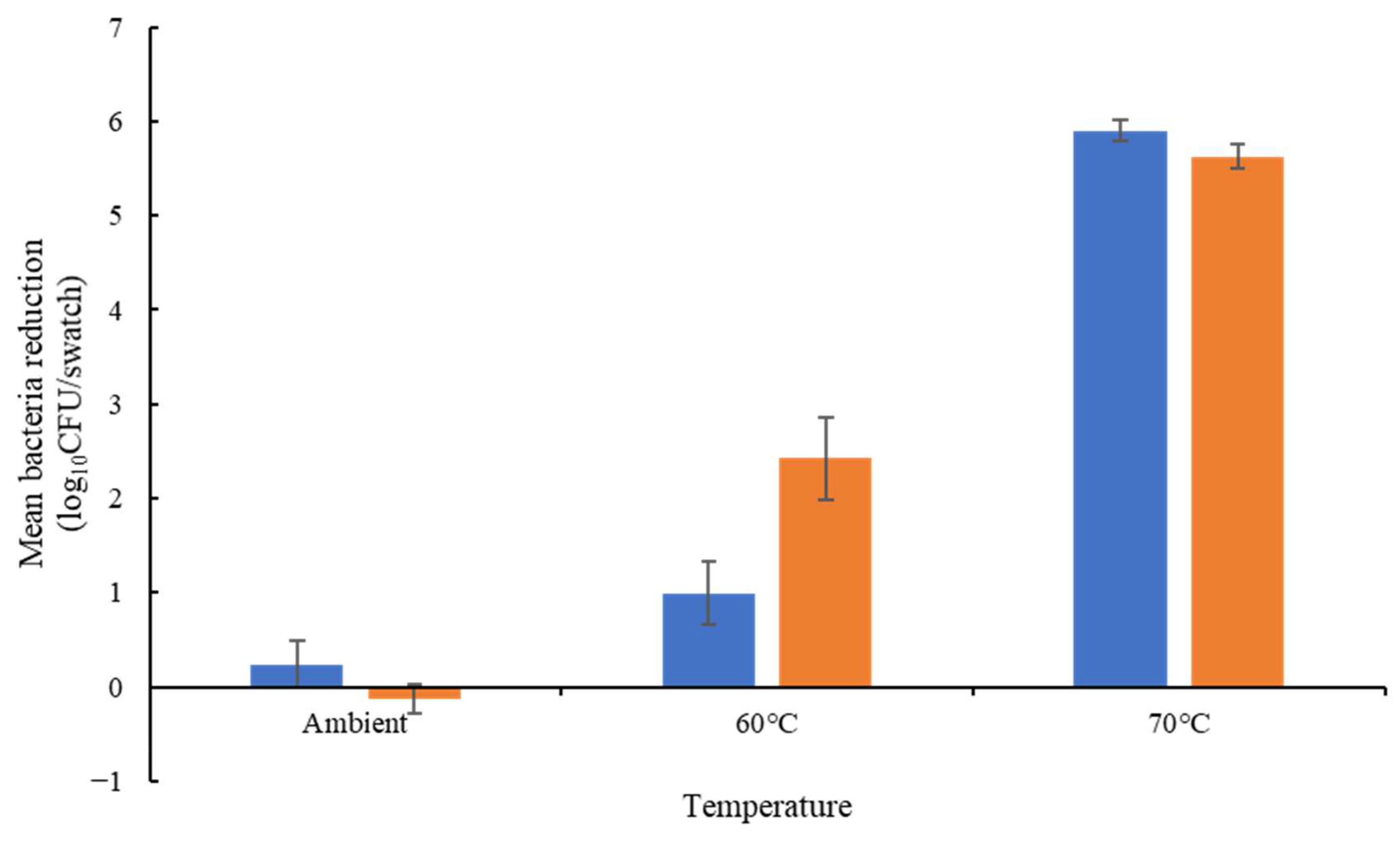
Among the three isolates tested, E. hirae exhibited less resistance to temperature and chemicals than the E. faecium isolates (Table S4). Considering only E. faecium, all isolates were able to survive 13 min at 60 °C in solution (Figure 1). Only the E. faecium ATCC 6057 was able to survive at 70 °C, with a 3.44 log10 CFU/mL reduction in the total bacteria concentration. On cotton, all isolates exhibited lower thermotolerance. E. faecium NCIMB 2699 showed a greater reduction at 60 °C (2.42 log10 CFU/mL). E. faecium ATCC 6057 showed a greater reduction on cotton (5.35 log10 CFU/swatch) at 70 °C compared to in solution (3.44 log10 CFU/mL reduction) at the same temperature (Figure 2).
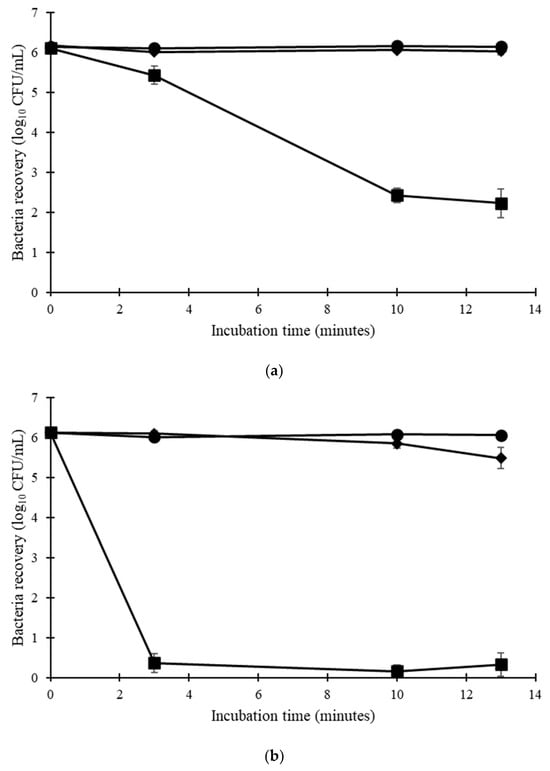
Figure 1.
Thermotolerance in solution of (a) E. faecium ATCC 6057 and (b) E. faecium NCIMB 2699 (n = 4 ± SEM). ● ambient temperature, ◆ 60 °C, ■ 70 °C.
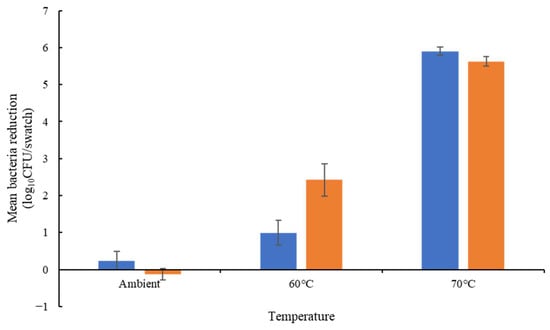
Figure 2.
Thermotolerance on cotton of E. faecium ATCC 6057 (blue bars) and E. faecium NCIMB 2699 (orange bars). Log10 reduction was calculated from the inoculum (n = 4 ± SEM); the results confirmed there was no significant difference between isolates at each temperature.
E. faecium NCIMB 2699 was less susceptible to peracetic acid than E. faecium ATCC 6057 (2.01 and 5.26 log10 CFU/swatch reductions, respectively); these isolates exhibited similar susceptibility to sodium hypochlorite (3.19 and 2.80 log10 CFU/swatch respectively) and SDS (3.22 to 3.33 log10 CFU/swatch reduction) (Figure 2).
3.7. Assessment of Stability of the PES Membrane
The results demonstrated that the PES membrane was stable through an industrial wash process and that E. faecium was unable to pass through the membrane (Method S5).
3.8. Flow Rate of Solution through the PES Membrane
The diffusion flow rate of the distilled water solution was determined on four types of membrane/fabric: 100% cotton, 100% polyester, PES and the DES controller bioindicator membrane. Cotton exhibited the highest flow rate, with an average of 11.96 mL/min at room temperature, 14.65 mL/min at 40 °C and 21.11 mL/min at 60 °C. Polyester showed lower flow rates between 0.27 and 0.3 mL/min, but temperature did not affect the flow rate. Similarly, the flow rate of the PES membrane was not affected by temperature but was lower than for polyester, with flow rates between 0.09 mL/min and 0.102 mL/min. Finally, the DES controller membrane showed the lowest flow rates of 0.045 mL/min at room temperature, 0.053 mL/min at 40 °C and 0.065 mL/min at 60 °C.
3.9. Detergent and Disinfectant Membrane Passage Assessment
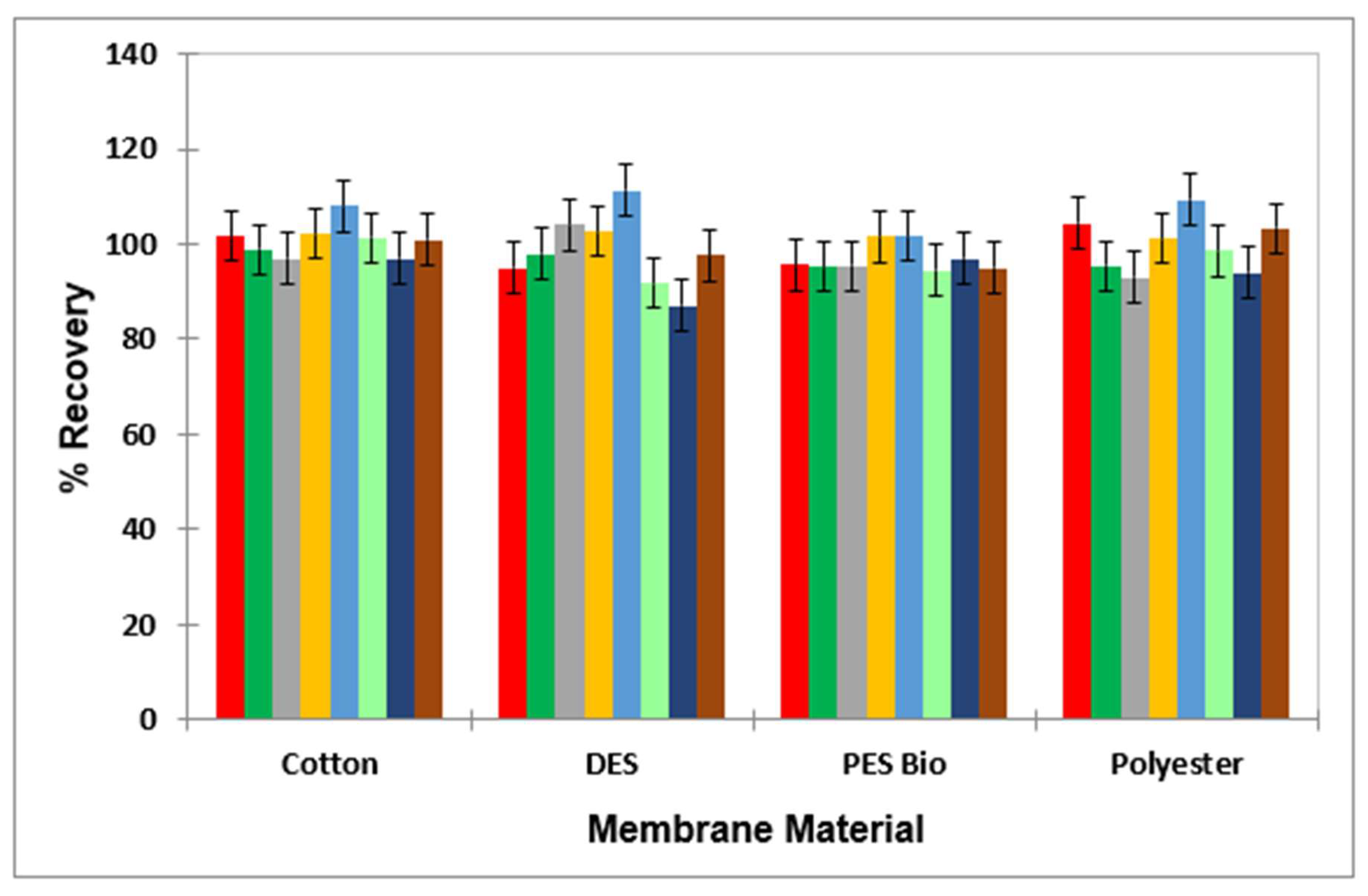
The mean ± 95% CI percentage values for each of the biocides through the membrane material were 99.1 ± 3.3% for BAC; 96.9 ± 2.5% for DDAC; 99.2 ± 2.5% for Tergitol; 107.6 ± 0.94.7% for peracetic acid; 106.9 ± 4.8% for peracetic acid’s decomposition product, acetic acid; 83.8 ± 2.7% for hypochlorite anion; 96.3 ± 3.1% for hypochlorous acid; and 101.9 ± 0.9% for hydrogen peroxide (Figure 3).
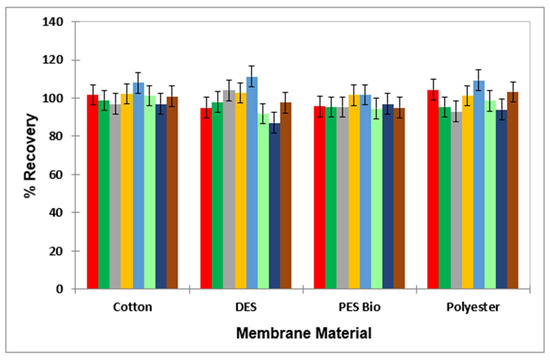
Figure 3.
Bar diagram for the mean ± 95% CI percentage recovery values (n = 3) for each laundry agent following passage through different membrane materials (cotton, DES controller bioindicator, PES and polyester), with the results confirming that there were no significant differences between the extent of passage for any agents evaluated for each of the materials involved. Colour key: benzalkonium chloride (red); didecyldimethylammonium chloride (dark green); hypochlorous acid (grey); hydrogen peroxide (yellow); peracetic acid (light blue); sodium dodecyl sulphate (light green); sodium hypochlorite (dark blue); and Tergitol (brown).
Coefficients of variation for the triplicate determinations made for each membrane tested ranged from as low as 0.14% to 7.12%, with only one of these values lying below recommended values of ≤5%. Therefore, with the exception of OCl−, all analytes successfully passed through the membrane with mean recoveries of >95%. The results obtained for OCl− are not unexpected, in view of its very high level of oxidant activities towards a range of organic substrates.
3.10. PES Bioindicator Permeability to Disinfectants and Detergents during the Wash Process
The permeability of the PES membrane to sodium hypochlorite and SDS was assessed during an ambient wash (20–24 °C) with E. faecium NCIMB 2699. Similar bacterial reductions were observed between the wash and in-solution controls. In the presence of chlorine, a mean reduction of 5.92 log10 CFU/swatch was observed during the wash (6.54 log10 CFU/swatch reduction in solution). The presence of blood did not interfere with the chlorine activity during the wash, with a mean bacteria reduction of 6.54 log10 CFU/swatch. During the wash with SDS, a similar bacteria reduction was observed between the wash (1.72 log10 CFU/swatch) and the in-solution control (1.93 log10 CFU/swatch). However, the presence of blood during the wash significantly reduced (p = 0.019) the antibacterial activity of SDS with a bacteria reduction of 0.80 log10 CFU/swatch (Table 7).

Table 7.
PES membrane chemical permeability during industrial test washing at room temperature (n = 4, ± SEM).
3.11. Comparison of PES Bioindicators and Swatch Methodologies within the Wash Process
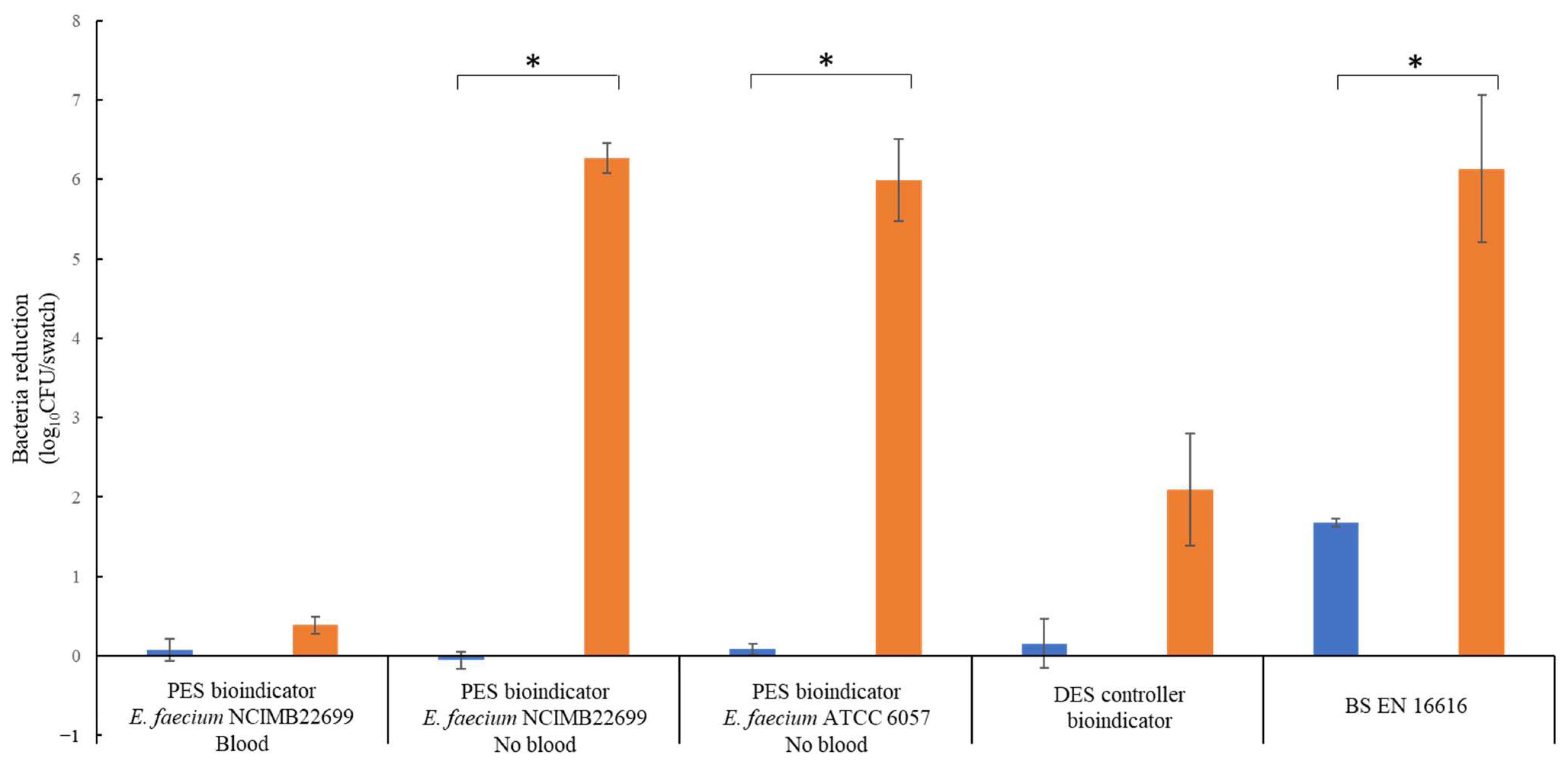
BS EN 16616 methodology and the bioindicators were compared at ambient temperature (20–24 °C). After the wash cycle without detergent, the DES controller and the PES bioindicators showed no bacteria reductions (between –0.06 and 0.08 log10 CFU/swatch reduction) due to the bacteria survival being similar to the inoculum (Figure 4). In comparison, a bacteria reduction of 1.7 log10 CFU/swatch was observed for the BS EN 16616-inoculated swatches. The non-inoculated swatches exhibited bacterial contamination with an average 3.8 log10 CFU/swatch. Except for the PES bioindicators containing sheep blood, all swatches exhibited reductions after the wash cycle with detergent, with 2.09, 6.27 and 5.99 log10 CFU/swatch reductions for the DES controller bioindicators and PES bioindicators (NCIMB 2699 and ATCC 6057 respectively) and a 6.13 log10 CFU/swatch reduction of the BS EN 16616 swatches. Uninoculated BS EN 16616 swatches exhibited non-significant bacterial contamination.
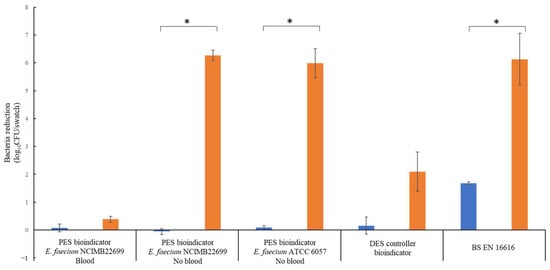
Figure 4.
Comparison of the efficacy of different swatch methods to detect bacteria killing during an ambient-temperature industrial wash. Washes were processed in an identical manner for all methods with (orange bar) or without industrial detergent (blue bar). The bacteria reduction was calculated from the inoculum concentration (n = 3 ± SEM).* p ≤ 0.05.
4. Discussion
There are currently no international standard methods for validating and monitoring the efficacy of industrial laundering processes. Variation in the methods and action levels employed may lead to differences in decontamination standards between laundries, both within and between countries. This study aimed to develop standard methodologies for establishing and validating the disinfection efficacy of laundering processes and for routine monitoring of post-laundering microbial bioburden, with a specific focus on demonstrating microbial kill within the wash process rather than a reliance on dilution. This is of particular importance for healthcare laundry due to the textiles being in contact with vulnerable patients and the increasing abundance of antibiotic-resistant microorganisms.
A comparison of currently used methods across the UK, Europe and the USA for recovering microorganisms from textiles demonstrated that surface testing methods such as RODAC plates were less sensitive than the agitation of samples in recovery diluent (Table 1). Rabuza et al. (2012) also reported that RODAC plates failed to recover approximately 5 log10 S. aureus and Klebsiella pneumoniae from cotton; this may be attributed to microorganisms absorbing into the textile, where they do not come into contact with the plate [25]. RODAC plates are commonly used for the microbial bioburden monitoring of textiles, including in Germany and the USA, with the advantage that they are easy to use and non-destructive in contrast to agitation methods [14,15]. Shaking by hand with PBS-T was marginally more effective than other agitation methods (Table 2), suggesting that this method is suitable for post-laundering microbial bioburden testing in order to monitor for potential recontamination of processed textiles following disinfection by the laundry process. Microbial recovery was not significantly different (p > 0.05) between selective and non-selective agars (Table 2) suggesting that the method can be adapted for total counts and/or the detection of specified microorganisms.
In Germany, and as best practice in England and Wales, the disinfection efficacy of industrial wash cycles must be validated using artificially contaminated textiles [15,16,17]. Following validation, input parameters can be monitored and controlled (e.g., temperature and time) to determine that disinfection has occurred. No such method is recommended in the USA [3,14]. In this study, the efficacy of laundering loose textile samples and bioindicators for monitoring the disinfection of microorganisms during laundering were compared. Laundering loose swatches resulted in a 1.83 log10 loss of E. faecium when laundered at ambient temperature with water only and a 4.86 log10 loss with detergent, and this loss was not ameliorated by enclosing within a range of cloth bags. The detergent was determined to be non-antimicrobial (Table S2), suggesting microorganisms were lost due to agitation and surfactant effects. This confounds the detection of disinfection alone, where a ≥5 log10 reduction must be achieved [16]. Cross-contamination of sterile textiles in a wash with contaminated samples has been observed; for example, 0–14 CFU/25 cm2 Clostridioides difficile spores cross-contaminated sterile textiles in an industrial wash with detergent, which may be an infection-control risk if disinfection is not achieved [26]. DES controller bioindicators retained E. faecium during the wash (Table 2), suggesting that microbial kill may be determined. A further advantage of bioindicators is increased safety for operators, with the test microorganism being enclosed within a membrane [27].
A validation protocol was developed to assess the permeability of the DES controller bioindicator membrane to disinfectants and detergents by comparing the antimicrobial efficacy of sublethal concentrations between loose samples in isolation compared to laundered bioindicators. The DES controller bioindicator produced equivalent or greater log10 reductions compared to loose samples for the tested disinfectants (peracetic acid, sodium hypochlorite, hydrogen peroxide and hypochlorous acid), and detergent (SDS, DDAC and BAC) permeability was limited (Table 3). Detergents may possess antimicrobial activity; thus, it is important for bioindicator membranes to be permeable to such components [19]. In accordance, Kagemann et al. (2008) reported that nylon membranes (0.45 μm) were not permeable to non-ionic or anionic detergents at 60 °C using chemical detection methods and resulted in a ~3.5 log10 lower reduction in E. faecium compared to loose textile samples [27]; it was hypothesised that surfactant molecules adhere to the charge membrane. Increasing the wash temperature to 60 °C in this study improved permeability to SDS (Table 3), conversely to the findings of Kagemann et al. (2008) for a nylon membrane, suggesting that the DES controller bioindicators may be suitable for validating thermal laundering processes but not for low-temperature laundering [27]. The composition of the DES controller bioindicator membrane used in this study is not stated by the manufacturer. Further research into the development of new membranes for monitoring microbial kill in the wash was required, and the use of a PES membrane demonstrated promising results. The investigations of all the detergents and disinfectants tested showed comparable permeation through the membrane for cotton, polyester, the DES controller bioindicator membrane and the PES membrane (Figure 3). Solution flow rate significantly varied, with the PES membrane exhibiting a constant flow rate across all temperatures tested (0.09–0.102 mL/min) and the DES controller membrane performing less well than the PES membrane (0.045–0.065 mL/min) but being most efficient at 60 °C, the temperature where the efficacy of the DES controller bioindicator took effect. The permeability of the PES membrane to SDS and sodium hypochlorite at low temperatures was confirmed by wash tests. Indeed, the bacteria reductions observed after the wash test were similar to the reductions observed when the swatches were treated directly with the chemical solution.
E. faecium ATCC 6057, used in the commercially available bioindicators, is a biosafety level 2 microorganism; E. hirae was explored as a surrogate, but its temperature and chemical tolerance were significantly different (Table S4). E. faecium NCIMB 2699 was also assessed due to its wide use in thermal process validation for food products, and it has a genome similar to commensal strains with no antibiotic-resistant genes [28]. In the USA, E. faecium NCIMB2699 is also a biosafety level 1 microorganism. In addition, E. faecium ATCC 6057, used in the commercially available DES controller bioindicators, demonstrated a lower chemical resistance than E. faecium NCIMB2699 to peracetic acid (5.26 log10 CFU/swatch reduction and 2.01 log10 CFU/swatch reduction, respectively) (Figure 2). Both strains exhibited similar chemical resistance to sodium hypochlorite and SDS (2.80 to 3.19 log10 CFU/swatch reduction and 3.22 to 3.33 log10 CFU/swatch reduction, respectively). Their thermotolerance on cotton was also similar.
Moreover, new laundry chemistries may be assessed for compatibility with the PES bioindicator membrane. Neutraliser validation tests were also developed for convenience when assessing other chemistries (Method S3).
The recovery of E. coli from untreated DES controller bioindicators was 0.54–1.93 log10 CFU lower than expected, with 4.55 log10 CFU recovered from the 6 log10 CFU swatch and 2.46 log10 CFU recovered from the 3 log10 CFU swatch (Table 4), and results were inconsistent between swatches, which could result in false negatives without quantification of untreated controls. B. subtilis spore reductions below 6 log10 cannot be calculated using this method, as only a 6 log10 CFU swatch is present in the bioindicator. This suggests that E. faecium DES controller bioindicators are most appropriate for use with the semi-quantitative method. The thermotolerance of E. faecium also enables chemo-thermal processes to be monitored for kill compared to E. coli, which was completely reduced by a 60 °C wash with water alone (Table 5). Further development of a B. subtilis spore bioindicator compatible with the semi-quantitative method would be advantageous, as there are currently no standards for the sporicidal activity of laundry processes, yet bacterial spores persisted during industrial laundering in a CBW and washer–extractor machine (Table 5 and Table 6), in accordance with previous work, and hospital outbreaks of B. cereus have been attributed to healthcare linen [26,29]. The results of the semi-quantitative method were generally concordant with the quantitative method, with some discrepancies observed for the domestic wash cycles with detergent against E. faecium, where a 4 log10 reduction was indicated compared to 4.8–6.35 log10 reductions calculated using the quantitative method (Table 5). Overall, the semi-quantitative method is easier to use, faster and has lower consumables costs than the quantitative method, which is advantageous for use in on-site laundry laboratories with limited resources.
5. Conclusions
The current use of multiple methodologies, based on different microbiological principles, such as contact plates, swatch testing and bioindicators, within the healthcare laundry industry, has given rise to incomparable disinfestation validation across this sector. In this study, methodologies were developed for validating the disinfection efficacy of microorganisms during laundering, based on the principles of microbial kill within wash systems. Overall, the proposed PES bioindicator is recommended for assessing disinfection within a wash process. Indeed, this method not only measures absolute kill within a wash process but can also be used effectively with a range of chemistries utilised within the laundry industry at both low and high temperatures. Traditional swatch testing and BS EN 16616 standard methods showed an underestimation of kill in view of the dilution of bacteria within the wash water; this methodology can also lead to microorganisms surviving in the wash and can therefore cross-contaminate other textiles. These effects are overcome by bioindicators, which enclose the microorganism within a semi-permeable membrane. However, the DES-controller bioindicator performance was reduced at temperatures below 60 °C. For recovery of microorganisms from textiles, agitation methods were more sensitive than surface sampling methods, and, therefore, a method based on shaking textiles by hand in PBS-T is recommended for post-laundering in bioburden testing. International standardisation of laundry validation tests is required to ensure consistency of disinfection throughout the global laundry sector to ensure that healthcare textiles are safe for patient use. This may be achieved by incorporating the developed methodology into risk management standards such as BS EN 14065, which focuses on laundry-processed textiles with biocontamination control systems.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/applmicrobiol4010014/s1, Methods; Method S1: Effect of Drying Time on the Recovery of Microorganisms from Cotton; Method S2: Disinfectant Suspension Test; Method S3: Swatch Suspension Test; Method S4: Swatch-Based Neutraliser Efficacy Validation Test; Method S5: Assessment of the Stability of the Polyethersulphone Membrane through a Wash System. Figure S1: Log10 CFU/swatch recovery of (a) B. cereus spores, (b) E. faecium, (c) E. coli and (d) S. aureus following incubation at room temperature (n = 4 ± standard error of the mean [SEM]) ▲ 108 CFU/swatch inoculum; ● 102 CFU/swatch inoculum. Figure S2: Log10 CFU/swatch recovery of E. faecium (8 and 2 log10 CFU/swatch) from (a) 25 cm2 and (b) 1 cm2 cotton swatches (6 log10 CFU/swatch) when shaken by hand in 30 mL neutraliser in comparison to PBS-T. Closed bars, Original inoculum; open bars, recovery from swatch. Percentage recoveries from the original inocula are shown as data labels. Table S1: Log10 reduction of E. faecium (BS EN 1040:2005, 8.02 log10 CFU; swatch test, 6.78 log10 CFU) by a range of disinfectants and detergents according to both the BS EN 1040:2005 suspension test and adapted swatch suspension test and time differences in the concentration of disinfectant required to produce an approximately equivalent log10 reduction. Table S2: Log10 CFU/swatch recovery of E. faecium from cotton swatches after treatment in solution without a membrane compared to an industrial wash without heating. * Significant (p ≤ 0.05) difference between loose swatch in solution and in the wash. § Significant (p ≤ 0.05) reduction compared to the water-only control. Table S3: Comparison of swatch-based neutraliser validation tests with the BS EN 1040:2005 suspension test against E. faecium (n = 4 ± SEM). Table S4: Log10 CFU/swatch survival of Enterococcus sp. treated with disinfectants for 13 min according to the adapted swatch suspension test (n = 4 ± SEM).
Author Contributions
Conceptualization, K.L. and L.O.; methodology, K.L., L.O., C.C., G.P. and M.G.; validation, K.L., L.O. and C.C.; formal analysis, K.L., L.O., C.C. and M.G.; investigation, L.O., C.C. and G.P.; resources, K.L.; data curation, K.L., L.O. and C.C.; writing—original draft preparation, K.L., L.O., C.C. and M.G.; writing—review and editing, K.L., L.O. and C.C.; visualization, K.L., C.C. and L.O.; supervision, K.L.; project administration, K.L.; funding acquisition, K.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by global industrial laundry trade bodies: Textile Services Association (UK), Textile Rental Services Association (USA), European Textile Services Association (Europe), Deutscher Textilreinigungs-Verband e.V. (Germany), De Federatie van de Belgische Textielverzorging vzw (Belgium), Norsk Renseri- og Vaskeriforening (Norway), Tekstiilihuoltoliitto ry (Finland) and Der Verband Textilpflege Schweiz (Switzerland).
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to commercial sensitivity in relation to the DES controller bioindicators.
Acknowledgments
The authors would like to thank Davy Stoker of Micronclean and Rachel Kahrman of Elis for their knowledge and input on laundry chemistry and microbiology. The authors would also like to thank all of the laundries and chemistry companies involved in supporting the research.
Conflicts of Interest
The authors declare no conflicts of interest. The sponsors had no role in the design, execution, interpretation or writing of the study.
References
- Glowicz, J.; Benowitz, I.; Arduino, M.J.; Li, R.; Wu, K.; Jordan, A.; Toda, M.; Garner, K.; Gold, J.A. Keeping Healthcare Linens Clean: Underrecognized Hazards and Critical Control Points to Avoid Contamination of Laundered Healthcare Textiles. Am. J. Infect. Control, 2022; in press. [Google Scholar]
- Bockmühl, D.P. Laundry hygiene—How to get more than clean. J. Appl. Microbiol. 2017, 122, 1124–1133. [Google Scholar] [CrossRef] [PubMed]
- CDC. Guidelines for Environmental Infection Control in Health-Care Facilities: G. Laundry and Bedding. 2003. Available online: https://www.cdc.gov/infectioncontrol/guidelines/environmental/background/laundry.html (accessed on 25 July 2022).
- Department of Health. Health Technical Memorandum 01-04: Decontamination of Linen for Health and Social Care: Management and Provision. 2016. Available online: https://www.england.nhs.uk/wp-content/uploads/2021/05/Mgmt_and_provision.pdf (accessed on 25 July 2022).
- Kampf, G. How long can nosocomial pathogens survive on textiles? A systematic review. GMS Hyg. Infect. Control 2020, 15, Doc10. [Google Scholar] [PubMed]
- Mallick, D.; Gupta, D.; Sharma, S. Transfer of bacteria between fabric and surrogate skin. Am. J. Infect. Control 2022, 50, 758–763. [Google Scholar] [CrossRef] [PubMed]
- Handorean, A.; Robertson, C.E.; Harris, J.K.; Frank, D.; Hull, N.; Kotter, C.; Stevens, M.J.; Baumgardner, D.; Pace, N.R.; Hernandez, M. Microbial aerosol liberation online soiled textiles isolated during routine residuals handling in a modern health care setting. Microbiome 2015, 3, 72. [Google Scholar] [CrossRef] [PubMed]
- Loveday, H.P.; Wilson, J.A.; Hoffman, P.N.; Pratt, R.J. Public perception and the social and microbiological significance of uniforms in the prevention and control of healthcare-associated infections: An evidence review. Br. J. Infect. Control 2007, 8, 10–21. [Google Scholar]
- Mitchell, A.; Spencer, M.; Edmiston Jr, C. Role of healthcare apparel and other healthcare textiles in the transmission of pathogens: A review of the literature. J. Hosp. Infect. 2015, 90, 285–292. [Google Scholar] [PubMed]
- Overcash, M.R.; Sehulster, L.M. Estimated incidence rate of healthcare-associated infections (HAIs) linked to laundered reusable healthcare textiles (HCTs) in the United States and United Kingdom over a 50-year period: Do the data support the efficacy of approved laundry practices? Infect. Control Hosp. Epidemiol. 2021, 43, 1510–1512. [Google Scholar] [PubMed]
- Sehulster, L.M. Healthcare laundry and textiles in the United States: Review and commentary on contemporary infection prevention issues. Infect. Control Hosp. Epidemiol. 2015, 36, 1073–1088. [Google Scholar] [PubMed]
- EN 14065; Textiles-Laundry Processed Textiles-Biocontamination Control System. CEN: Brussels, Belgium, 2016.
- BS EN 14065:2016; Textiles-Laundry Processed Textiles-Biocontamination Control System. British Standards Institute (BSI): London, UK, 2016.
- TRSA. Standard for Producing Hygienically Clean Reusable Textiles for Use in the Healthcare Industry. 2021. Available online: https://hygienicallyclean.org/wp-content/uploads/2021/01/HCH_Standard_011421.pdf (accessed on 25 July 2022).
- Fijan, S.; Cencic, A.; Turk, S.Š. Hygiene monitoring of textiles used in the food industry. Braz. J. Microbiol. 2006, 37, 356–361. [Google Scholar]
- Department of Health. Health Technical Memorandum 01-04: Decontamination of Linen for Health and Social Care: Engineering, Equipment and Validation. 2016. Available online: https://www.england.nhs.uk/wp-content/uploads/2021/05/Engineering.pdf (accessed on 25 July 2022).
- NHS Wales Shared Services Partnership–Specialist Estates Services. WHTM 01-04–Decontamination of Linen for Health and Social Care -Engineering, Equipment and Validation. 2017. Available online: https://nwssp.nhs.wales/a-wp/all-wales-laundry-service-review/all-wales-laundry-service-documents/useful-documents/whtm-01-04-2017-linen-engineering/ (accessed on 7 July 2023).
- Owen, L.; Laird, K. The role of textiles as fomites in the healthcare environment: A review of the infection control risk. PeerJ 2020, 8, e9790. [Google Scholar] [PubMed]
- Riley, K.; Williams, J.; Owen, L.; Shen, J.; Davies, A.; Laird, K. The effect of low-temperature laundering and detergents on the survival of Escherichia coli and Staphylococcus aureus on textiles used in healthcare uniforms. J. Appl. Microbiol. 2017, 123, 280–286. [Google Scholar] [PubMed]
- BS EN ISO 6330:2012; Textiles. Domestic Washing and Drying Procedures for Textile Testing. British Standards Institute (BSI): London, UK, 2012.
- BS EN 1040:2005; Chemical Disinfectants and Antiseptics. Quantitative Suspension Test for the Evaluation of Basic Bactericidal Activity of Chemical Disinfectants and Antiseptics. Test Method and Requirements (Phase 1). British Standards Institute (BSI): London, UK, 2005.
- Horváth, A.K.; Nagypál, I. Kinetics and mechanism of the reaction between hypochlorous acid and tetrathionate ion. Int. J. Chem. Kinet. 2000, 32, 395–402. [Google Scholar] [CrossRef]
- Beers, R.F.; Sizer, I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140. [Google Scholar] [PubMed]
- BS EN 16616:2022; Chemical Disinfectants and Antiseptics. Chemical-Thermal Textile Disinfection. Test Method and Requirements (Phase 2, Step 2). British Standards Institute (BSI): London, UK, 2022; 2.
- Rabuza, U.; Šostar Turk, S.; Fijan, S. Efficiency of four sampling methods used to detect two common nosocomial pathogens on textiles. Text. Res. J. 2012, 82, 2099–2105. [Google Scholar] [CrossRef]
- Tarrant, J.; Jenkins, R.O.; Laird, K.T. Online ward to washer: The survival of Clostridium difficile spores on hospital bed sheets through a commercial UK NHS healthcare laundry process. Infect. Control Hosp. Epidemiol. 2018, 39, 1406–1411. [Google Scholar] [CrossRef] [PubMed]
- Kagemann, G.; Hilgenberg, B.; Rech, J.; Heintz, M.; Vossebein, L. Use of Biomonitors for the Validation of Chemo-thermal Disinfecting Washing Procedures. Tenside Surfactants Deterg. 2008, 45, 334–339. [Google Scholar] [CrossRef]
- Kopit, L.M.; Kim, E.B.; Siezen, R.J.; Harris, L.J.; Marco, M.L. Safety of the Surrogate Microorganism Enterococcus faecium NRRL B-2354 for Use in Thermal Process Validation. Appl. Environ. Microbiol. 2014, 80, 1899–1909. [Google Scholar] [PubMed]
- Cheng, V.C.; Chen, J.H.; Leung, S.S.; So, S.Y.; Wong, S.C.; Wong, S.C.; Tse, H.; Yuen, K.Y. Seasonal outbreak of Bacillus bacteremia associated with contaminated linen in Hong Kong. Clin. Infect. Dis. 2017, 64, S91–S97. [Google Scholar] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).