Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity
Abstract
1. Introduction
2. Materials and Methods
2.1. Oomycete Pathogen
2.2. Isolation of Rhizobacteria from the Rhizospheric Soil
2.3. Screening for Antagonistic Bacteria
2.4. Effects of Bacterial Isolates on the Mycelial Structure of P. vexans
2.5. Identification of Antagonistic Bacteria
2.6. Antifungal Effect of Volatile Organic Compounds (VOCs)
2.7. Antibiosis by Bacterial Supernatant
2.8. Biochemical Characterizations
2.8.1. Proteolytic Activity
2.8.2. Amylase Activity
2.8.3. Cellulase Degradation
2.8.4. Pectinase Production
2.8.5. Phosphate Solubilization
2.8.6. Production of Indole Acetic Acid (IAA)
2.8.7. Production of Hydrogen Cyanide (HCN)
2.8.8. Detection of Lipopeptides by the PCR Method
2.9. PGPR Test on Sorghum Bicolor
2.10. Statistical Analysis
3. Results
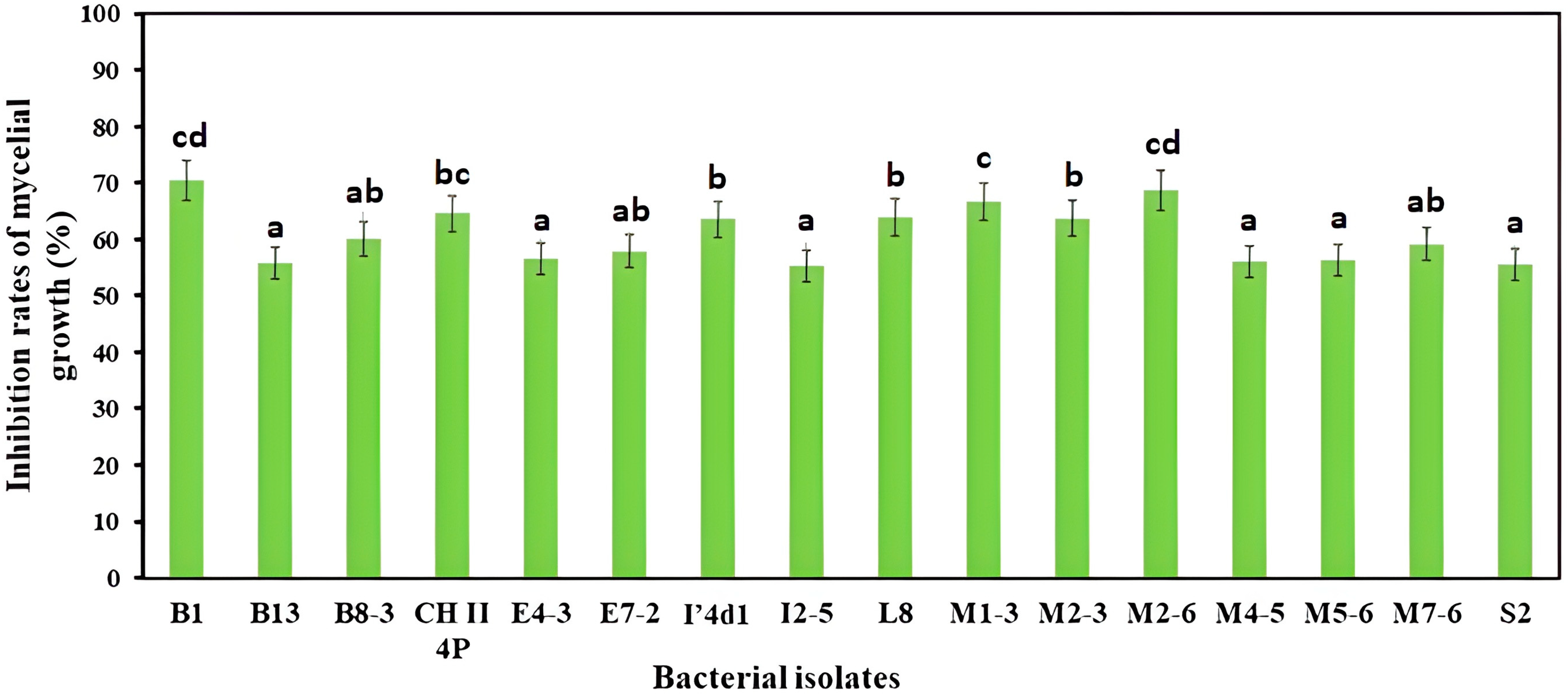
3.1. Dual Culture Bioassay
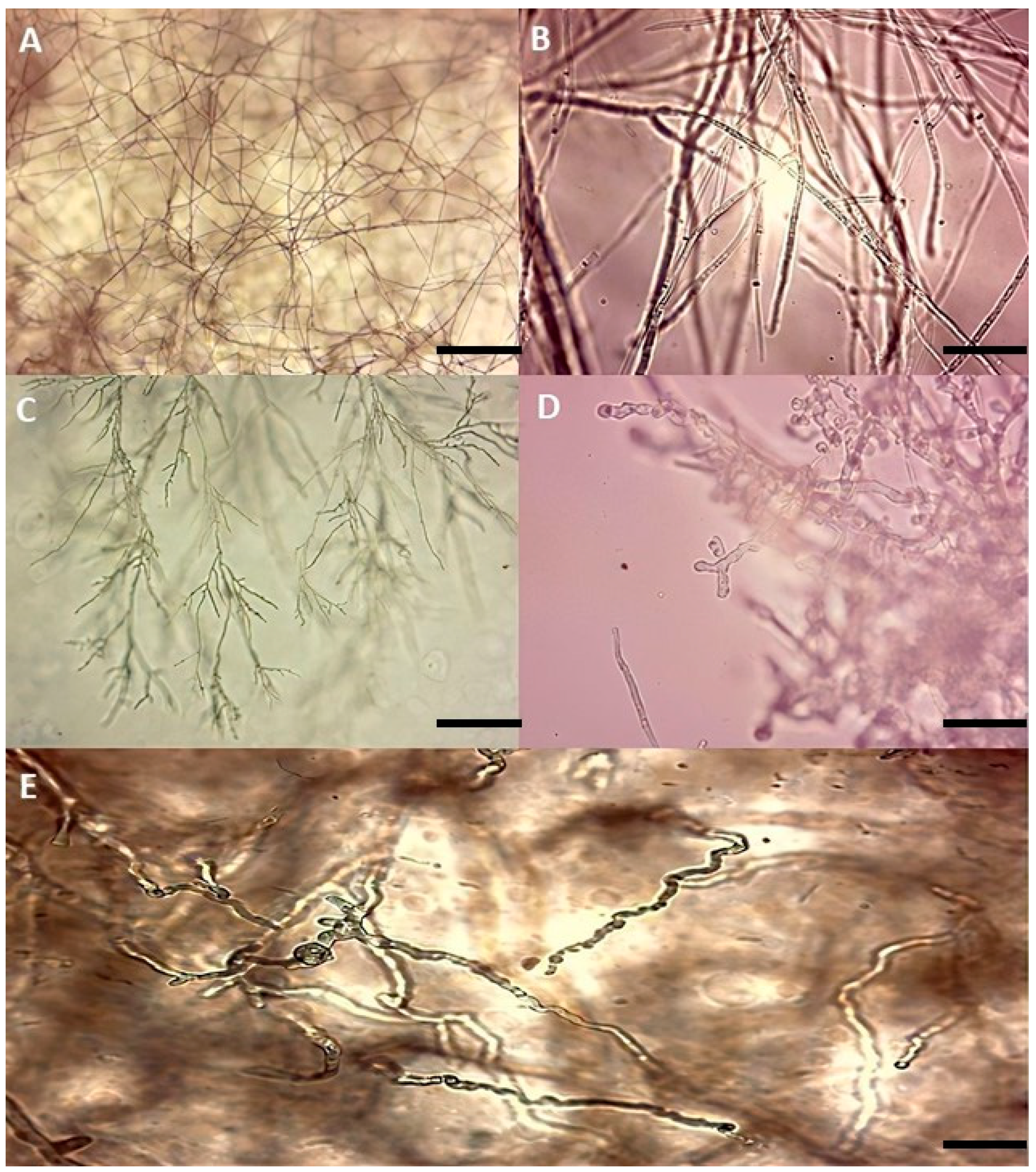
3.2. Microscopic Observation of Mycelium in the Presence of Antagonistic Isolates
3.3. Identification of Bacterial Isolates Using 16S rDNA Amplicon Sequencing
3.4. Volatile Organic Compound Effects
3.5. Antibiosis via Bacterial Supernatant
3.6. Morphological Characterization of Antagonistic Isolates
3.7. Biochemical Characterization
3.7.1. Lytic Enzymes and Plant-Growth Promoting Production
3.7.2. Detection of Antifungal Lipopeptide Genes by PCR
3.8. PGPR Effect of Antagonistic Bacteria In Vivo
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moussadek, R. Impacts de l’agriculture de Conservation Sur Les Propriétés et La Productivité Des Vertisols Du Maroc Central. Afr. Focus 2012, 25, 1–16. [Google Scholar] [CrossRef]
- El Jaouhari, N.; Abouabdillah, A.; Bouabid, R.; Bourioug, M.; Chaoui, M. Assessment of Sustainable de Fi Cit Irrigation in a Moroccan Apple Orchard as a Climate Change Adaptation Strategy. Sci. Total Environ. 2018, 642, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Nabyl, B.; Hanane, L.; Houda, E.K.; Zakaria, A.; Otman, H.; Rahali, K.; Aouane, E.M. Evaluation of the Physico-Chemical Properties of Soil and Apple Leaves (Malus Domestica) in Beni Mellal-Khenifra Region, Morocco. Adv. Sci. Technol. Eng. Syst. 2020, 5, 1103–1108. [Google Scholar] [CrossRef]
- Moinina, A.; Lahlali, R.; Boulif, M. Important Pests, Diseases and Weather Conditions Affecting Apple Production in Morocco: Current State and Perspectives. Rev. Marocaine Sci. Agron. Vétérinaires 2019, 7, 71–87. [Google Scholar]
- El Yaacoubi, A.; El Jaouhari, N.; Bourioug, M.; El Youssfi, L.; Cherroud, S.; Bouabid, R.; Chaoui, M.; Abouabdillah, A. Potential Vulnerability of Moroccan Apple Orchard to Climate Change–Induced Phenological Perturbations: Effects on Yields and Fruit Quality. Int. J. Biometeorol. 2020, 64, 377–387. [Google Scholar] [CrossRef]
- Prencipe, S.; Savian, F.; Nari, L.; Ermacora, P.; Spadaro, D.; Martini, M. First Report of Phytopythium vexans Causing Decline Syndrome of Actinidia Deliciosa ‘Hayward’in Italy. Plant Dis. 2020, 104, 2032. [Google Scholar] [CrossRef]
- Thao, L.; Hien, L.; Liem, N.; Thanh, H.; Khanh, T.; Binh, V.; Trang, T.; Anh, P.; Tu, T. First Report of Phytopythium vexans Causing Root Rot Disease on Durian in Vietnam. New Dis. Rep. 2020, 41, 588–2044. [Google Scholar] [CrossRef]
- Boari, A.J.; Cunha, E.M.; Quadros, A.F.F.; Barreto, R.W.; Fernandes, A.F. First Report of Phytopythium Sp. Causing Storage Root Rot and Foliage Blight of Cassava in Brazil. Plant Dis. 2018, 102, 1042. [Google Scholar] [CrossRef]
- Jabiri, S.; Bahra, C.; Maclean, D.; Radouane, N.; Barka, E.A. Phytopythium vexans Associated with Apple and Pear Decline in the Saïss Plain of Morocco. Microorganisms 2021, 9, 1916. [Google Scholar] [CrossRef]
- Jabiri, S.; Lahlali, R.; Bahra, C.; Bendriss Amraoui, M.; Tahiri, A.; Amiri, S. First Report of Phytopythium vexans Associated with Dieback Disease of Apple Trees in Morocco. J. Plant Pathol. 2020, 102, 1319. [Google Scholar] [CrossRef]
- Zhou, C.; Pan, X.; Kong, B.; Cun, H.; Li, N.; He, Y.; Ma, J.; Zhang, Y.; Ma, Y.; Cao, K. First Report of Apple Root Rot Caused by Phytopythium vexans in China. Plant Dis. 2022, 106, 3002. [Google Scholar] [CrossRef] [PubMed]
- Williamson-Benavides, B.A.; Dhingra, A. Understanding Root Rot Disease in Agricultural Crops. Horticulturae 2021, 7, 33. [Google Scholar] [CrossRef]
- Legrifi, I.; Al Figuigui, J.; Radouane, N.; Ezrari, S.; Belabess, Z.; Tahiri, A.; Amiri, S.; Lahlali, R. First Report of Pythium Schmitthenneri on Olive Trees and in Morocco. Australas. Plant Dis. Notes 2022, 17, 3. [Google Scholar] [CrossRef]
- Panth, M.; Baysal-Gurel, F.; Avin, F.A.; Simmons, T. Identification and Chemical and Biological Management of Phytopythium vexans, the Causal Agent of Phytopythium Root and Crown Rot of Woody Ornamentals. Plant Dis. 2021, 105, 1091–1100. [Google Scholar] [CrossRef]
- Bellini, A.; Ferrocino, I.; Cucu, M.A.; Pugliese, M.; Garibaldi, A.; Gullino, M.L. A Compost Treatment Acts as a Suppressive Agent in Phytophthora Capsici–Cucurbita Pepo Pathosystem by Modifying the Rhizosphere Microbiota. Front. Plant Sci. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Sun, Z.B.; Yuan, X.F.; Zhang, H.; Wu, L.F.; Liang, C.; Feng, Y.J. Isolation, Screening and Identification of Antagonistic Downy Mildew Endophytic Bacteria from Cucumber. Eur. J. Plant Pathol. 2013, 137, 847–857. [Google Scholar] [CrossRef]
- Scott, K.; Eyre, M.; McDuffee, D.; Dorrance, A.E. The Efficacy of Ethaboxam as a Soybean Seed Treatment toward Phytophthora, Phytopythium, and Pythium in Ohio. Plant Dis. 2020, 104, 1421–1432. [Google Scholar] [CrossRef]
- Marin, M.V.; Seijo, T.E.; Zuchelli, E.; Peres, N.A. Detection and Characterization of Quinone Outside Inhibitor-Resistant Phytophthora Cactorum and p. Nicotianae Causing Leather Rot in Florida Strawberry. Plant Dis. 2022, 106, 1203–1208. [Google Scholar] [CrossRef]
- Carbú, M.; González-Rodríguez, V.E.; Garrido, C.; Husaini, A.M.; Cantoral, J.M. New Biocontrol Strategies for Strawberry Fungal Pathogens. In Strawberry: Growth, Development and Diseases; CABI: Wallingford, UK, 2016; pp. 196–211. [Google Scholar]
- Maridueña-Zavala, M.G.; Freire-Peñaherrera, A.; Cevallos-Cevallos, J.M.; Peralta, E.L. GC-MS Metabolite Profiling of Phytophthora Infestans Resistant to Metalaxyl. Eur. J. Plant Pathol. 2017, 149, 563–574. [Google Scholar] [CrossRef]
- Matić, S.; Gilardi, G.; Gisi, U.; Gullino, M.L.; Garibaldi, A. Differentiation of Pythium Spp. from Vegetable Crops with Molecular Markers and Sensitivity to Azoxystrobin and Mefenoxam. Pest Manag. Sci. 2019, 75, 356–365. [Google Scholar] [CrossRef]
- Martins, P.M.M.; Merfa, M.V.; Takita, M.A.; De Souza, A.A. Persistence in Phytopathogenic Bacteria: Do We Know Enough? Front. Microbiol. 2018, 9, 1099. [Google Scholar] [CrossRef] [PubMed]
- Pacios-Michelena, S.; Aguilar Gonzalez, C.N.; Alvarez-Perez, O.B.; Rodriguez-Herrera, R.; Chávez-González, M.; Arredondo Valdes, R.; Ascacio Valdes, J.A.; Govea Salas, M.; Ilyina, A. Application of Streptomyces Antimicrobial Compounds for the Control of Phytopathogens. Front. Sustain. Food Syst. 2021, 5, 696518. [Google Scholar] [CrossRef]
- Verma, M.; Mishra, J.; Arora, N.K. Plant Growth-Promoting Rhizobacteria: Diversity and Applications. In Environmental Biotechnology: For Sustainable Future; Springer: Berlin, Germany, 2019; ISBN 9789811072840. [Google Scholar]
- Kazan, K.; Gardiner, D.M. Targeting Pathogen Sterols: Defence and Counterdefence? PLoS Pathog. 2017, 13, e1006297. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, S.C.; Nakasone, A.K.; do Nascimento, S.M.C.; de Oliveira, D.A.; Siqueira, A.S.; Cunha, E.F.M.; de Castro, G.L.S.; de Souza, C.R.B. Isolation and Characterization of Cassava Root Endophytic Bacteria with the Ability to Promote Plant Growth and Control the in Vitro and in Vivo Growth of Phytopythium Sp. Physiol. Mol. Plant Pathol. 2021, 116. [Google Scholar] [CrossRef]
- Cheffi, M.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A. Olea europaea L. Root Endophyte Bacillus velezensis OEE1 Counteracts Oomycete and Fungal Harmful Pathogens and Harbours a Large Repertoire of Secreted and Volatile Metabolites and Beneficial Functional Genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef]
- Panth, M.; Witcher, A.; Baysal-Gurel, F. Response of Cover Crops to Phytopythium vexans, Phytophthora nicotianae, and Rhizoctonia solani, Major Soilborne Pathogens of Woody Ornamentals. Agriculture 2021, 11, 742. [Google Scholar] [CrossRef]
- Savian, F.; Ginaldi, F.; Musetti, R.; Sandrin, N.; Tarquini, G.; Pagliari, L.; Firrao, G.; Martini, M.; Ermacora, P. Studies on the Aetiology of Kiwifruit Decline: Interaction between Soil-Borne Pathogens and Waterlogging. Plant Soil 2020, 456, 113–128. [Google Scholar] [CrossRef]
- Benfradj, N.; Tounsi, S.; Hamdi, N.B. In-Vitro Evaluation of Antagonists and Fungicides in Controlling Citrus Gummosis Caused by Phytophthora, Phytopythium and Pythium Species in Tunisia. Br. Microbiol. Res. J. 2016, 16, 1–14. [Google Scholar] [CrossRef]
- Legrifi, I.; Al Figuigui, J.; El Hamss, H.; Lazraq, A.; Belabess, Z.; Tahiri, A.; Amiri, S.; Barka, E.A.; Lahlali, R. Potential for Biological Control of Pythium Schmitthenneri Root Rot Disease of Olive Trees (Olea europaea L.) by Antagonistic Bacteria. Microorganisms 2022, 10, 1635. [Google Scholar] [CrossRef]
- Wang, S.; Ruan, C.; Yi, L.; Deng, L.; Yao, S.; Zeng, K. Biocontrol Ability and Action Mechanism of Metschnikowia Citriensis against Geotrichum Citri-Aurantii Causing Sour Rot of Postharvest Citrus Fruit. Food Microbiol. 2020, 87, 103375. [Google Scholar] [CrossRef]
- Tariq, M.; Noman, M.; Ahmed, T.; Hameed, A.; Manzoor, N.; Zafar, M. Antagonistic Features Displayed by Plant Growth Promoting Rhizobacteria (PGPR): A Review. J. Plant Sci. Phytopathol. 2017, 1, 38–43. [Google Scholar]
- El-sayed, W.S.; Akhkha, A.; El-naggar, M.Y.; Elbadry, M. In Vitro Antagonistic Activity, Plant Growth Promoting Traits and Phylogenetic Affiliation of Rhizobacteria Associated with Wild Plants Grown in Arid Soil. Front. Microbiol. 2014, 5, 651. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Bajii, M.; Jijakli, M.H. Isolation and evaluation of bacteria and fungi as biological control agents. Commun. Agric. Appl. Biol. Sci. 2007, 72, 973–982. [Google Scholar] [PubMed]
- Farhaoui, A.; Adadi, A.; Tahiri, A.; El Alami, N.; Khayi, S.; Mentag, R.; Ezrari, S.; Radouane, N.; Mokrini, F.; Belabess, Z.; et al. Biocontrol Potential of Plant Growth-Promoting Rhizobacteria (PGPR) against Sclerotiorum Rolfsii Diseases on Sugar Beet (Beta vulgaris L.). Physiol. Mol. Plant Pathol. 2022, 119, 101829. [Google Scholar] [CrossRef]
- Llop, P.; Bonaterra, A.; Peñalver, J.; María, M.; Llop, P.; Bonaterra, A.; Pen, J. Development of a Highly Sensitive Nested-PCR Procedure Using a Single Closed Tube for Detection of Erwinia Amylovora in Asymptomatic Plant Material Development of a Highly Sensitive Nested-PCR Procedure Using a Single Closed Tube for Detection of Erwinia. Appl. Environ. Microbiol. 2000, 66, 2071–2078. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Herrera, S.D.; Grossi, C.; Zawoznik, M.; Groppa, M.D. Wheat Seeds Harbour Bacterial Endophytes with Potential as Plant Growth Promoters and Biocontrol Agents of Fusarium Graminearum. Microbiol. Res. 2016, 186, 37–43. [Google Scholar] [CrossRef]
- Guevara-Avendan, E.; Carrillo, J.D.; Moreno, K.; Méndez-Bravo, A.; Guerrero-Analco, J.A.; Reverchon, F. Antifungal Activity of Avocado Rhizobacteria against Fusarium euwallaceae and Graphium Spp., Associated with Euwallacea Spp. Nr. fornicatus, and Phytophthora cinnamomi. Antonie Van Leeuwenhoek 2018, 111, 563–572. [Google Scholar] [CrossRef]
- Li, Z.; Guo, B.; Wan, K.; Cong, M.; Huang, H.; Ge, Y. Effects of Bacteria-Free Filtrate from Bacillus megaterium Strain L2 on the Mycelium Growth and Spore Germination of Alternaria alternata. Biotechnol. Biotechnol. Equip. 2015, 29, 1062–1068. [Google Scholar] [CrossRef]
- Syed-Ab-Rahman, S.F.; Carvalhais, L.C.; Chua, E.; Xiao, Y.; Wass, T.J.; Schenk, P.M. Identification of Soil Bacterial Isolates Suppressing Different Phytophthora Spp. And Promoting Plant Growth. Front. Plant Sci. 2018, 9, 1502. [Google Scholar] [CrossRef]
- Dinesh, R.; Anandaraj, M.; Kumar, A.; Bini, Y.K.; Subila, K.P.; Aravind, R. Isolation, Characterization, and Evaluation of Multi-Trait Plant Growth Promoting Rhizobacteria for Their Growth Promoting and Disease Suppressing Effects on Ginger; Elsevier GmbH: Amsterdam, The Netherlands, 2015; Volume 173, ISBN 9104952731. [Google Scholar]
- Etesami, H.; Alikhani, H.A.; Mirseyed Hosseini, H. Evaluation of Halotolerant Endophytic Bacteria Isolated from the Halophyte Suaeda for Biological Control of Fungal Rice Pathogens. Arch. Phytopathol. Plant Prot. 2019, 52, 560–581. [Google Scholar] [CrossRef]
- Slama, H.B.; Cherif-silini, H.; Bouket, A.C.; Qader, M. Screening for Fusarium Antagonistic Bacteria from Contrasting Niches Designated the Endophyte Bacillus Halotolerans as Plant Warden Against Fusarium. Front. Microbiol. 2019, 9, 3236. [Google Scholar] [CrossRef] [PubMed]
- Yuttavanichakul, W.; Lawongsa, P.; Wongkaew, S.; Teaumroong, N.; Boonkerd, N.; Nomura, N.; Tittabutr, P. Improvement of Peanut Rhizobial Inoculant by Incorporation of Plant Growth Promoting Rhizobacteria (PGPR) as Biocontrol against the Seed Borne Fungus, Aspergillus niger. Biol. Control 2012, 63, 87–97. [Google Scholar] [CrossRef]
- Devi, A.R.; Sharma, G.D.; Majumdar, P.B.; Pandey, P. A Multispecies Consortium of Bacteria Having Plant Growth Promotion and Antifungal Activities, for the Management of Fusarium Wilt Complex Disease in Potato (Solanum tuberosum L.). Biocatal. Agric. Biotechnol. 2018, 16, 614–624. [Google Scholar] [CrossRef]
- Bahroun, A.; Jousset, A.; Mhamdi, R.; Mrabet, M.; Mhadhbi, H. Anti-Fungal Activity of Bacterial Endophytes Associated with Legumes against Fusarium Solani: Assessment of Fungi Soil Suppressiveness and Plant Protection Induction. Appl. Soil Ecol. 2018, 124, 131–140. [Google Scholar] [CrossRef]
- Lahlali, R.; Mchachti, O.; Radouane, N.; Ezrari, S.; Belabess, Z.; Khayi, S.; Mentag, R.; Tahiri, A.; Barka, E.A. The Potential of Novel Bacterial Isolates from Natural Soil for the Control of Brown Rot Disease (Monilinia fructigena) on Apple Fruits. Agronomy 2020, 10, 1814. [Google Scholar] [CrossRef]
- Lahlali, R.; Aksissou, W.; Lyousfi, N.; Ezrari, S.; Blenzar, A.; Tahiri, A.; Ennahli, S.; Hrustić, J.; MacLean, D.; Amiri, S. Biocontrol Activity and Putative Mechanism of Bacillus amyloliquefaciens (SF14 and SP10), Alcaligenes faecalis ACBC1, and Pantoea agglomerans ACBP1 against Brown Rot Disease of Fruit. Microb. Pathog. 2020, 139, 103914. [Google Scholar] [CrossRef]
- Syed Ab Rahman, S.F.; Singh, E.; Pieterse, C.M.J.; Schenk, P.M. Emerging Microbial Biocontrol Strategies for Plant Pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Solanki, M.K.; Singh, R.K.; Srivastava, S.; Kumar, S. Isolation and Characterization of Siderophore Producing Antagonistic Rhizobacteria against Rhizoctonia Solani. J. Basic Microbiol. 2013, 54, 585–597. [Google Scholar] [CrossRef]
- Al-daghari, D.S.S.; Al-abri, S.A.; Al-mahmooli, I.H. Efficacy of Native Antagonistic Rhizobacteria in the Biological Control of Pythium Aphanidermatum -Induced Damping-off of Cucumber in Oman. J. Plant Pathol. 2019, 102, 305–310. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological Control of Plant Pathogens by Bacillus Species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Brígido, C.; Singh, S.; Menéndez, E.; Tavares, M.J.; Glick, B.R.; Do Rosário Félix, M.; Oliveira, S.; Carvalho, M. Diversity and Functionality of Culturable Endophytic Bacterial Communities in Chickpea Plants. Plants 2019, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Nair, D.N.; Padmavathy, S. Impact of Endophytic Microorganisms on Plants, Environment and Humans. Sci. World J. 2014, 2014, 250693. [Google Scholar] [CrossRef] [PubMed]
- Effmert, U.; Kalderás, J.; Warnke, R.; Piechulla, B. Volatile Mediated Interactions between Bacteria and Fungi in the Soil. J. Chem. Ecol. 2012, 38, 665–703. [Google Scholar] [CrossRef]
- Lyousfi, N.; Letrib, C.; Legrifi, I.; Blenzar, A.; El Khetabi, A.; El Hamss, H.; Belabess, Z.; Barka, E.A.; Lahlali, R. Combination of Sodium Bicarbonate (SBC) with Bacterial Antagonists for the Control of Brown Rot Disease of Fruit. J. Fungi 2022, 8, 636. [Google Scholar] [CrossRef]
- Aloo, B.N.; Makumba, B.A.; Mbega, E.R. The Potential of Bacilli Rhizobacteria for Sustainable Crop Production and Environmental Sustainability. Microbiol. Res. 2019, 219, 26–39. [Google Scholar] [CrossRef]
- Çakmakçi, R.; Dönmez, F.; Aydin, A.; Sahin, F. Greenhouse and Two Different Field Soil Conditions Growth Promotion of Plants by Plant Growth-Promoting Rhizobacteria under Greenhouse and Two Different Field Soil Conditions. Soil Biol. Biotechnol. 2006, 38, 1482–1487. [Google Scholar] [CrossRef]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.M.; Lee, I.J. Inoculation of Abscisic Acid-Producing Endophytic Bacteria Enhances Salinity Stress Tolerance in Oryza Sativa. Environ. Exp. Bot. 2017, 136, 68–77. [Google Scholar] [CrossRef]
- Liu, R.; Li, J.; Zhang, F.; Zheng, D.; Chang, Y.; Xu, L.; Huang, L. Biocontrol Activity of Bacillus velezensis D4 against Apple Valsa Canker. Biol. Control 2021, 163, 104760. [Google Scholar] [CrossRef]
- Huang, M.H.; Zhang, S.Q.; Di, G.L.; Xu, L.K.; Zhao, T.X.; Pan, H.Y.; Li, Y.G. Determination of a Bacillus velezensis Strain for Controlling Soybean Root Rot. Biocontrol Sci. Technol. 2017, 27, 696–701. [Google Scholar] [CrossRef]
- Kadiri, M.; Sevugapperumal, N.; Nallusamy, S.; Ragunathan, J.; Ganesan, M.V.; Alfarraj, S.; Ansari, M.J.; Sayyed, R.Z.; Lim, H.R.; Show, P.L. Pan-Genome Analysis and Molecular Docking Unveil the Biocontrol Potential of Bacillus velezensis VB7 against Phytophthora Infestans. Microbiol. Res. 2023, 268, 127277. [Google Scholar] [CrossRef] [PubMed]
- Kanjanamaneesathian, M.; Wiwattanapatapee, R.; Rotniam, W.; Wongpetkhiew, W. Spraying Hydroponic Lettuce Roots with a Suspension Concentrate Formulation of Bacillus velezensis to Suppress Root Rot Disease and Promote Plant Growth. N. Z. Plant Prot. 2014, 67, 213–219. [Google Scholar] [CrossRef][Green Version]
- Martínez-Raudales, I.; De La Cruz-Rodríguez, Y.; Alvarado-Gutiérrez, A.; Vega-Arreguín, J.; Fraire-Mayorga, A.; Alvarado-Rodríguez, M.; Balderas-Hernández, V.; Fraire-Velázquez, S. Genome Sequence of Bacillus velezensis 2A-2B Strain: A Rhizospheric Inhabitant of Sporobolus Airoides (Torr.) Torr., with Antifungal Activity against Root Rot Causing Phytopathogens. Stand. Genom. Sci. 2017, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Shen, D.; Xiong, Q.; Bao, B.; Zhang, W.; Dai, T.; Zhao, Y.; Borriss, R.; Fan, B. The Plant-Beneficial Rhizobacterium Bacillus velezensis FZB42 Controls the Soybean Pathogen Phytophthora Sojae Due to Bacilysin Production. Appl. Environ. Microbiol. 2021, 87, e01601-21. [Google Scholar] [CrossRef]
- Ye, M.; Tang, X.; Yang, R.; Zhang, H.; Li, F.; Tao, F.; Li, F.; Wang, Z. Characteristics and Application of a Novel Species of Bacillus: Bacillus velezensis. ACS Chem. Biol. 2018, 13, 500–505. [Google Scholar] [CrossRef]
- Kanjanamaneesathian, M.; Wiwattanapatapee, R.; Rotniam, W.; Pengnoo, A.; Wongpetkhiew, W.; Tanmala, V. Application of a Suspension Concentrate Formulation of Bacillus velezensis to Control Root Rot of Hydroponicallygrown Vegetables. N. Z. Plant Prot. 2013, 66, 229–234. [Google Scholar] [CrossRef]
- Petatán-Sagahón, I.; Anducho-Reyes, M.A.; Silva-Rojas, H.V.; Arana-Cuenca, A.; Tellez-Jurado, A.; Cárdenas-Álvarez, I.O.; Mercado-Flores, Y. Isolation of Bacteria with Antifungal Activity against the Phytopathogenic Fungi Stenocarpella maydis and Stenocarpella macrospora. Int. J. Mol. Sci. 2011, 12, 5522–5537. [Google Scholar] [CrossRef]
- Robinson, P.M.; McKee, N.D.; Thompson, L.A.A.; Harper, D.B.; Hamilton, J.T.G. Autoinhibition of Germination and Growth in Geotrichum Candidum. Mycol. Res. 1989, 93, 214–222. [Google Scholar] [CrossRef]
- Ramette, A.; Moënne-Loccoz, Y.; Défago, G. Prevalence of Fluorescent Pseudomonads Producing Antifungal Phloroglucinols and/or Hydrogen Cyanide in Soils Naturally Suppressive or Conducive to Tobacco Black Root Rot. FEMS Microbiol. Ecol. 2003, 44, 35–43. [Google Scholar] [CrossRef]
- Bardin, S.D.; Huang, H.C.; Moyer, J.R. Control of Pythium Damping-off of Sugar Beet by Seed Treatment with Crop Straw Powders and a Biocontrol Agent. Biol. Control 2004, 29, 453–460. [Google Scholar] [CrossRef]
- Meena, K.R.; Tandon, T.; Sharma, A.; Kanwar, S.S. Lipopeptide Antibiotic Production by Bacillus velezensis KLP2016. J. Appl. Pharm. Sci. 2018, 8, 91–98. [Google Scholar]
- Vahidinasab, M.; Adiek, I.; Hosseini, B.; Akintayo, S.O.; Abrishamchi, B.; Pfannstiel, J.; Henkel, M.; Lilge, L.; Voegele, R.T.; Hausmann, R. Characterization of Bacillus velezensis UTB96, Demonstrating Improved Lipopeptide Production Compared to the Strain B. velezensis FZB42. Microorganisms 2022, 10, 2225. [Google Scholar] [CrossRef]
- Théatre, A.; Cano-Prieto, C.; Bartolini, M.; Laurin, Y.; Deleu, M.; Niehren, J.; Fida, T.; Gerbinet, S.; Alanjary, M.; Medema, M.H. The Surfactin-like Lipopeptides from Bacillus Spp.: Natural Biodiversity and Synthetic Biology for a Broader Application Range. Front. Bioeng. Biotechnol. 2021, 9, 623701. [Google Scholar] [CrossRef]
- Fazle Rabbee, M.; Baek, K.-H. Antimicrobial Activities of Lipopeptides and Polyketides of Bacillus velezensis for Agricultural Applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef] [PubMed]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus Subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Wang, H.; Tan, Z.; Xuan, Z.; Dahar, G.Y.; Li, Q.X.; Miao, W.; Liu, W. Antifungal Mechanism of Bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum Gloeosporioides Penz. Pestic. Biochem. Physiol. 2020, 163, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Qin, Y.; Li, P. Complete Genome Sequence of Bacillus velezensis LPL-K103, an Antifungal Cyclic Lipopeptide Bacillomycin L Producer from the Surface of Lemon. 3 Biotech 2020, 10, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef]
- Nasraoui, B. Pathogenic Fungi and Pseudo-Fungi of Cultivated Plants: Biology, New Systematic, Pathological Interaction; Editions Universitaires Européennes, Schaltungsdienst Lange O.H.G.: Berlin, Germany, 2006. [Google Scholar]
- Wilson, D.B. Three Microbial Strategies for Plant Cell Wall Degradation. Ann. N. Y. Acad. Sci. 2008, 1125, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-H.; Shim, H.; Shin, J.-H.; Kim, K.S. Antagonistic Activities of Bacillus Spp. Strains Isolated from Tidal Flat Sediment towards Anthracnose Pathogens Colletotrichum Acutatum and C. Gloeosporioides in South Korea. Plant Pathol. J. 2015, 31, 165. [Google Scholar] [CrossRef]
- Sadeghi, A.; Koobaz, P.; Azimi, H.; Karimi, E.; Akbari, A.R. Plant Growth Promotion and Suppression of Phytophthora Drechsleri Damping-off in Cucumber by Cellulase-Producing Streptomyces. Biocontrol 2017, 62, 805–819. [Google Scholar] [CrossRef]
- Kikot, G.E.; Hours, R.A.; Alconada, T.M. Contribution of Cell Wall Degrading Enzymes to Pathogenesis of Fusarium Graminearum: A Review. J. Basic Microbiol. 2009, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Thallinger, B.; Prasetyo, E.N.; Nyanhongo, G.S.; Guebitz, G.M. Antimicrobial Enzymes: An Emerging Strategy to Fight Microbes and Microbial Biofilms. Biotechnol. J. 2013, 8, 97–109. [Google Scholar] [CrossRef]
- Moon, J.; Won, S.; Ei, C.; Maung, H.; Choi, J.; Choi, S.; Ajuna, H.B.; Ahn, Y.S. Bacillus velezensis CE 100 Inhibits Root Rot Diseases (Phytophthora Spp.) and Promotes Growth of Japanese Cypress (Chamaecyparis obtusa Endlicher) Seedlings. Microorganisms 2021, 9, 821. [Google Scholar] [CrossRef]
- Mota, M.S.; Gomes, C.B.; Souza, I.T.; Moura, A.B. Bacterial Selection for Biological Control of Plant Disease: Criterion Determination and Validation. Braz. J. Microbiol. 2017, 48, 62–70. [Google Scholar] [CrossRef]
- Neeraja, C.; Anil, K.; Purushotham, P.; Suma, K.; Sarma, P.; Moerschbacher, B.M.; Podile, A.R. Biotechnological Approaches to Develop Bacterial Chitinases as a Bioshield against Fungal Diseases of Plants. Crit. Rev. Biotechnol. 2010, 30, 231–241. [Google Scholar] [CrossRef]
- Gajera, H.P.; Vakharia, D.N. Production of Lytic Enzymes by Trichoderma Isolates during in Vitro Antagonism with Aspergillus niger, the Causal Agent of Collar Rot of Peanut. Braz. J. Microbiol. 2012, 43, 43–52. [Google Scholar] [CrossRef]
- MohaMMadi, P.; Tozlu, E.; KoTan, R.; Kotan, M. Potential of Some Bacteria for Biological Control of Postharvest Citrus Green Mould Caused by Penicillium Digitatum. Plant Prot. Sci. 2017, 53, 134–143. [Google Scholar] [CrossRef]
- Dutta, V.; Sharma, U.; Iqbal, K.; Kumar, R.; Pathak, A.K. Impact of River Channelization and Riverfront Development on Fluvial Habitat: Evidence from Gomti River, a Tributary of Ganges, India. Environ. Sustain. 2018, 1, 167–184. [Google Scholar] [CrossRef]
- Kumar, P.; Dubey, R.C.; Maheshwari, D.K. Bacillus Strains Isolated from Rhizosphere Showed Plant Growth Promoting and Antagonistic Activity against Phytopathogens. Microbiol. Res. 2012, 167, 493–499. [Google Scholar] [CrossRef]
- Breedt, G.; Labuschagne, N.; Coutinho, T.A. Seed Treatment with Selected Plant Growth-promoting Rhizobacteria Increases Maize Yield in the Field. Ann. Appl. Biol. 2017, 171, 229–236. [Google Scholar] [CrossRef]
- Reetha, S.; Bhuvaneswari, G.; Thamizhiniyan, P.; Mycin, T.R. Isolation of Indole Acetic Acid (IAA) Producing Rhizobacteria of Pseudomonas Fluorescens and Bacillus Subtilis and Enhance Growth of Onion (Allim cepa L.). Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 568–574. [Google Scholar]
- Gerbore, J.; Benhamou, N.; Vallance, J.; Le Floch, G.; Grizard, D.; Regnault-Roger, C.; Rey, P. Biological Control of Plant Pathogens: Advantages and Limitations Seen through the Case Study of Pythium Oligandrum. Environ. Sci. Pollut. Res. 2014, 21, 4847–4860. [Google Scholar] [CrossRef]
- Mitrofanova, O.; Mardanova, A.; Evtugyn, V.; Bogomolnaya, L.; Sharipova, M. Effects of Bacillus Serine Proteases on the Bacterial Biofilms. Biomed Res. Int. 2017, 2017, 8525912. [Google Scholar] [CrossRef]
- Dunne, C.; Crowley, J.J.; Mo, Y.; Dowling, D.N.; De Bruijn, F.J.; Gara, F.O. Biological Control of Pythium Ultimum by Stenotrophomonas Maltophilia W81 Is Mediated by an Extracellular Proteolytic Activity. Microbiology 1997, 143, 3921–3931. [Google Scholar] [CrossRef]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol Potentiality of Plant Growth Promoting Bacteria (PGPR)-Pseudomonas Fluorescens and Bacillus Subtilis: A Review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]


| Bacterial Isolate Code | Region | Species | Accession Numbers | VOCs | Cell-Free Filtrates |
|---|---|---|---|---|---|
| B1 | Meknes | Bacillus velezensis | ON738666 | 17.92 h | 31.37 g |
| B13 | Meknes | B. subtilis | ON746648 | 20.67 j | 8.48 a |
| B8-3 | Azrou | B. velezensis | ON746644 | 19.34 i | 18.63 c |
| CH II 4P | Chelihat | B. amyloliquefaciens | ON73668 | 13.79 g | 24.84 d |
| E4-3 | El Hajeb | Serratia odifera | ON740660 | 00.00 a | 11.35 b |
| E7-2 | El Hajeb | B. velezensis | ON73669 | 23.17 l | 52.84 o |
| I’4d1 | Imouzzer | B. velezensis | ON746649 | 4.59 b | 51.43 m |
| I2-5 | Imouzzar | Stenotrophomas matipholia | ON738715 | 9.21 e | 51.66 n |
| L8 | Imouzzar | B. velezensis | ON738718 | 5.48 d | 18.72 c |
| M1-3 | Meknes | B. velezensis | ON738671 | 00.00 a | 40.40 k |
| M2-3 | Meknes | B. amyloliquefaciens | ON738672 | 10.19 f | 31.03 f |
| M2-6 | Meknes | B. velezensis | ON746646 | 4.74 c | 30.51 e |
| M4-5 | Meknes | B. subtilis | ON746647 | 00.00 a | 36.43 h |
| M5-6 | Meknes | B. siamensis | ON746650 | 32.17 m | 39.21 j |
| M7-6 | Meknes | B. amyloliquefaciens | ON746645 | 21.36 k | 41.38 l |
| S2 | Sefrou | B. atrophaeus | ON738674 | 4.74 c | 37.23 i |
| Isolate | Colony Color | Shape | Surface | Relief | Opacity | Consistency | Microscopic Shape and Grouping | Gram |
|---|---|---|---|---|---|---|---|---|
| B1 | yellow | circular | smooth, shiny | convex | translucent | viscous | Cocci, diplococci, streptococci | + |
| B13 | white | Round with wavy edge | matte | concave in the center | translucent | viscous | Bacilli, diplobacilli, streptobacilli | - |
| B8-3 | yellow | circular | smooth, shiny | convex | translucent | viscous | Coccobacilli, in clusters, in chains | - |
| CH II 4P | white | circular | matte | convex | opaque | viscous | Sporulated bacilli | + |
| E4-3 | yellow | circular | smooth, shiny | convex | translucent | viscous | Coccobacilli | - |
| E7-2 | yellow | circular | smooth, shiny | convex | translucent | viscous | Cocci, diplococci | + |
| I’4d1 | white | round with irregular edge | matte | flat | opaque | viscous | Bacilli, streptobacilli | + |
| I2-5 | yellow | circular | smooth, shiny | convex | translucent | viscous | Sporulated bacilli, streptobacilli | + |
| L8 | brown | irregular with wavy edge | matte | flat | opaque | granular | Coccobacilli in clusters | - |
| M1-3 | whitish | circular | matte | flat | opaque | viscous | Bacilli, diplobacilli, streptobacilli | - |
| M2-3 | yellow | circular | smooth, shiny | convex | translucent | viscous | Cocci, diplococci, streptococci | + |
| M2-6 | white | circular | smooth, shiny | domed | opaque | viscous | Bacilli in chains, in clusters | - |
| M4-5 | yellow | circular | smooth, shiny | convex | translucent | viscous | Bacilli, diplobacilli | - |
| M5-6 | yellow | circular | smooth, shiny | convex | translucent | viscous | Isolated cocci, staphylococci | - |
| M7-6 | yellow | circular | smooth, shiny | convex | translucent | viscous | Coccobacilli in clusters | - |
| S2 | yellow | circular | smooth, shiny | convex | translucent | viscous | Isolated coccobacilli, in chains | - |
| Isolates | AI | CI | PI | PrI | HCN | PSI | IAA |
|---|---|---|---|---|---|---|---|
| B1 | 0.66 a | 0.061 abcd | 0.000 a | 0.514 e | − | − | − |
| B13 | 3.09 e | 0.075 bcd | 3.769 k | 0.160 a | − | − | − |
| B8-3 | 1.36 bc | 0.145 ef | 2.087 i | 1.409 h | − | − | + |
| CH II 4P | 2.83 de | 0.615 h | 0.000 a | 0.722 f | − | − | − |
| E4-3 | 6.20 i | 0.125 def | 0.753 d | 0.842 g | − | − | − |
| E7-2 | 4.34 fg | 1.277 i | 1.582 g | 0.251 b | − | − | − |
| I’4d1 | 5.27 h | 0.168 f | 0.952 e | 0.241 b | − | − | − |
| I2-5 | 3.79 f | 0.42 g | 0.546 c | 0.304 bc | − | − | + |
| L8 | 3.11 e | 0.019 ab | 0.417 f | 0.300 bc | − | − | − |
| M1-3 | 4.63 g | 0.014 ab | 0.000 a | 0.271 b | − | − | − |
| M2-3 | 3.23 e | 0.036 abc | 0.467 b | 0.368 cd | − | − | − |
| M2-6 | 2.27 d | 0.066 abcd | 0.000 a | 0.294 bc | − | − | − |
| M4-5 | 1.14 abc | 0.000 a | 2.401 j | 0.412 d | − | − | − |
| M5-6 | 1.24 bc | 0.000 a | 0.000 a | 0.583 e | − | − | − |
| M7-6 | 1.61 c | 0.000 a | 1.686 h | 0.367 cd | − | − | + |
| S2 | 0.85 ab | 0.092 cde | 0.000 a | 0.274 b | − | − | + |
| Isolates | Bacillomycin | Fengycin | Iturin | Surfactin |
|---|---|---|---|---|
| B1 | + | − | + | − |
| B13 | − | − | + | − |
| B8-3 | + | − | − | − |
| CH II 4P | + | − | + | − |
| E4-3 | + | − | + | − |
| E7-2 | + | − | + | − |
| I’4d1 | − | − | + | − |
| I2-5 | − | − | + | − |
| L8 | + | − | + | − |
| M1-3 | + | − | + | − |
| M2-3 | + | − | + | − |
| M2-6 | + | − | + | − |
| M4-5 | − | − | + | − |
| M5-6 | − | − | − | − |
| M7-6 | − | − | − | − |
| S2 | − | + | − | − |
| Isolates | Length of Aerial Part | Fresh Plant Weight | Fresh Root Weight |
|---|---|---|---|
| B1 | 32.01 bc | 0.537 abcd | 0.199 bcd |
| B13 | 33.60 def | 0.461 a | 0.172 abc |
| B8-3 | 33.50 de | 0.524 abc | 0.187 abc |
| CH II 4P | 30.73 ab | 0.511 abc | 0.168 ab |
| E4-3 | 33.23 cde | 0.710 ef | 0.317 g |
| E7-2 | 36.70 hi | 0.613 cde | 0.243 e |
| I’4d1 | 32.50 cd | 0.604 bcde | 0.242 e |
| I2-5 | 29.73 a | 0.45 a | 0.177 abc |
| L8 | 30.74 ab | 0.496 ab | 0.226 de |
| M1-3 | 38.97 j | 0.741 f | 0.231 de |
| M2-3 | 35.57 gh | 0.714 ef | 0.283 f |
| M2-6 | 37.63 ij | 0.651 ef | 0.203 cd |
| M4-5 | 38.01 ij | 0.646 def | 0.224 de |
| M5-6 | 34.25 efg | 0.610 cde | 0.238 e |
| M7-6 | 35.04 fg | 0.664 ef | 0.350 g |
| S2 | 34.20 efg | 0.486 a | 0.164 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jabiri, S.; Legrifi, I.; Benhammou, M.; Laasli, S.-E.; Mokrini, F.; Amraoui, M.B.; Lahlali, R. Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity. Appl. Microbiol. 2023, 3, 948-967. https://doi.org/10.3390/applmicrobiol3030065
Jabiri S, Legrifi I, Benhammou M, Laasli S-E, Mokrini F, Amraoui MB, Lahlali R. Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity. Applied Microbiology. 2023; 3(3):948-967. https://doi.org/10.3390/applmicrobiol3030065
Chicago/Turabian StyleJabiri, Salma, Ikram Legrifi, Majda Benhammou, Salah-Eddine Laasli, Fouad Mokrini, Mohammed Bendriss Amraoui, and Rachid Lahlali. 2023. "Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity" Applied Microbiology 3, no. 3: 948-967. https://doi.org/10.3390/applmicrobiol3030065
APA StyleJabiri, S., Legrifi, I., Benhammou, M., Laasli, S.-E., Mokrini, F., Amraoui, M. B., & Lahlali, R. (2023). Screening of Rhizobacterial Isolates from Apple Rhizosphere for Their Biocontrol and Plant Growth Promotion Activity. Applied Microbiology, 3(3), 948-967. https://doi.org/10.3390/applmicrobiol3030065









