Skin Microbiota of Salmonids: A Procedure to Examine Active Bacterial Populations Using an RNA-Based Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
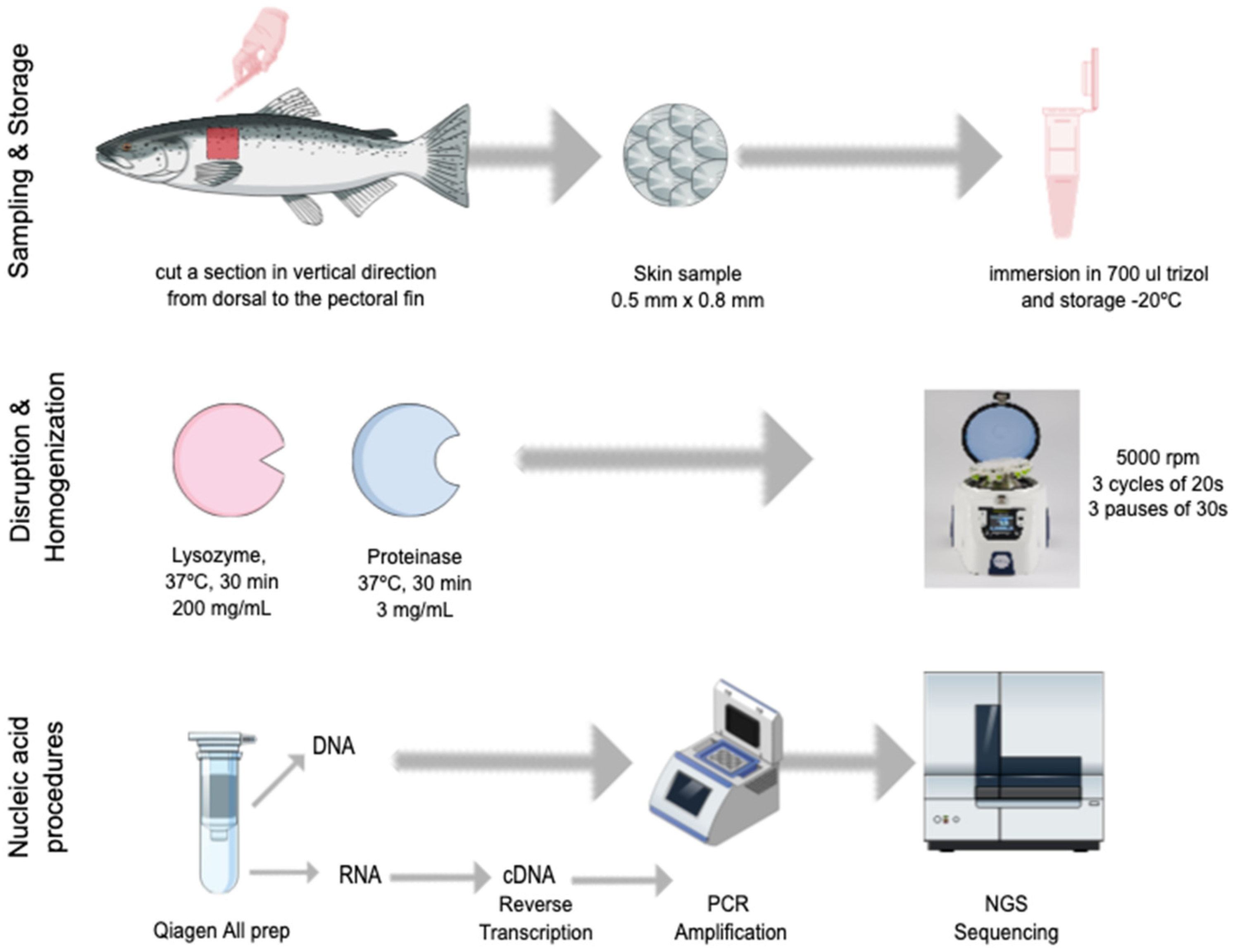
2.2. Skin Sampling
2.3. Nucleic Acid Extraction
2.4. Bioinformatic Analysis
3. Results
3.1. DNA and RNA Extractions and PCR Products
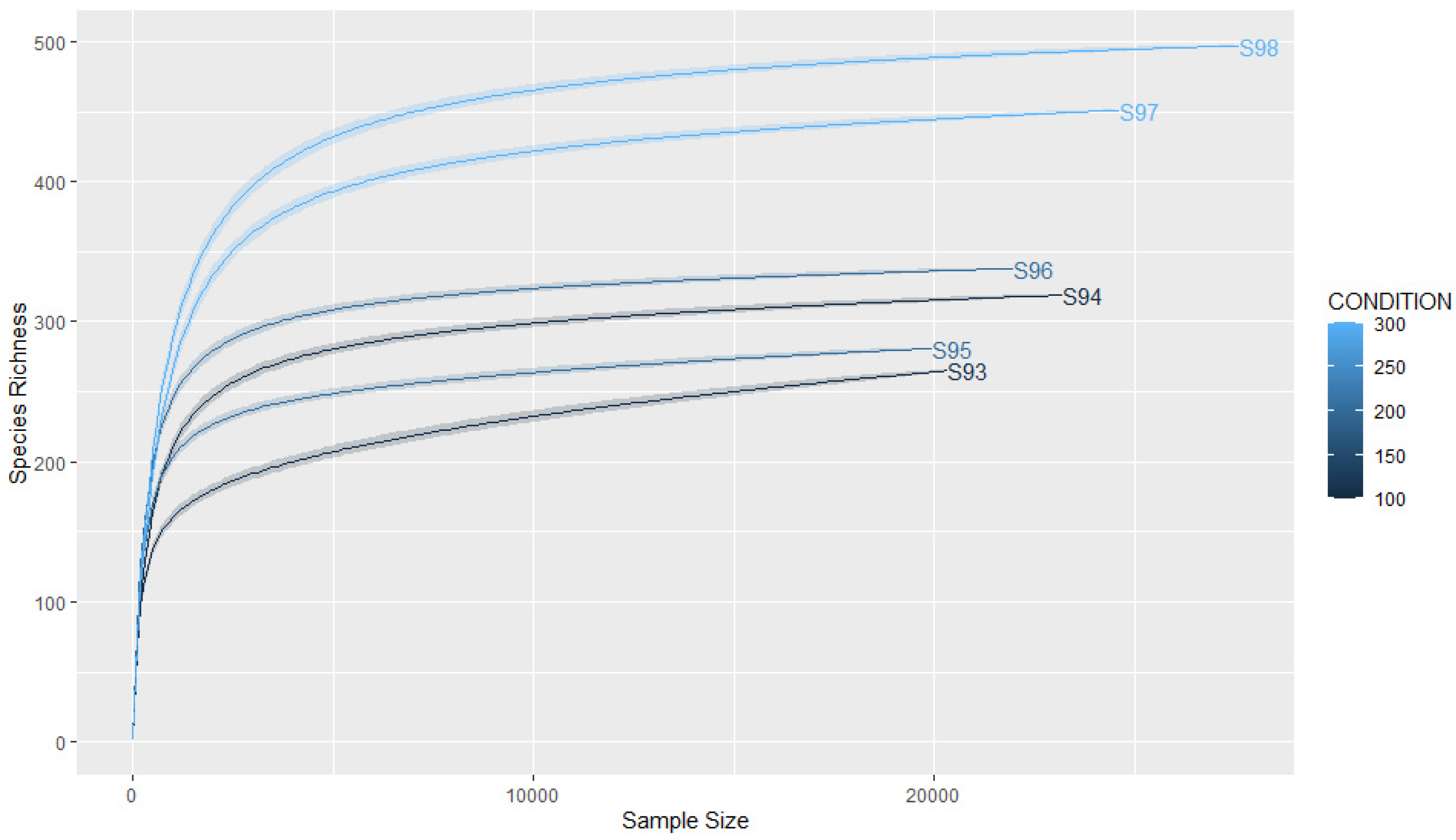
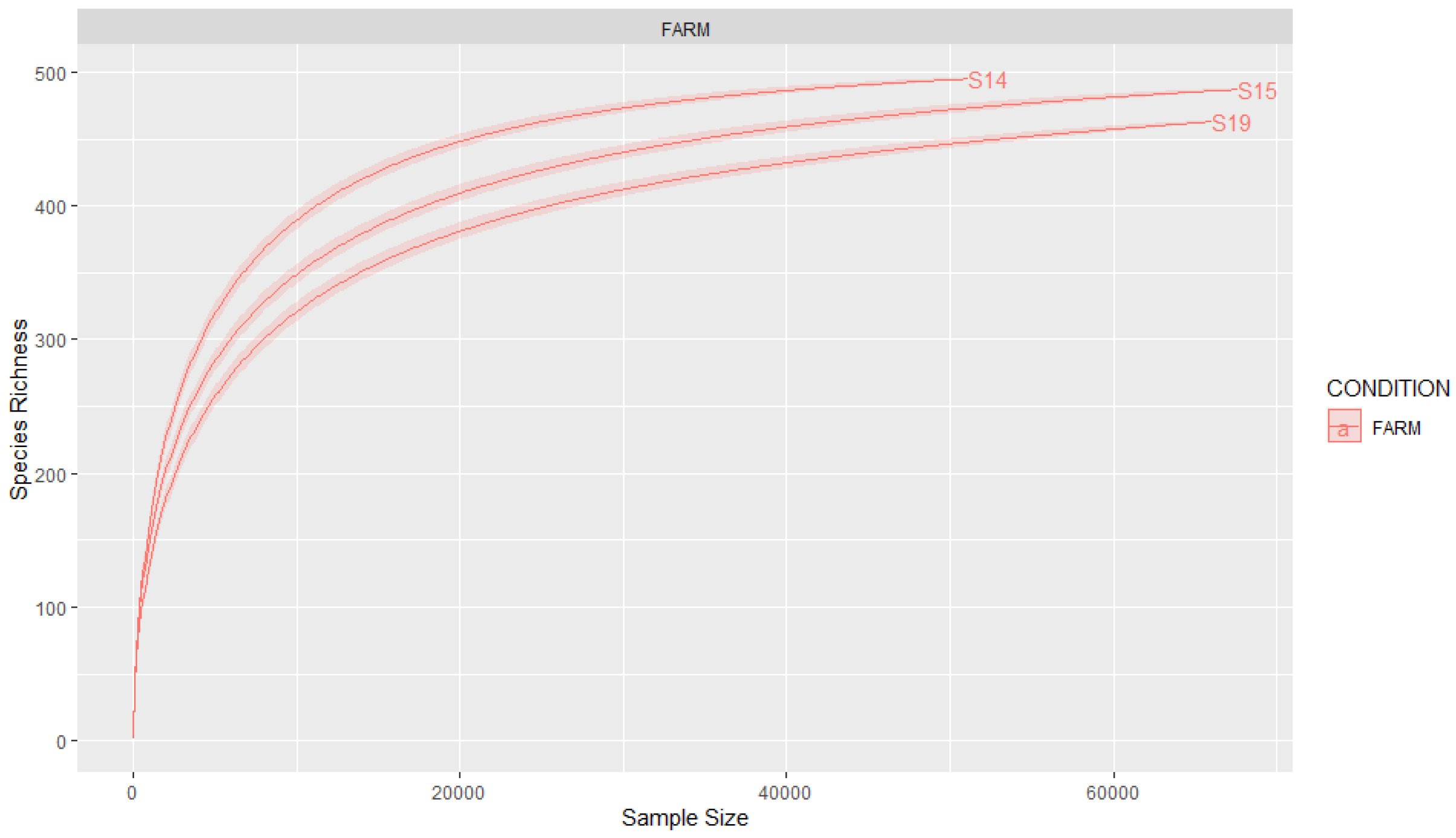
3.2. Coverage and Sample Size
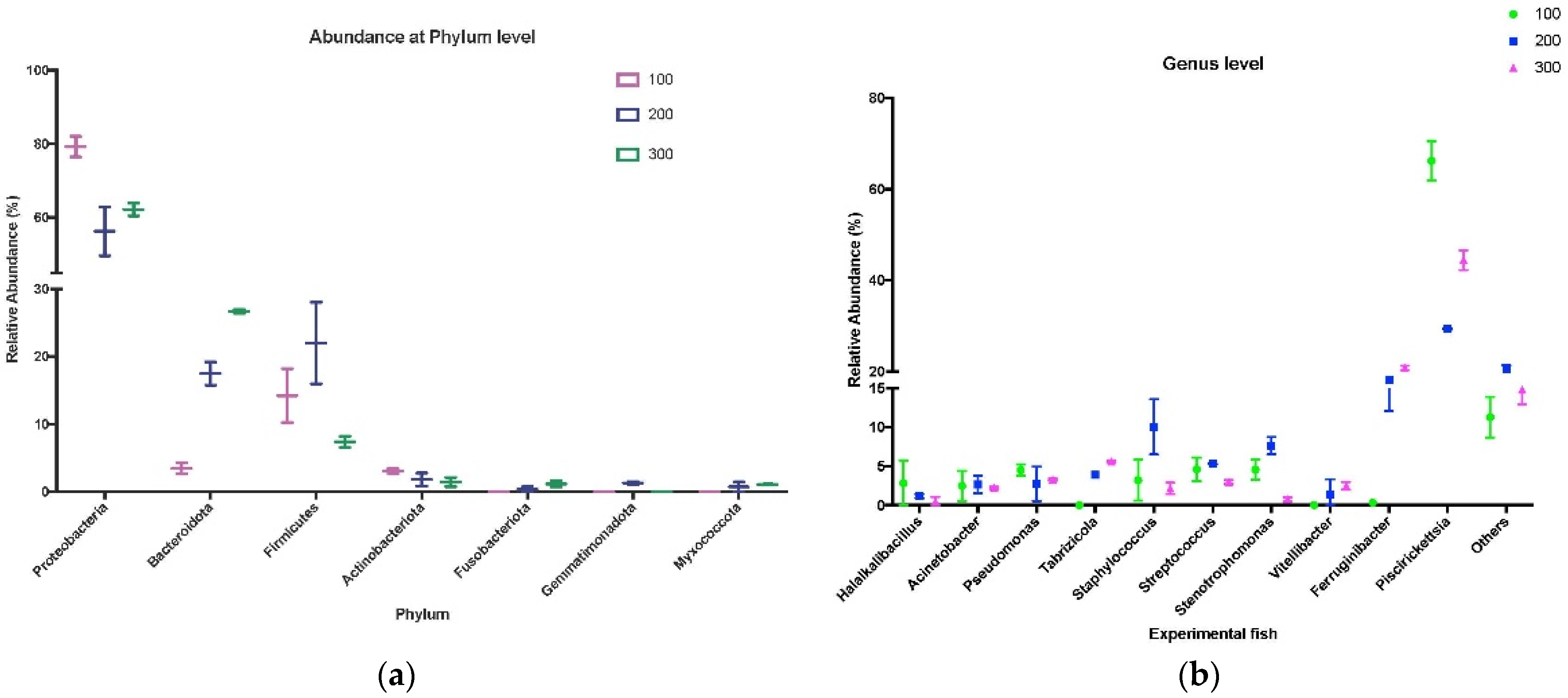
3.3. Taxonomic Analysis of Skin Microbiota from Experimental Trout
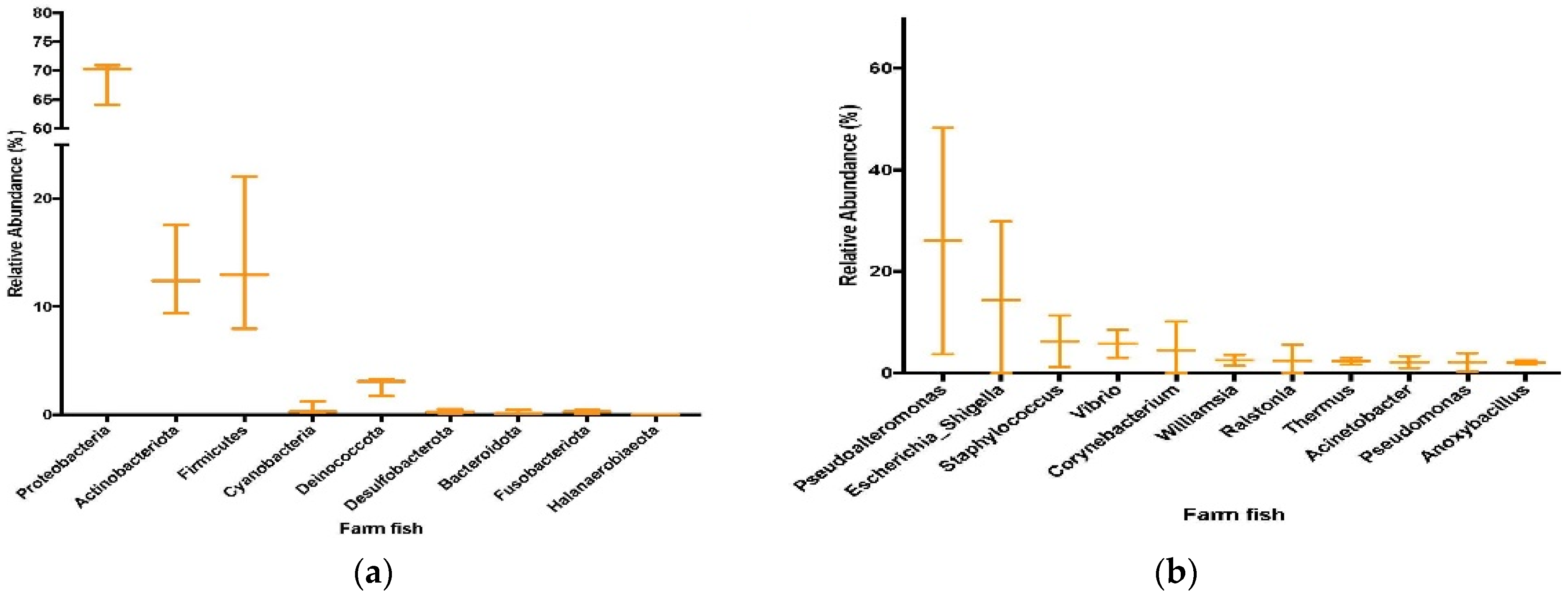
3.4. Sequencing and Taxonomic Results from Farmed Fish
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benhamed, S.; Guardiola, F.A.; Mars, M.; Esteban, M.Á. Pathogen bacteria adhesion to skin mucus of fishes. Vet. Microbiol. 2014, 171, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Terova, G.; Gini, E.; Gasco, L.; Moroni, F.; Antonini, M.; Rimoldi, S. Effects of full replacement of dietary fishmeal with insect meal from Tenebrio molitor on rainbow trout gut and skin microbiota. J. Anim. Sci. Biotechnol. 2021, 12, 30. [Google Scholar] [CrossRef]
- Easy, R.H.; Ross, N.W. Changes in Atlantic salmon (Salmo salar) epidermal mucus protein composition profiles following infection with sea lice (Lepeophtheirus salmonis). Comp. Biochem. Physiol. D Genom. Proteom. 2009, 4, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and ecological roles of external fish mucus: A review. Fishes 2018, 3, 41. [Google Scholar] [CrossRef]
- Gomez, J.A.; Primm, T.P. A slimy business: The future of fish skin microbiome studies. Microb. Ecol. 2021, 82, 275–287. [Google Scholar] [CrossRef]
- Larsen, A.; Tao, Z.; Bullard, S.A.; Arias, C.R. Diversity of the skin microbiota of fishes: Evidence for host species specificity. FEMS Microbiol. Ecol. 2013, 85, 483–494. [Google Scholar] [CrossRef] [PubMed]
- Berggren, H.; Tibblin, P.; Yıldırım, Y.; Broman, E.; Larsson, P.; Lundin, D.; Forsman, A. Fish skin microbiomes are highly variable among individuals and populations but not within individuals. Front. Microbiol. 2021, 12, 767770. [Google Scholar] [CrossRef]
- Ingerslev, H.C.; Von Gersdorff Jørgensen, L.; Strube, M.L.; Larsen, N.; Dalsgaard, I.; Boye, M.; Madsen, L. The development of the gut microbiota in rainbow trout (Oncorhynchus mykiss) is affected by first feeding and diet type. Aquaculture 2014, 424–425, 24–34. [Google Scholar] [CrossRef]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef]
- Karlsen, C.; Ottem, K.F.; Brevik, Ø.J.; Davey, M.; Sørum, H.; Winther-Larsen, H.C. The environmental and host-associated bacterial microbiota of Arctic seawater-farmed Atlantic salmon with ulcerative disorders. J. Fish Dis. 2017, 40, 1645–1663. [Google Scholar] [CrossRef]
- Larsen, A.M.; Bullard, S.A.; Womble, M.; Arias, C.R. Community structure of skin microbiome of gulf killifish, fundulus grandis, is driven by seasonality and not exposure to oiled sediments in a louisiana salt marsh. Microb. Ecol. 2015, 70, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Jurado, J.; Fuentes-Almagro, C.A.; Guardiola, F.A.; Cuesta, A.; Esteban, M.; Prieto-Álamo, M.J. Proteomic profile of the skin mucus of farmed gilthead seabream (Sparus aurata). J. Proteom. 2015, 120, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Lokesh, J.; Kiron, V. Transition from freshwater to seawater reshapes the skin-associated microbiota of Atlantic salmon. Sci. Rep. 2016, 6, 19707. [Google Scholar] [CrossRef] [PubMed]
- Austin, B. The bacterial microflora of fish, revised. Sci. World J. 2006, 6, 931–945. [Google Scholar] [CrossRef] [PubMed]
- Carbajal-González, M.T.; Fregeneda-Grandes, J.M.; Suárez-Ramos, S.; Cadenas, F.R.; Aller-Gancedo, J.M. Bacterial skin flora variation and in vitro inhibitory activity against Saprolegnia parasitica in brown and rainbow trout. Dis. Aquat. Organ 2011, 96, 125–135. [Google Scholar] [CrossRef]
- Diler, Ö.; Altun, S.; Çalikuşu, F.; Diler, A. A study on qualitative and quantitative bacterial flora of the rainbow trout (Oncorhynchus mykiss) living in different fish farms. Turk. J. Vet. Anim. Sci. 2000, 24, 251–259. [Google Scholar]
- Elekwachi, C.O.; Wang, Z.; Wu, X.; Rabee, A.; Forster, R.J. Total rRNA-seq analysis gives insight into bacterial, fungal, protozoal and archaeal communities in the rumen using an optimized RNA isolation method. Front. Microbiol. 2017, 8, 1814. [Google Scholar] [CrossRef]
- Griffiths, R.I.; Whiteley, A.S.; O’Donnell, A.G.; Bailey, M.J. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 2000, 66, 5488–5491. [Google Scholar] [CrossRef]
- Lowrey, L.; Woodhams, D.C.; Tacchi, L.; Salinas, I. Topographical mapping of the rainbow trout (Oncorhynchus mykiss) microbiome reveals a diverse bacterial community with antifungal properties in the skin. Appl. Environ. Microbiol. 2015, 81, 6915–6925. [Google Scholar] [CrossRef]
- Blazewicz, S.J.; Barnard, R.L.; Daly, R.A.; Firestone, M.K. Evaluating rRNA as an indicator of microbial activity in environmental communities: Limitations and uses. ISME J. 2013, 7, 2061–2068. [Google Scholar] [CrossRef]
- Caballero, S.; Galeano, A.M.; Lozano, J.D.; Vives, M. Description of the microbiota in epidermal mucus and skin of sharks (Ginglymostoma cirratum and Negaprion brevirostris) and one stingray (Hypanus americanus). PeerJ 2020, 8, e10240. [Google Scholar] [CrossRef]
- Legrand, T.; Catalano, S.R.; Wos-Oxley, M.L.; Stephens, F.; Landos, M.; Bansemer, M.S.; Stone, D.A.J.; Qin, J.G.; Oxley, A.P.A. The inner workings of the outer surface: Skin and gill microbiota as indicators of changing gut health in yellowtail kingfish. Front. Microbiol. 2017, 8, 2664. [Google Scholar] [CrossRef]
- Committee for the Update of the Guide for the Care and Use of Laboratory Animals. In Guide for the Care and Use of Laboratory Animals; National Academies Press: Washington, DC, USA, 2011.
- Villasante, A.; Ramírez, C.; Rodríguez, H.; Dantagnan, P.; Hernández, A.; Figueroa, E.; Romero, J. Dietary carbohydrate-to-protein ratio influences growth performance, hepatic health and dynamic of gut microbiota in atlantic salmon (Salmo salar). Anim. Nutr. 2022, 10, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using microbiomeanalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Jung, T.S.; Del Castillo, C.S.; Javaregowda, P.K.; Dalvi, R.S.; Nho, S.W.; Park, S.B.; Jang, H.B.; Cha, I.S.; Sung, H.W.; Hikima, J.; et al. Seasonal variation and comparative analysis of non-specific humoral immune substances in the skin mucus of olive flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 2012, 38, 295–301. [Google Scholar] [CrossRef]
- Esteban, M.Á. An overview of the immunological defenses in fish skin. ISRN Immunol. 2012, 2012, 853470. [Google Scholar] [CrossRef]
- Kramer, M.F.; Coen, D.M. Enzymatic amplification of DNA by PCR: Standard procedures and optimization. In Current Protocols in Molecular Biology; Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., Struh, K., Eds.; John Wiley & Sons, Ltd.: New York, NY, USA, 2001; Volume 56, pp. 230–257. [Google Scholar]
- Lorenz, T.C. Polymerase chain reaction: Basic protocol plus troubleshooting and optimization strategies. J. Vis. Exp. 2012, 63, e3998. [Google Scholar] [CrossRef]
- Gaggero, A.; Castro, H.; Sandino, A.M. First isolation of Piscirickettsia salmonis from coho salmon, Oncorhynchus kisutch (Walbaum), and rainbow trout, Oncorhynchus mykiss (Walbaum), during the freshwater stage of their life cycle. J. Fish Dis. 1995, 18, 277–280. [Google Scholar] [CrossRef]
- Miranda, C.D.; Godoy, F.A.; Lee, M.R. Current status of the use of antibiotics and the antimicrobial resistance in the chilean salmon farms. Front. Microbiol. 2018, 9, 1284. [Google Scholar] [CrossRef]
- Uren Webster, T.M.; Rodriguez-Barreto, D.; Castaldo, G.; Gough, P.; Consuegra, S.; Garcia de Leaniz, C. Environmental plasticity and colonisation history in the Atlantic salmon microbiome: A translocation experiment. Mol. Ecol. 2020, 29, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Sylvain, F.É.; Holland, A.; Bouslama, S.; Audet-Gilbert, É.; Lavoie, C.; Val, A.L.; Derome, N. Fish Skin and Gut Microbiomes Show Contrasting Signatures of Host Species and Habitat. Appl. Environ. Microbiol. 2020, 86, e00789-20. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pardo, A.; Villasante, A.; Romero, J. Skin Microbiota of Salmonids: A Procedure to Examine Active Bacterial Populations Using an RNA-Based Approach. Appl. Microbiol. 2023, 3, 485-492. https://doi.org/10.3390/applmicrobiol3020034
Pardo A, Villasante A, Romero J. Skin Microbiota of Salmonids: A Procedure to Examine Active Bacterial Populations Using an RNA-Based Approach. Applied Microbiology. 2023; 3(2):485-492. https://doi.org/10.3390/applmicrobiol3020034
Chicago/Turabian StylePardo, Alda, Alejandro Villasante, and Jaime Romero. 2023. "Skin Microbiota of Salmonids: A Procedure to Examine Active Bacterial Populations Using an RNA-Based Approach" Applied Microbiology 3, no. 2: 485-492. https://doi.org/10.3390/applmicrobiol3020034
APA StylePardo, A., Villasante, A., & Romero, J. (2023). Skin Microbiota of Salmonids: A Procedure to Examine Active Bacterial Populations Using an RNA-Based Approach. Applied Microbiology, 3(2), 485-492. https://doi.org/10.3390/applmicrobiol3020034





