Abstract
Lactobacillus plantarum is a catalase-negative species and distributes in human intestinal tracts. However, the cytoprotective effects of the catalase-activated L. plantarum strain have yet to be exploited against reactive oxygen species (ROS). Here, a catalase-activated L. plantarum CGMCC 6888 (CatA+) was obtained using exogenous added heme. The scavenging free radical abilities of this strain were obviously increased. Moreover, the activated catalase A in L. plantarum CGMCC 6888 endowed the intestinal epithelium NCM460 with lower ROS content after degrading H2O2. In addition, the transcription levels of Nrf2 and Nrf2-related antioxidant enzyme genes (HO-1, GCLC, NQO-1 and TXNRD1) and tight junction protein genes (ZO-1, OCLN, and JAM-1) were upregulated significantly when co-incubated with CGMCC 6888/CatA+. This work confirmed that the catalase A conferred L. plantarum with the strong protection effects in the intestinal epithelial cells against ROS.
1. Introduction
Oxidative stress, a deleterious process resulting from an imbalance between the formation of reactive oxygen species (ROS) and the endogenous antioxidant defense systems, is considered to play a critical role in the pathogenesis of a number of diseases such as cardiovascular disease, cancer, aging, and mucosal inflammation [1,2]. To deal with oxidative damage, most living organisms are equipped with different defense systems to eliminate damage from ROS [2,3]. Nuclear factor-erythroid 2-related factor 2 (Nrf2) is a primary intracellular regulator for the response to the state of cellular oxidative stress. It can bind to its suppressor keap1 in cytoplasm but is free from the latter during oxidative stress [2,3,4,5]. When the free Nrf2 binds to antioxidant response element (ARE) in nucleus, it immediately drives the transcription of the genes of downstream antioxidant enzymes, including heme oxygenase-1 (HO-1), NAD(P)H: quinone oxidoreductase 1 (NQO-1), thioredoxin reductase 1 (TXNRD1), glutamate-cystine ligase catalytic (GCLC), etc. [6,7]. At the same time, intestinal epithelial cells could scavenge cellular ROS using three major antioxidant enzymes: catalase (CAT), superoxide dismutase (SOD) and glutathione peroxidase (GSH-px). However, when these endogenous antioxidant systems are inadequate to resist the oxidative stress, antioxidant additives or small functional molecules originating from foods and other functional dietary supplements (such as glutathione (GSH), vitamin E (VE), ferulic acid, probiotics) which delay or prevent the oxidation of cellular substances were demonstrated to be functional in the protection of organisms from oxidative damage [8,9,10].
Lactic acid bacteria (LAB) are the most widely used industrial strains in food processing. They can convert soluble carbohydrates to lactic acid as main end products and contribute to flavor, texture, and the shelf life of fermented products [11,12]. Lactobacillus plantarum is a versatile lactic acid bacterium that distributes in a range of niches (human host, vegetable, meat and dairy products), and has the ability to survive during gastric transit and colonize the intestinal tract of both humans and other animals in the community [13,14,15,16].
Numbers of L. plantarum strains are well-known probiotics owing to their beneficial effects on hosts, including antioxidant effects [17,18], anti-infective effects [19,20], modulation of the gut microbiota [16,21], and enhancement of immunity [14,22]. Those properties make this species attractive for the development of new functional foods and several genetic and physiological studies [23,24,25]. It was demonstrated that L. plantarum executed aerobic and respiratory growth in the presence of heme and/or menaquinone in the medium, which contributed to new applications in food technology and probiotic development [26]. Several studies have proved that an aerobic culture of L. plantarum showed a higher scavenging rate for DPPH radical than an anaerobic culture [26]. Additionally, exogenous heme supplementation may promote the synthesis of a heme-dependent catalase and a bd-type cytochrome oxidase in several Lactobacillus strains [27,28]. Catalase protects cells against oxidative stress by degrading hydrogen peroxide (H2O2), while cytochrome bd oxidase contributes to energy supply (through extra ATP generation) and the depletion of intracellular oxygen [27,28]. The tolerance of oxidative stresses may be important for LAB survival in the gut. However, the beneficial effects of the catalase-activated L. plantarum strain have yet to be exploited in the protection against reactive oxygen species (ROS) in intestinal epithelial cells. In this study, we detected the catalase A activities of L. plantarum CGMCC 6888 in the presence of heme in anaerobic and aerobic conditions, and then evaluated the enhanced capacities of the bacterial cells or cell lysates to eliminate ROS. Furthermore, the cytoprotective effects of L. plantarum CGMCC 6888 with catalase activity on intestinal epithelial cells against oxidative stress were investigated. The aim of this study was to strengthen the antioxidant effects of L. plantarum conferred by catalase.
2. Materials and Methods
2.1. Strains, Cells and Culture Conditions
L. plantarum CGMCC 6888 was isolated from a traditional yogurt in Xinjiang, China. Briefly, 1 g of yogurt sample was added to 10 mL of sterile phosphate-buffered saline (PBS) and the mixture was serially diluted in PBS, followed by surface spreading on de Man, Rogosa and Sharpe (MRS) agar and cultivation at 37 °C for 48 h. Single colonies were picked from the MRS agar and incubated statically in MRS broth at 37 °C for 48 h. Then, cells were harvested from the above cultures and the genomic DNA was extracted to be used as a template for the PCR amplification of 16S rRNA gene with primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The 16S rRNA gene of CGMCC6888 showed 99% identities to those of L. plantarum by sequence alignment.
L. plantarum CGMCC6888 was routinely cultured in MRS broth, statically or with shaking, at 37 °C for 18 h, generating the CatA− cultures. For the activation of catalase, heme (Sigma) at a final concentration of 20 µM was added to the MRS broth, generating the CatA+ cultures.
The intestinal epithelial cell line NCM460 was grown in RPMI-1640 medium (SparkJade, Jinan, China) containing 10% (vol/vol) fetal bovine serum (Gibco, Shanghai, China) and 1% (vol/vol) penicillin-streptomycin solution (100 U/mL of penicillin and 100 µg/mL of streptomycin, SparkJade, Jinan, China) at 37 °C in a saturated humidity atmosphere containing 5% CO2.
2.2. Preparation of Bacterial Cells and Cell Lysates
Cells pellets from 20 mL of L. plantarum CGMCC 6888/CatA− or CatA+ cultures were collected after centrifugation at 6000 rpm for 5 min, washed three times using sterile saline solution, and resuspended with sterile saline solution to 108 CFU/mL. For cell lysate preparation, the cell suspension was subjected to ultrasonic disruption (450 W, 5-s stroke on ice with 5-s intervals), followed by centrifuging at 10,000 rpm for 10 min to remove the cell debris. The resulting cell lysate was filtered (sterile 0.22 μm pore size).
2.3. Analysis of Catalase Activity
To qualitatively analyze the catalase activities of the CatA− and CatA+ cultures, 30% H2O2 solution was dropped into the 100 µL of cell suspensions and the formation of bubbles was observed.
The catalase activities of the cultures and cell lysates were determined according to the instructions of the CAT kit (Nanjing Jiancheng Biological Engineering Research Institute, Nanjing, China). A unit of enzyme was defined as the enzyme amount that breaks down 1 µM of H2O2 at 37 °C per minute. The protein concentration of the cell lysates was assayed using a NanoDrop 2000 (Thermo Fisher Scientific, Shanghai, China).
2.4. Scavenging Free Radical Abilities of L. plantarum CGMCC 6888
The scavenging activities of the L. plantarum CGMCC 6888 on 1,1-diphenyl-2-picrylhydrazyl (DPPH) radicals were analyzed according to a previously described method with some modifications [29]. Briefly, 350 µL of the sample or saline buffer as a control and 350 µL of 0.2 mM DPPH (Macklin, Shanghai, China) dissolved in methanol were mixed and incubated in the dark at room temperature. After incubation for 30 min, the mixture was centrifuged at 6000 rpm for 5 min, and the supernatant was recovered to measure the reduction in absorbance at 517 nm. The DPPH radical scavenging activity was calculated as follows: (1 − Asample/Acontrol) × 100%.
The scavenging activity of the superoxide anion radical was analyzed as described previously by Tang et al. [29]. Briefly, 200 µL of the sample or buffer as a control was added to 600 µL of 50 mM Tris-HCl (pH 8.5) containing 10 mM EDTA and incubated at room temperature for 20 min. Then, 80 µL of 25 mM pyrogallol in 0.1 M HCl was added to the mixture and incubated accurately for 10 min, subsequently the reaction was terminated with the addition of 100 µL of HCl. The mixture was centrifuged at 6000 rpm for 5 min and the supernatant was recovered to measure the reduction in absorbance at 325 nm. The superoxide anion scavenging activity was calculated as follows: (1 − Asample/Acontrol) × 100%.
The scavenging activity of the hydroxyl radical was analyzed using the Fenton reaction [30]. A reaction system containing 200 µL of the sample or buffer as control, 200 µL of 5 mM salicylic acid ethanol solution, 200 µL of 5 mM FeSO4 and 200 µL of 3 mM H2O2 was incubated at room temperature for 30 min, followed by centrifugation at 6000 rpm for 5 min. The supernatant was recovered to measure the absorbance at 510 nm. The hydroxyl radical scavenging activity was calculated as follows: (1 − Asample/Acontrol) × 100%.
2.5. The Cytoprotective Effects of L. plantarum CGMCC 6888 against Oxidative Stress
2.5.1. Intestinal Epithelial Cell Viability Assay
The viability of the NCM460 cells was assayed after exposure to H2O2 using Cell Counting Kit 8 according to the instructions (CCK-8. US Everbright® Inc., Suzhou, China). A total of 20 mL of the intestinal epithelial cell NCM460 detached with 0.25% trypsin-EDTA solution (SparkJade, Jinan, China) was centrifuged at 1000 rpm for 5 min. The cells were collected and suspended in 20 mL of RPMI 1640 medium, and seeded in a 96-well plate at a concentration of 1 × 104 cells/well and incubated for 24 h at 37 °C in 5% CO2. After washing with phosphate-buffered saline (PBS, pH 7.2), the NCM460 cells were co-incubated with 100 µL of the bacterial cells/cell lysates prepared according to 2.2 method but washed with phosphate-buffered saline (PBS, pH 7.2) and then resuspended with RPMI-1640 medium of L. plantarum CGMCC 6888/CatA− or CGMCC 6888/CatA+ for 2 h. Then, the NCM460 cells were washed three times with PBS buffer and treated with 100 µL of 0.35 mM H2O2 for 30 min. Subsequently, the cells were washed three times with PBS buffer, and 100 µL of 10% CCK-8 was added and incubated for 3 h. The optical density (OD) at 450 nm was measured to evaluate the cell viability. The cells free of H2O2 treatment were used as normal group, and the cells treated with H2O2 alone were used as control group.
2.5.2. Intracellular ROS Detection in NCM460 Cells
NCM460 cells were seeded in 6-well plates at a concentration of 1×106 cells/well and incubated for 24 h. After washing the cells with PBS buffer, 2 mL of the bacterial cells/cell lysates according to 2.5.1 method of the L. plantarum CGMCC 6888/CatA− or CGMCC 6888/CatA+ was added to the NCM460 cells and co-incubated for 2 h. Then, the medium was removed, and the cells were washed three times with PBS buffer and treated with 2 mL of 0.35 mM H2O2 in RPMI-1640 medium for 30 min. After removing the H2O2, 2′,7′-dichlorofluorescin diacetate (DCFH-DA) (Beijing Solarbio Science & Technology, Beijing, China) at a final concentration of 10 µM was added and incubated with the NCM460 cells at 37 °C for 20 min. After washing the cells with PBS buffer, the intracellular ROS level in the NCM460 cells was observed using fluorescence microscopy.
2.5.3. Determination of CAT, GSH-px and SOD Activities in NCM460 Cells
The NCM460 cells co-incubated with the bacterial cells/cell lysates according to 2.5.1 method of L. plantarum CGMCC 6888 were exposed to H2O2 to induce oxidative stress as described in 2.5.2. Subsequently, 100 µL of NP-40 lysis buffer (Beyotime Biotechnology, Shanghai, China) with PMSF (Beyotime Biotechnology, Shanghai, China) at a final concentration of 1 mM was added to each well on ice, with repeated pipetting for complete cell lysis. The NCM460 cell lysates were recovered after centrifugation at 10,000 rpm for 5 min at 4 °C. The determination of intracellular CAT, GSH-px and SOD activities in the lysates was carried out according to the protocols of the corresponding kit’s instructions (Nanjing Jiancheng Biological Engineering Research Institute, Nanjing, China). A unit of CAT was defined as above. A unit of GSH-px was defined as the enzyme amount that catalyzes 1 nmol of GSH oxidation per minute. A unit of SOD was defined as the enzyme amount that inhibits the reaction rate of xanthine oxidase by 50% per minute. The protein concentration was assayed using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA).
2.5.4. Analysis of the Gene Transcription Levels of the Antioxidant Enzyme Genes and Tight Junction Protein Genes
Primers used in this study are listed in Table 1. NCM460 cells were washed three times with PBS buffer and collected after centrifugation at 1000 rpm for 5 min and dissociation with 0.25% trypsin-EDTA solution. Total RNA was obtained with a SPARKeasy Cell Kit (SparkJade, Jinan, China) and the cDNA was synthesized using the PrimeScript RT reagent kit (Takara, Dalian, China). The gene transcription levels of antioxidant enzymes including Nrf2, HO-1, GCLC, NQO-1 and TXNRD1, and tight junction proteins including ZO-1, OCLN, CLDN-1 and JAM-1, were analyzed using RT-qPCR, which was carried out with a 2×Universal SYBR Green Fast qPCR Mix (ABclonal, Wuhan, China). The following amplification conditions were used: 95 °C for 3 min, followed by 40 cycles at 95 °C for 5 s, 55 °C for 30 s and 72 °C for 30 s. Real-time quantification Ct values were collected, and the target gene expression was normalized to β-actin using the 2−∆∆Ct method [31,32].

Table 1.
Real–time qPCR primers used for quantification of human mRNAs.
2.5.5. Statistical Analysis
The experiments were repeated three times, and the results are expressed as mean ± standard deviation. Statistical significance between the normal/control and experimental groups was analyzed using GraphPad Prism 5, and p < 0.05 was considered statistically significant. p < 0.01 was considered highly statistically significant.
3. Results
3.1. Identification and Activity Analysis of Catalase A in L. plantarum CGMCC 6888
Normally, L. plantarum lacks catalase activity and respiration, and encountering toxic reactive oxygen species is a challenge. Here, a putative catalase gene (catA) was identified in the genome of L. plantarum CGMCC 6888 through comparative genomic analysis using Clustal W and ESPript 3.0. The catA was 1455 bp in length, encoding 484 amino acids residues. The deduced amino acid sequence showed 99.79%, 69.67% and 56.03% identities to the catalases of L. plantarum SK151 (NZ_CP030105), L. casei JCM 1134 (NZ_AP612544) and Enterococcus faecalis V583 (NZ_004668), respectively. A conserved domain search showed that CatA belongs to the KatE superfamily, which catalyzes the conversion of hydrogen peroxide to water and molecular oxygen. Tertiary structure prediction showed that the polypeptide chain of CatA forms three regions: the N-terminal helix, the β-barrel and the C-terminal helical, similar to that of catalase from E. faecalis V583 (PDB: 1Si8). Sequence alignment revealed the amino acids Arg51, His54, Arg91, Phe140, Arg333, Tyr337 and Arg344 [33] are important for heme and ligand interaction in CatA (Figure 1a). The results suggested that the CatA of L. plantarum CGMCC 6888 was a heme-dependent catalase. However, heme cannot be produced by LAB [34,35], which lead to the heme-dependent catalase in L. plantarum CGMCC 6888 operating silently in the absence of heme and no bubbles were observed after dropping H2O2 (CatA−). When heme was supplemented to the medium, the CatA was activated to degrade H2O2 into H2O and O2, with the observation of the bubble formation showing a catalase positive phenotype (CatA+) (Figure 1b).
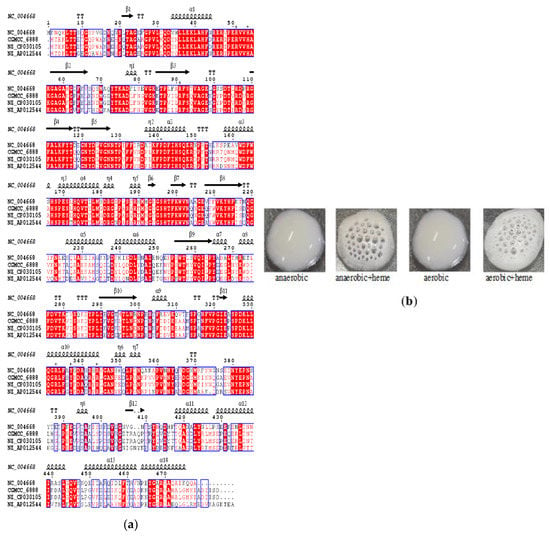
Figure 1.
Identification of heme-dependent catalase of L. plantarum CGMCC 6888. (a) Sequence alignment of catalase from Enterococcus faecalis V583 (NZ_004668), L. plantarum CGMCC 6888, L. plantarum SK151 (NZ_CP030105) and L. casei JCM 1134 (NZ_AP612544). All sequences are displayed as full-length sequences before processing. Abbreviations: α, α-helix; β, β-sheet; η, 310-helix; TT, β-turn. The important amino acids for the heme ligand are indicated by stars. (b) Qualitative analysis of the catalase activity of L. plantarum CGMCC 6888. * represents some amino acids including Arg51, His54, Arg91, Phe140, Arg333, Tyr337 and Arg344 that are important for heme and ligand interaction CatA.
As result, 8.74 ± 0.39 U/mg of catalase was obtained in the cell lysates in the presence of heme, but not for the absence of heme.
3.2. Scavenging Free Radical Activities of L. plantarum CGMCC 6888
To evaluate the antioxidant activity of L. plantarum CGMCC 6888, the scavenging free radical activities of the bacterial cells and cell lysates against DPPH, •OH and O2− radicals were determined. As shown in Table 2, both of the bacterial cells and cell lysates from CatA− and CatA+ strains could scavenge three kinds of free radicals. Interestingly, the scavenging rates of CatA+ bacterial cells/cell lysates towards O2− radicals were 1.63- and 2.07-fold higher than those of the CatA− strain, and for •OH radicals, they were 1.29- and 1.37-fold higher, respectively. These results indicated that the antioxidant activity was significantly enhanced after the activation of catalase A in L. plantarum CGMCC 6888.

Table 2.
Free radical scavenging activities of L. plantarum CGMCC 6888.
3.3. Effects of L. plantarum CGMCC 6888 on Oxidative Stress in NCM460 Cells
Firstly, the cytotoxicity of L. plantarum CGMCC 6888 to NCM460 cells was tested. The results showed the survival rates of the NCM460 cells co-incubated with the CGMCC 6888/CatA+ bacterial cell/cell lysate strain or CGMCC 6888/CatA− bacterial cells/cell lysates were higher than the NCM460 cells alone (Figure S1), suggesting that L. plantarum CGMCC 6888 and the activated CatA had no cytotoxicity against the intestinal epithelial cells.
As shown in Figure 2a, when NCM460 cells were exposed to H2O2 alone, the cell viability decreased to 46.48%. However, the bacterial cells of L. plantarum CGMCC 6888 CatA− or CatA+ could attenuate the damage from H2O2, and significantly increased the viability of the NCM460 cells by 18.54% and 53.96%, respectively. In particular, the bacterial cells of CatA+ showed obvious protection against H2O2, leading to the viability of NCM460 cells being the same as the normal group without H2O2 treatment. Additionally, the cell lysates of the CatA− and CatA+ strains showed similar cytoprotective effects as bacterial cells against H2O2. The results indicated that the activation of CatA endowed L. plantarum CGMCC 6888 with stronger antioxidant activity, thereby alleviating the oxidative stress of intestinal epithelial cells.
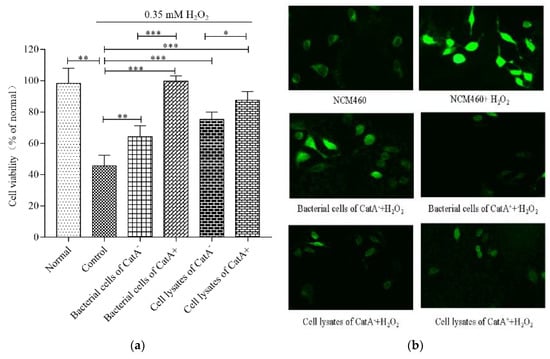
Figure 2.
Effects of L. plantarum CGMCC 6888 on oxidative stress in NCM460 cells. (a) Viabilities of the NCM460 cells. The normal or control represented the cells without or with H2O2 treatment, respectively. * p < 0.05, ** p < 0.01, *** p < 0.001. (b) H2O2–induced ROS levels in NCM460 cells observed using a fluorescence microscope.
To assess the effects of L. plantarum CMCC 6888 on the intracellular ROS levels induced by H2O2, the fluorescence intensity was detected. As shown in Figure 2b, the fluorescence intensity of NCM460 cells treated with H2O2 was significantly higher than that of untreated cells, indicating that H2O2 increased the intracellular ROS content. However, when co-incubated with bacterial cells or cell lysates of CGMCC 6888 CatA− or CatA+ before H2O2 treatment, the fluorescence intensities of NCM460 cells were obviously reduced. In particular, the bacterial cells or cell lysates of CGMCC 6888 CatA+ showed stronger abilities to eliminate ROS, leading to the fluorescence intensities that were the same as NCM460 cells without H2O2 treatment. These results were consistent with the scavenging activities for free radicals.
3.4. Antioxidant Enzyme Activities in NCM460 Cells Promoted by L. plantarum CGMCC 6888
To confirm the effects of L. plantarum/CatA+ on the ROS reduction, the activities of the intracellular antioxidant enzymes activities including CAT, GSH-px and SOD in NCM460 cells were assayed (Table 3). When NCM460 cells were exposed to H2O2 alone, the activity of CAT sharply decreased from 0.55 U/mg protein to 0.11 U/mg protein, as did the activities of GSH-px and SOD. After co-incubation with the bacterial cells of CatA− and CatA+, the activities of CAT and GSH-px were restored to the same levels as control, and the SOD activity increased by 21.62% and 18.92%, respectively, compared with H2O2 alone. In addition, the cell lysates of CatA− and CatA+ showed stronger abilities to promote the activities of these antioxidant enzymes. However, there were no significant differences in the activities of the three antioxidant enzymes promoted by bacterial cells of CatA− and CatA+ or their cell lysates. These results showed that L. plantarum could recover the intracellular antioxidant enzyme activities damaged by H2O2. These results also indicated that the difference in ROS levels in NCM460 cells came from the CatA of L. plantarum CGMCC 6888.

Table 3.
Effects of L. plantarum CGMCC 6888 on the activities of antioxidant enzymes in NCM460 cells.
3.5. L. plantarum CGMCC 6888 Enhanced the Transcription Level of Antioxidant Enzyme Genes
To further investigate the protection of L. plantarum CGMCC 6888 on the intestinal epithelial cells, the gene transcription levels of Nrf2, a key pathway of intestinal epithelial cells to resist oxidative stress, and the four antioxidant enzymes HO-1, GCLC, NQO-1 and TXNRD1 regulated by Nrf2, were detected using RT-qPCR. The results in Figure 3a,b showed that the transcription levels of the Nrf2 gene in NCM460 cells co-incubated with bacterial cells of CatA− or CatA+ were 1.24~1.51-fold higher than the normal group, and the corresponding cell lysates increased 2.37~2.69-fold. Similarly, the gene transcription levels of two or three of the antioxidant enzymes HO-1, GCLC, NQO-1 and TXNRD1 were significantly upregulated. In particular, following CatA+ lysate treatment, the mRNA levels of HO-1 and TXNRD1 were elevated 5.71- and 4.63-fold compared with the normal group.
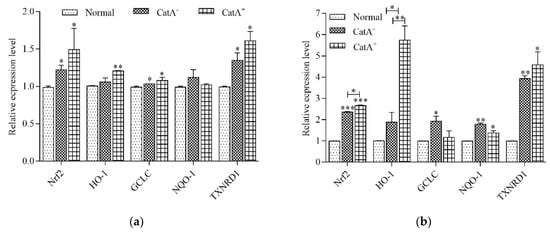
Figure 3.
Transcriptional levels of Nrf2 and the four genes regulated by Nrf2 in NCM460 cells treated with (a) bacterial cells of CatA– or CatA+ and (b) cell lysates of CatA– or CatA+. NCM460 cells were treated with bacterial cells or cell lysates for 2 h. Normal represented the cells without bacterial cells or cell lysate treatment. * p < 0.05, ** p < 0.01, *** p < 0.001.
The results verified that L. plantarum CGMCC 6888 could stimulate the transcription of Nrf2 and Nrf2-related antioxidant enzyme genes to attenuate cellular oxidative stress, and the activation of CatA in L. plantarum CGMCC 6888 had more significant cytoprotective effects.
3.6. CatA-Mediated Protection of L. plantarum CGMCC 6888 on the Intestinal Barrier
Tight junction proteins play crucial roles in maintaining barrier integrity and function. The abundances of ZO-1, OCLN, CLDN and JAM-1 transcripts in NCM460 cells were examined using RT-qPCR (Figure 4). Compared with the normal group, the NCM460 cells stimulated with H2O2 alone for 30 min showed significant downregulation (p < 0.01) for the expression of these four genes. L. plantarum CGMCC 6888/CatA+ bacterial cells/cell lysates or CGMCC 6888/CatA− bacterial cells/cell lysates alone had no significant influence on the transcription levels of tight junction proteins gene compared with the NCM460 cells without H2O2 treatment. However, co-incubation of NCM460 cells with the CGMCC 6888/CatA+ bacterial cell/cell lysate strain or CGMCC 6888/CatA− bacterial cells/cell lysates before H2O2 addition could recover the gene transcription levels of the above-mentioned tight junction proteins, and even promote ZO-1 and JAM-1. Moreover, the bacterial cells of CatA+ had obvious effects, especially on the gene transcription levels of ZO-1, OCLN and JAM-1, which were 1.40-, 1.23- and 1.55-fold higher, respectively, than those of CatA−. Similarly, the ZO-1, OCLN and JAM-1 genes were upregulated 2.47-, 1.67- and 1.26-fold by CatA+ lysate treatment than CatA− lysates. These results indicated that L. plantarum CGMCC 6888 could improve the intestinal barrier function by increasing the transcription levels of tight junction proteins, and the activation of catalase could effectively enhance the defense function of intestinal epithelial cells.
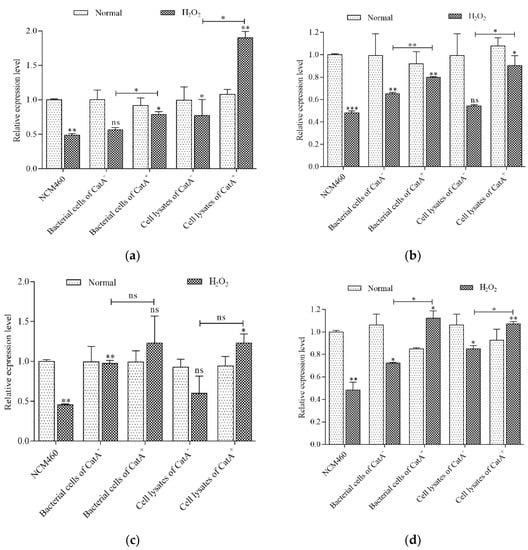
Figure 4.
Effects of L. plantarum CGMCC 6888 on the gene transcription levels of (a) ZO–1, (b) OCLN, (c) CLDN–1 and (d) JAM–1 in NCM460 cells. The normal group represented the cells treated with bacterial cells or cell lysates for 2 h but without H2O2 treatment. The H2O2 group represented cells treated with bacterial cells or cell lysates for 2 h, and then with 0.35 mM H2O2 for 30 min. * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Discussion
A number of L. plantarum strains inhabit human intestinal tracts and are widely used as probiotics. The production of ROS compounds may be harmful for intestinal mucosal cells. Therefore, the identification of L. plantarum strains with intrinsic ROS degrading activities could contribute to human health. In this study, a heme-dependent catalase gene catA was identified in the genome of L. plantarum CGMCC 6888, and its enzymatic activity was obtained in the presence of heme. The activation of the CatA was proved to endow the bacterial cells and cell lysates of L. plantarum CGMCC 6888 stronger scavenging abilities against free radicals. For the intestinal epithelial cell NCM460, the activated CatA in L. plantarum CGMCC 6888 also promoted the upregulation of the transcription levels of Nrf2 and Nrf2-related antioxidant enzymes genes, indicating the enhanced protection of the strain against ROS damage. In the similar way, the transcription levels of the tight junction proteins of ZO-1, OCLN and JAM-1 in NCM460 cells co-incubated with CatA+ bacterial cells/cell lysates were obviously increased compared to those co-incubated with CatA− bacterial cells/cell lysates, suggesting the enhanced intestinal barrier function of activated catalase A of L. plantarum.
LAB are facultative anaerobic microorganisms, usually negative for catalase phenotype, but a number of them, such as L. plantarum, L. casei, L. pentosus, equipped with genes encoding for catalase, require cofactors to activate [34,36]. After sequence alignment, it was found that L. plantarum CGMCC 6888 carried the heme-dependent catA gene (Figure 1a), but it was negative for catalase activity under normal growth conditions, owing to a lack of heme production by L. plantarum [35,36]. Activated-catalase in the presence of heme can decompose hydrogen peroxide to H2O2 and O2 (Figure 1b) and relieve ROS damage in intestinal epithelial cells. Previous studies on the catalase activity of L. plantarum mainly focused on the improvement of the oxidative stress tolerance for bacterial cell survival [37], while the effect of catalase on the probiotic effects in L. plantarum was limited.
Except for the catalase, other antioxidant mechanisms may be involved in the ROS degrading, such as chelating metal ions, antioxidant enzymes and reducing compounds [38]. Free radical scavenging abilities and intestinal epithelial cell models are usually adopted to evaluate the antioxidant activity of LAB and their metabolites in vitro [39,40,41]. Here, the catalase A in L. plantarum CGMCC 6888 was activated by exogenous heme, leading to the remarkable enhancement of antioxidant activity. As shown in Table 1, L. plantarum CatA+ bacterial cells/cell lysates exhibited higher scavenging free radical abilities than those of CatA− bacterial cells/cell lysates. The same phenomenon was observed through a determination of the cell viability, ROS level and antioxidant enzyme activities in NCM460 cells (Figure 2 and Figure 3 and Table 3), indicating that activated-catalase endowed L. plantarum CGMCC 6888 with stronger protection capacities against ROS for resistance to oxidative stress.
The intestinal barrier plays critical roles in the defense against external toxic factors. Faulty intestinal defense renders epithelial cells more susceptible to oxidative stress, impairing the intestinal mucosal barrier and increasing the effect of inflammatory bowel diseases [42]. The tight junction proteins ZO-1, OCLN, CLDN-1 and JAM-1 are usually used as indicators to evaluate the integrity of the intestinal mucosal barrier [42]. Here, the treatment by H2O2 markedly downregulated the transcription of above four genes in NCM460 cells. However, L. plantarum CGMCC 6888 pretreatment negated the reduction in gene transcription levels in ZO-1, OCLN and JAM-1, and the activation of CatA in L. plantarum CGMCC 6888 exhibited more prominent upregulation effects on these three genes. These results showed that L. plantarum CGMCC 6888 played a critical role in promoting the formation of tight junctions, which was consistent with the report that probiotics alleviated the damage to the intestinal barrier by pathogenic microorganisms [43,44].
Table 3 showed that there were no significant differences between the CGMCC 6888/CatA+ bacterial cells/cell lysates and CGMCC 6888/CatA− bacterial cells/cell lysates in the activities of the three antioxidant enzymes including CAT, GSH-px and SOD, indicating that the reduction in ROS levels in NCM460 cells resulted from the activation of CatA in the CGMCC 6888strain. Thus, we speculated that the presence of heme may confer other physiological characteristics to this strain, such as changing the cell robustness and cell surface proteins. Therefore, this provided a new application in the development of probiotic products, based on in-depth analysis of the physiological and genetic characteristics, optimization of the culture medium, and preparation of probiotic adjuvants.
Here, L. plantarum GCMCC 6888 cells were lysed using ultrasonic disruption to release intracellular substances. The cell lysates exhibited higher activity than bacterial cells in promoting antioxidant enzyme activities and increasing the gene transcription levels of antioxidant enzymes. We speculated that more active components (such as GSH, amino acids and fatty acids) involved in the cell lysates took part in the antioxidant responses. These phenomena were in agreement with the previous literature [45,46,47], although there are several proteins displayed in the cell surface, including S-layer proteins, peptidoglycans, teichoic acids and lipoproteins, to crosstalk with the intestinal epithelial cells to activate antioxidant pathways [48,49,50]. However, the specific ingredient is yet to be identified [51,52,53]. The results also provided theoretical support for the development of postbiotics.
5. Conclusions
In this study, the heme-dependent catalase of L. plantarum CGMCC 6888 was activated by exogenous added heme, which contributed to the enhancement of the free radical scavenging abilities and the cytoprotective effects of bacterial cells/cell lysates in intestinal epithelial cells against oxidative stress responses. The study provided a new idea to improve the antioxidant capacities of L. plantarum.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/applmicrobiol3010011/s1. Figure S1 Cytotoxic effects of L. plantarum CGMCC 6888 on NCM460 cells evaluated by the CCK-8 method.
Author Contributions
X.T.: Resources, Writing—original draft; X.Z.: Data curation, Software; M.W.: Validation; T.G.: Funding acquisition, Supervision, Writing—review and editing; J.K.: Project administration, Supervision, Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Natural Science Foundation of Shandong Province (ZR2020MC014).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Acknowledgments
We would like to thank Xiang Gao for providing the NCM460 epithelial cells, and Jingyao Qu, Jing Zhu, Yan Wang and Zhifeng Li from State Key Laboratory of Microbial Technology of Shandong University for help and guidance in qPCR.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lykkesfeldt, J.; Svendsen, O. Oxidants and antioxidants in disease: Oxidative stress in farm animals. Vet. J. 2007, 173, 502–511. [Google Scholar] [CrossRef] [PubMed]
- Mishra, V.; Shah, C.; Mokashe, N.; Chavan, R.; Yadav, H.; Prajapati, J. Probiotics as Potential Antioxidants: A Systematic Review. J. Agric. Food Chem. 2015, 63, 3615–3626. [Google Scholar] [CrossRef] [PubMed]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Pulford, D.J. The glutathione S-transferase supergene family: Regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit. Rev. Biochem. Mol. Biol. 1995, 30, 445–600. [Google Scholar] [CrossRef] [PubMed]
- Townsend, D.M.; Tew, K.D. The role of glutathione-S-transferase in anti-cancer drug resistance. Oncogene 2003, 22, 7369–7375. [Google Scholar] [CrossRef]
- Barrera, G.; Cucci, M.; Grattarola, M.; Dianzani, C.; Muzio, G.; Pizzimenti, S. Control of Oxidative Stress in Cancer Chemoresistance: Spotlight on Nrf2 Role. Antioxidants 2021, 10, 510. [Google Scholar] [CrossRef]
- He, F.; Ru, X.; Wen, T. NRF2, a Transcription Factor for Stress Response and Beyond. Int. J. Mol. Sci. 2020, 21, 4777. [Google Scholar] [CrossRef]
- Nowak, A.; Paliwoda, A.; Błasiak, J. Anti-proliferative, pro-apoptotic and anti-oxidative activity of Lactobacillus and Bifidobacterium strains: A review of mechanisms and therapeutic perspectives. Crit. Rev. Food Sci. Nutr. 2019, 59, 3456–3467. [Google Scholar] [CrossRef]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Zerrouki, M.; Benkaci-Ali, F. DFT study of the mechanisms of nonenzymatic DNA repair by phytophenolic antioxidants. J. Mol. Model. 2018, 24, 78. [Google Scholar] [CrossRef]
- De Boeck, I.; Spacova, I.; Vanderveken, O.M.; Lebeer, S. Lactic acid bacteria as probiotics for the nose? Microb. Biotechnol. 2021, 14, 859–869. [Google Scholar] [CrossRef]
- De Filippis, F.; Pasolli, E.; Ercolini, D. The food-gut axis: Lactic acid bacteria and their link to food, the gut microbiome and human health. FEMS Microbiol. Rev. 2020, 44, 454–489. [Google Scholar] [CrossRef]
- Amanatidou, A.; Smid, E.J.; Bennik, M.H.; Gorris, L.G. Antioxidative properties of Lactobacillus sake upon exposure to elevated oxygen concentrations. FEMS Microbiol. Lett. 2001, 203, 87–94. [Google Scholar] [CrossRef]
- Hernández-Delgado, N.C.; Torres-Maravilla, E.; Mayorga-Reyes, L.; Martín, R.; Langella, P.; Pérez-Pastén-Borja, R.; Bermúdez-Humarán, L.G. Antioxidant and Anti-Inflammatory Properties of Probiotic Candidate Strains Isolated during Fermentation of Agave (Agave angustifolia Haw). Microorganisms 2021, 9, 1063. [Google Scholar] [CrossRef]
- Pan, Y.; Ning, Y.; Hu, J.; Wang, Z.; Chen, X.; Zhao, X. The Preventive Effect of Lactobacillus plantarum ZS62 on DSS-Induced IBD by Regulating Oxidative Stress and the Immune Response. Oxidative Med. Cell. Longev. 2021, 2021, 9416794. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Y.; Wang, X.; Wang, S.; Bi, D. The Impact of Lactobacillus plantarum on the Gut Microbiota of Mice with DSS-Induced Colitis. Biomed. Res. Int. 2019, 2019, 3921315. [Google Scholar]
- Ge, Q.; Yang, B.; Liu, R.; Jiang, D.; Yu, H.; Wu, M.; Zhang, W. Antioxidant activity of Lactobacillus plantarum NJAU-01 in an animal model of aging. BMC Microbiol. 2021, 21, 182. [Google Scholar] [CrossRef]
- Mu, G.; Li, H.; Tuo, Y.; Gao, Y.; Zhang, Y. Antioxidative effect of Lactobacillus plantarum Y44 on 2,2’-azobis(2-methylpropionamidine) dihydrochloride (ABAP)-damaged Caco-2 cells. J. Dairy Sci. 2019, 102, 6863–6875. [Google Scholar] [CrossRef]
- Scillato, M.; Spitale, A.; Mongelli, G.; Privitera, G.F.; Mangano, K.; Cianci, A.; Stefani, S.; Santagati, M. Antimicrobial properties of Lactobacillus cell-free supernatants against multidrug-resistant urogenital pathogens. Microbiologyopen 2021, 10, e1173. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, Y.; Zhou, H.; Tian, F.; Ni, Y. Antimicrobial activities and in vitro properties of cold-adapted Lactobacillus strains isolated from the intestinal tract of cold water fishes of high latitude water areas in Xinjiang, China. BMC Microbiol. 2019, 19, 247. [Google Scholar] [CrossRef]
- Qiao, Y.; Qiu, Z.; Tian, F.; Yu, L.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. Pediococcus acidilactici Strains Improve Constipation Symptoms and Regulate Intestinal Flora in Mice. Front. Cell. Infect. Microbiol. 2021, 11, 655258. [Google Scholar] [CrossRef] [PubMed]
- Song, M.W.; Jang, H.J.; Kim, K.-T.; Paik, H.-D. Probiotic and Antioxidant Properties of Novel Lactobacillus brevis KCCM 12203P Isolated from Kimchi and Evaluation of Immune-Stimulating Activities of Its Heat-Killed Cells in RAW 264.7 Cells. J. Microbiol. Biotechnol. 2019, 29, 1894–1903. [Google Scholar] [CrossRef] [PubMed]
- Bayat, E.; Moosavi-Nasab, M.; Fazaeli, M.; Majdinasab, M.; Mirzapour-Kouhdasht, A.; Garcia-Vaquero, M. Wheat Germ Fermentation with Saccharomyces cerevisiae and Lactobacillus plantarum: Process Optimization for Enhanced Composition and Antioxidant Properties In Vitro. Foods 2022, 11, 1125. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, N.; Li, D.; Wang, M.; Li, C.; Tian, H. Mechanism of gastrointestinal adaptability and antioxidant function of infant-derived Lactobacillus plantarum BF_15 through genomics. Food Sci. Biotechnol. 2022, 31, 1451–1462. [Google Scholar] [CrossRef]
- Wang, Z.; Feng, Y.; Yang, N.; Jiang, T.; Xu, H.; Lei, H. Fermentation of kiwifruit juice from two cultivars by probiotic bacteria: Bioactive phenolics, antioxidant activities and flavor volatiles. Food Chem. 2022, 373, 131455. [Google Scholar] [CrossRef]
- Kostelac, D.; Gerić, M.; Gajski, G.; Frece, J. Probiotic and paraprobiotic derivates exhibit anti-inflammatory and genoprotective effects during induced stress. J. Appl. Microbiol. 2022, 133, 819–829. [Google Scholar] [CrossRef]
- Lechardeur, D.; Cesselin, B.; Fernandez, A.; Lamberet, G.; Garrigues, C.; Pedersen, M.; Gaudu, P.; Gruss, A. Using heme as an energy boost for lactic acid bacteria. Curr. Opin. Biotechnol. 2011, 22, 143–149. [Google Scholar] [CrossRef]
- Pedersen, M.B.; Gaudu, P.; Lechardeur, D.; Petit, M.-A.; Gruss, A. Aerobic Respiration Metabolism in Lactic Acid Bacteria and Uses in Biotechnology. Annu. Rev. Food Sci. Technol. 2012, 3, 37–58. [Google Scholar] [CrossRef]
- Tang, W.; Xing, Z.; Li, C.; Wang, J.; Wang, Y. Molecular mechanisms and in vitro antioxidant effects of Lactobacillus plantarum MA2. Food Chem. 2017, 221, 1642–1649. [Google Scholar] [CrossRef]
- Quatravaux, S.; Remize, F.; Bryckaert, E.; Colavizza, D.; Guzzo, J. Examination of Lactobacillus plantarum lactate metabolism side effects in relation to the modulation of aeration parameters. J. Appl. Microbiol. 2006, 101, 903–912. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Z.; Gu, X.; Zhao, J.; Guo, T.; Kong, J. Probiotic Bacillus subtilis LF11 Protects Intestinal Epithelium Against Salmonella Infection. Front. Cell. Infect. Microbiol. 2022, 12, 837886. [Google Scholar] [CrossRef]
- Sun, Z.; Huang, L.; Kong, J.; Hu, S.; Zhang, X.; Kong, W. In vitro evaluation of Lactobacillus crispatus K313 and K243: High-adhesion activity and anti-inflammatory effect on Salmonella braenderup infected intestinal epithelial cell. Vet. Microbiol. 2012, 159, 212–220. [Google Scholar] [CrossRef]
- Håkansson, K.O.; Brugna, M.; Tasse, L. The three-dimensional structure of catalase from Enterococcus faecalis. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1374–1380. [Google Scholar] [CrossRef]
- Igarashi, T.; Kono, Y.; Tanaka, K. Molecular Cloning of Manganese Catalase from Lactobacillus plantarum. J. Biol. Chem. 1996, 271, 29521–29524. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ricciardi, A. Aerobic metabolism in the genus Lactobacillus: Impact on stress response and potential applications in the food industry. J. Appl. Microbiol. 2017, 122, 857–869. [Google Scholar] [CrossRef]
- Barynin, V.V.; Whittaker, M.M.; Antonyuk, S.V.; Lamzin, V.S.; Harrison, P.M.; Artymiuk, P.J.; Whittaker, J.W. Crystal Structure of Manganese Catalase from Lactobacillus plantarum. Structure 2001, 9, 725–738. [Google Scholar] [CrossRef]
- Fu, L.; Kong, J.; Sun, Z.; Zhang, L.; Zhang, X.; Guo, T. Enhancing the oxidative resistance of yoghurt starter bacteria with heterologous catalase expression in Streptococcus thermophilus. Int. Dairy J. 2013, 30, 68–72. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Finamore, A.; Ambra, R.; Nobili, F.; Garaguso, I.; Raguzzini, A.; Serafini, M. Redox Role of Lactobacillus casei Shirota Against the Cellular Damage Induced by 2,2’-Azobis (2-Amidinopropane) Dihydrochloride-Induced Oxidative and Inflammatory Stress in Enterocytes-Like Epithelial Cells. Front Immunol. 2018, 9, 1131. [Google Scholar] [CrossRef]
- Min, W.-H.; Fang, X.-B.; Wu, T.; Fang, L.; Liu, C.-L.; Wang, J. Characterization and antioxidant activity of an acidic exopolysaccharide from Lactobacillus plantarum JLAU103. J. Biosci. Bioeng. 2019, 127, 758–766. [Google Scholar] [CrossRef]
- Hou, Y.; Li, X.; Liu, X.; Zhang, Y.; Zhang, W.; Man, C.; Jiang, Y. Transcriptomic responses of Caco-2 cells to Lactobacillus rhamnosus GG and Lactobacillus plantarum J26 against oxidative stress. J. Dairy Sci. 2019, 102, 7684–7696. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T. Regulation of the intestinal barrier by nutrients: The role of tight junctions. Anim. Sci. J. 2020, 91, e13357. [Google Scholar] [CrossRef] [PubMed]
- Adora, W.; Pinaki, P. Effects of Lactobacillus plantarum on Caco-2 cell tight junction proteins after infection with enteric bacteria. Gastroenterology 2003, 124, A479. [Google Scholar]
- Yu, Q.; Yuan, L.; Deng, J.; Yang, Q. Lactobacillus protects the integrity of intestinal epithelial barrier damaged by pathogenic bacteria. Front. Cell. Infect. Microbiol. 2015, 5, 26. [Google Scholar] [CrossRef]
- Cuevas-González, P.; Aguilar-Toalá, J.; García, H.; González-Córdova, A.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. Protective Effect of the Intracellular Content from Potential Probiotic Bacteria against Oxidative Damage Induced by Acrylamide in Human Erythrocytes. Probiotics Antimicrob. Proteins 2020, 12, 1459–1470. [Google Scholar] [CrossRef]
- Lin, M.-Y.; Chang, F.-J. Antioxidative Effect of Intestinal Bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig. Dis. Sci. 2000, 45, 1617–1622. [Google Scholar] [CrossRef]
- Saide, J.; Gilliland, S. Antioxidative Activity of Lactobacilli Measured by Oxygen Radical Absorbance Capacity. J. Dairy Sci. 2005, 88, 1352–1357. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Hall, F.G.; Urbizo-Reyes, U.C.; Garcia, H.S.; Vallejo-Cordoba, B.; González-Córdova, A.F.; Hernández-Mendoza, A.; Liceaga, A.M. In Silico Prediction and In Vitro Assessment of Multifunctional Properties of Postbiotics Obtained from Two Probiotic Bacteria. Probiotics Antimicrob. Proteins 2020, 12, 608–622. [Google Scholar] [CrossRef]
- Aguilar-Toalá, J.E.; Estrada-Montoya, M.C.; Liceaga, A.M.; Garcia, H.S.; González-Aguilar, G.A.; Vallejo-Cordoba, B.; Hernández-Mendoza, A. An insight on antioxidant properties of the intracellular content of Lactobacillus casei CRL-431. LWT 2018, 102, 58–63. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Y.; Zhang, L.; Zhang, X.; Huang, L.; Li, D.; Wang, Q. Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 2012, 135, 1914–1919. [Google Scholar] [CrossRef]
- Ahire, J.J.; Mokashe, N.U.; Patil, H.J.; Chaudhari, B.L. Antioxidative potential of folate producing probiotic Lactobacillus helveticus CD6. J. Food Sci. Technol. 2013, 50, 26–34. [Google Scholar] [CrossRef]
- Endo, H.; Niioka, M.; Kobayashi, N.; Tanaka, M.; Watanabe, T. Butyrate-Producing Probiotics Reduce Nonalcoholic Fatty Liver Disease Progression in Rats: New Insight into the Probiotics for the Gut-Liver Axis. PLoS ONE 2013, 8, e63388. [Google Scholar] [CrossRef]
- Kullisaar, T.; Zilmer, M.; Mikelsaar, M.; Vihalemm, T.; Annuk, H.; Kairane, C.; Kilk, A. Two antioxidative lactobacilli strains as promising probiotics. Int. J. Food Microbiol. 2002, 72, 215–224. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).