Abstract
The bacterial foodborne enteropathogen Escherichia albertii, despite enjoying increased attention paid to its pathogenesis, global dissemination, and antimicrobial resistance capacity, remains difficult to identify from human foods. The primary objective of this study was to develop and test a selective and differential plating medium for the isolation of E. albertii from enteric pathogens commonly transmitted via fresh poultry meat, namely E. coli and Salmonella enterica. MacConkey agar supplemented with α-D-+-melibiose and the lactose analogue X-gal was prepared and used to differentially enumerate E. albertii, Salmonella, and E. coli from inoculated ground chicken meat. The medium, MXgMac agar, differentiated the inoculated pathogens with a greater degree of efficiency than did the previously developed E. albertii-selective medium xylose–rhamnose–melibiose (XRM) MacConkey agar, based on differential usage of the lactose analogue and melibiose. Chicken-derived feces and litter samples were subsequently tested using the medium and found not to contain E. albertii by 16S rRNA gene amplification. MXgMac agar facilitates improved differential recovery of E. albertii and other enteric pathogens from poultry meat versus other E. albertii selective/differential media.
1. Introduction
The bacterium Escherichia albertii, a Gram-negative facultatively anaerobic bacillus, has been previously identified and/or implicated in the occurrence of multiple foodborne disease outbreaks in various locations around the globe [1,2,3]. Muchaamba et al. [4] recently reviewed critical aspects of its global distribution, microbial physiology, the various identified and possible routes of its transmission into the human food supply. These authors also summarized the unique biochemical properties of this pathogen that have facilitated its early misidentification after its first report of human disease, as well as more recent attempts to develop culture-dependent and -independent tools to differentiate E. albertii from members of the genus Escherichia and family Enterobacteriaceae [5,6]. The pathogen has been previously recovered from human fecal specimens collected from those suffering diarrheal disease [6,7], human blood from a bacteremic patient [8], ground and surface waters [9], wild birds [10], poultry GI tracts [11], and poultry carcass rinse fluids from commercially harvested chickens [12]. Isolates of E. albertii recovered and characterized by these and other studies have been demonstrated to possess multiple pathogenesis effectors, including intimin and other components of the locus of enterocyte effacement (LEE) [2,13], cytolethal distending toxin B subunit (CDT), and Shiga toxin 2 variants 2a and 2f [2,13,14,15,16].
Various plating media have been reported in the literature in recent years providing differing degrees of utility for distinguishing E. albertii from E. coli and other enteric Gram-negative bacteria, as well as yielding colonies for subsequent molecular identification (e.g., multiplex PCR, multilocus sequence typing (MLST)) [12,16,17]. Maheux et al. [18] reported mEA agar for recovery of lactose-positive and -negative E. albertii isolates from human feces and differentiation from other Escherichia spp. The authors reported 19/19 E. albertii isolates were able to be recovered on the medium, and demonstrated indole-positive results, followed by E. albertii confirmation with PCR. Nonetheless, many other isolates belonging to various genera within the family Enterobacteriaceae, as well as other Gram-negatives, also demonstrated growth on the medium with no differences in appearance. Additionally, 13 isolates with E. albertii-typical appearance and indole test results from human-recovered diarrheal stool samples could not be confirmed as E. albertii [18]. Hinenoya et al. [7] reported the development of xylose–rhamnose–melibiose MacConkey (XRM-MacConkey) agar for the selective differentiation of E. albertii from clinical specimens, though the authors reported some isolates of Shigella could not be visually differentiated from E. albertii due to similar fermentation capabilities. E. albertii were consistently reported as unable to utilize the supplemented carbohydrates, whereas E. coli and Salmonella enterica routinely used at least one of the carbohydrates, producing red-tinted colonies. The authors further compared XRM-MacConkey to mEA and MacConkey agars, reporting 100% specificity of XRM-MacConkey for E. albertii presumptive identification versus other tested plating media. However, these research reports did not provide any characterization of the developed medium’s utility for E. albertii differential detection from human food samples.
The U.S. Department of Agriculture Food Safety Inspection Service (USDA-FSIS) previously identified need for assessment of the distribution of E. albertii in the US meat and poultry supply [19]. To that end, the primary purpose of this study was to develop and evaluate a microbiological medium for the selective differentiation of E. albertii from E. coli and Salmonella from fresh non-intact poultry meat inoculated with a blend of isolates belonging to the three pathogens. The medium was then screened for the isolation and presumptive recovery of E. albertii-typical colonies from poultry animal feces samples for subsequent 16S rRNA-based identification, to gain preliminary assessment of its utility for pathogen isolation during poultry animal production.
2. Materials and Methods
2.1. Bacterial Culture Preparation
Bacterial organisms used in the current study are reported in Table 1 and were obtained from various sources. E. albertii isolates were either revived or obtained via material transfer agreement (MTA) from the US Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) and stored upon receipt at –80 °C in the Texas A&M University Food Microbiology Laboratory (FML). Other isolates were either obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) or from the Texas A&M University FML culture collection. Isolates from the ATCC or CDC were revived according to instructions provided by the organism source. Other organisms were revived from –80 °C cryo-preservation in tryptic soy broth supplemented with 0.6% (w/v) yeast extract (TSB-YE; Becton, Dickinson and Co., Sparks, MD, USA) with 24 h of incubation at –37 °C. All cultures were then passed a second time in TSB-YE with a second 24 h incubation at 37 °C prior to further experimentation.

Table 1.
Bacterial strains used, sources, and typical appearance on MXgMac agar.
2.2. Melibiose–X-Gal–MacConkey (MXgMac) Agar Formulation
Preliminary experiments designed to identify carbohydrate(s) and/or their analogues giving useful differentiation of E. albertii from E. coli and Salmonella indicated melibiose and the lactose analogue 5-bromo-4-chloro-indolyl-β-D-galactopyranoside (X-gal) provided good differentiation of the three organisms from one another. E. albertii has been reported as unable to metabolize melibiose due to no production of an α-galactosidase to cleave the α(1→6) glycosidic bond between the galactose and glucose moieties [7,20]. Lactose non-utilization by E. albertii, like Salmonella, has been reported to occur by multiple research groups [5,6]. Unlike E. albertii, Salmonella and E. coli utilize melibiose and lactose, respectively. Consequently, X-gal (Teknova, Inc., Hollister, CA, USA) and α-D-+-melibiose (Thermo Fisher Scientific, Inc., Waltham, MA, USA), each at 0.5% (w/v), were filter sterilized (0.22 μm) and added to already sterilized, tempered (48–50 °C) MacConkey agar base. The medium was stirred for 1 min to homogenize and then Petri dishes (100 × 15 mm) were filled prior to experimental use. Individual isolates of cultures were grown up in tryptic soy broth (TSB; Becton, Dickinson and Co.) for 24 h at 37 °C and then streaked for isolation onto surfaces of MXgMac agar. Inoculated plates were incubated thereafter for 24–36 h at 37 °C and inspected at 24 and 36 h for colony development, appearance, and any changes occurring in colony formation or appearance between 24 and 36 h of incubation.
2.3. Preparation of Microorganisms for Inoculated Chicken Meat Experiments
To determine the capacity of the experimental medium to facilitate differentiation of E. albertii from E. coli and Salmonella organisms when all were present in a food sample, individual isolates of each pathogen were revived and grown as described in Section 2.2, and then mixtures of isolates for each organism were subsequently prepared. E. albertii isolates were mixed in equal volumes, as were E. coli and Salmonella enterica isolates, in sterile 50 mL conical tubes. Tubes were then centrifuged at 2191× g for 15 min at 25 °C, after which the supernatant was carefully poured off and one volume of phosphate buffered saline (PBS) was added. The bacterial pellet was then vortexed vigorously in applied PBS for 1–2 min, after which the cells were centrifuged again in identical fashion to wash cells of any remaining biomatter and provide for inoculum preparation. Following the second centrifugation, the supernatant was again poured off gently and discarded. The resulting pellet was hydrated with one volume of PBS and mixed thoroughly by vortexing. Cell preparations were then placed in ice to suspend growth prior to subsequent dilution and chicken meat sample inoculation.
To determine the capacity of MXgMac to differentiate isolated E. albertii from E. coli and Salmonella from an inoculated “spiked” chicken meat sample when E. coli and Salmonella were present at higher counts than E. albertii, three differing cocktails of the three pathogens were prepared. Cocktail 1 contained all three organisms, each at a target of 102 CFU/g of chicken meat following inoculation. Cocktail 2 was devised to deliver a final count of 103 each of E. coli and Salmonella, while Cocktail 3 was devised to produce counts of E. coli and Salmonella of ~104 each in inoculated chicken. E. albertii target counts were kept at 102 CFU/g chicken meat for Cocktails 2 and 3, yielding 10- and 100-fold higher numbers of other pathogens versus E. albertii, respectively. Each pathogen mixture was serially diluted in PBS and counts enumerated on TSA following 24 h incubation at 37 °C to quantify the ingoing load of each organism (E. albertii, E. coli, or Salmonella) for each cocktail that was applied to a chicken sample and allow for subsequent comparison of a pathogen’s recovery from spiked chicken versus inoculated numbers.
2.4. Preparation of Ground Chicken Meat Samples and Inoculation with Pathogens
Refrigerated ground chicken meat (97% lean) was purchased from a College Station, TX, USA retail grocer and immediately returned to the FML. Upon return, 250 g aliquots of chicken were aseptically weighed and placed in polyethylene refrigerator/freezer bags and flattened. Bags were transported to the National Center for Electron Beam Research (Texas A&M AgriLife, College Station, TX, USA) and subjected to electron beam pasteurization to reduce numbers of background microorganisms prior to inoculation of pathogen mixtures/cocktails. Two hundred and fifty-gram samples packed in Ziploc pouches were arrayed as depicted in Figure 1. Chicken packages were irradiated to a target of at least 10.0 kGy via the Tower accelerator (10.0 MeV), positioning the electron beam horn above the chicken samples; samples were passed through the accelerator once. A dose absorption study was completed using three alanine pellets positioned at differing locations within the packaged chicken array. Resulting minimum and maximum dose absorptions were 10.09 and 11.39 kGy, respectively. The mean absorbed dose was 10.71 ± 0.51 kGy, and the dose uniformity ratio (DUR) was 1.13 (11.39 kGy/10.09 kGy).

Figure 1.
Orientation and array of ground chicken pouches for electron beam device processing/irradiation to pasteurize chicken meat prior to inoculation with microorganisms.
Following irradiation, sample bags were returned to the FML and placed under frozen (−20 °C) storage or prepared for immediate use. Chicken sample portions (25 g each) were aseptically weighed from irradiated aliquots of chicken meat and inoculated with 1.0 mL of Cocktails 1, 2, or 3 of E. albertii, E. coli, and Salmonella. Samples were hand-massaged for 1 min and then allowed to rest for 30 min to facilitate microbial attachment to meat. Thereafter, inoculated samples were serially diluted in PBS and pathogens selectively/differentially enumerated on MXgMac and XRM-Mac [11] agars in order to compare the two media for their ability to presumptively discriminate E. albertii from E. coli and Salmonella. Inoculated plates were incubated at 36 + 1 °C for 24–30 h prior to inspection and counting.
2.5. Evaluation of MXgMac Agar to Assist Identification of E. albertii from Chicken Fecal/Litter Sample
Dropped fecal grab samples were collected (50–100 g each) from four cages (n = 16 hens) containing white leghorn egg-laying hens at the Texas A&M University Department of Poultry Science Research, Teaching, and Extension Center (College Station, TX, USA), placed in sterile whirl-pack plastic bags, and then returned to the Food Microbiology Laboratory. Chickens were fed a standard diet and managed according to the Texas A&M University Institutional Animal Care and Use Committee (IACUC) Animal Use Permit 2019-0171 (Principal Investigator: M. Farnell, Department of Poultry Science, Texas A&M AgriLife Research, College Station, TX, USA). Upon return to the laboratory, 10 g sample material was diluted in 90 mL sterile PBS and 10 μL streaked onto surfaces of MXgMac agar-containing Petri dishes. Following a 24–36 h incubation at 37 °C, plates were removed and visually inspected for the presence of colorless colonies without haloes, typical of E. albertii. Colonies with this appearance were picked onto MXgMac plates for reisolation. From these plates, following incubation, colonies were picked and transferred to TSA slants for subsequent identification via 16S rRNA gene amplification.
2.6. Identification of E. albertii-typical colonies picked by 16S rRNA sequence typing
2.6.1. DNA Extraction
A pure culture of each isolate from a TSA slant (Section 2.5) was grown overnight in TSB at 36 ± 1 °C. Each tube was then centrifuged at 5000× g for 10 min at 4 °C and the resulting pellet resuspended in 2 mL PBS, and then transferred to two microcentrifuge tubes (1.0 mL each) and centrifugated at 10,000× g for 15 min at 4°C. One of the microtubes was stored at −80 °C and the other was used for DNA extraction and purification (Quick-DNA™ Fungal/Bacterial Miniprep Kit, Zymo Research Co., Orange, CA, USA) per manufacturer instructions. For each bacterial isolate, the obtained pellet was resuspended in 200 µL of PBS and transferred to a labeled ZR Bashing Bead™ Lysis Tube (0.1 × 0.5 mm) to which 750 μL Bashing Bead™ Buffer was added. Samples were vortexed for 2 min, sonicated for 1 min at 25 °C, vortexed for 2 min, sonicated again for 2 min at 25 °C, and vortexed individually for 30 s. The samples were then centrifuged at 10,000× g for 1 min at 4 °C, and 400 µL of the supernatant was transferred to a Zymo-Spin™ III-F Filter in a collection tube and then centrifuged at 8000× g for 1 min. A volume of 1.2 mL of Genomic Lysis Buffer was then added to the filtrate in the collection tube and mixed thoroughly.
Eight hundred microliters of the mixture were then transferred to a Zymo-Spin™ IICR Column in a collection tube and centrifuged at 10,000× g for 1 min at 4 °C. After this, the flow through in the collection tube was discarded and the remaining 800 μL of the mixture was transferred to the Zymo-Spin™ IICR and centrifuged at 10,000× g for 1 min. The Zymo-Spin™ IICR Column was then transferred to a new collection tube, where 200 μL of the DNA Pre-Wash Buffer was added and the microtubes were centrifuged at 10,000 × g for 1 min, followed by the addition of 500 μL of DNA Wash Buffer and centrifugation at 10,000× g for 1 min, both times at 4 °C.
For elution of DNA, each Zymo-Spin™ IICR Column was transferred to a clean 1.5 mL microcentrifuge tube, 100 μL of DNA Elution Buffer was added directly to the column matrix and centrifuged at 10,000× g for 30 s at 4 °C. The DNA concentration of the samples was then quantified using a Qubit® dsDNA HS Assay kit (Thermo Fisher Scientific).
2.6.2. PCR, Sequencing, and Organism Identification
All DNA samples for PCR were stored at −20 °C and thawed on ice before use. Amplification of 16S rRNA genes was conducted in a total reaction volume of 25 µL, using universal primers 27F (5′ AGA GTT TGA TCC TGG CTC AG 3′) and 1492R (5′ ACG GCT ACC TTG TTA CGA CTT 3′) (Integrated DNA Technologies, Coralville, IA, USA).
The 25 µL PCR mixture was prepared by mixing 12.5 µL of 2× KAPA2G Fast Hot Start ready mix (Sigma-Aldrich Co., St. Louis, MO, USA), 1.25 µL of the forward and reverse primers, 8 µL PCR-grade water, and 2 µL sample DNA. Amplification was undertaken in a programmable thermocycler (Biometra Tone 96 G, 230 V, Analytik Jena, Konrad-Zuse-Str. Germany), under the following conditions: 1 initial cycle of denaturation at 95 °C for 3 min; 35 cycles of denaturation at 95 °C for 15 sec, annealing at 60 °C for 15 sec, and extension at 72 °C for 1 min; and a final extension at 72 °C for 1 min.
To corroborate the molecular weight of the PCR products, agarose gel electrophoresis was performed using 1% agarose (CulGenex Agarose LE, Molecular Biology Grade, Hardy Diagnostics, Santa Monica, CA, USA). For this, 2 µL of each PCR product was mixed with dye (Gel Loading Dye, Purple (6×), no SDS, New England Biolabs, Ipswich, MA, USA) and electrophoresed in 1× TAE buffer (Omega Bio-Tek, Inc., Norcross, GA, USA) through a 1% agarose gel containing 5 µL GelGreen® Nucleic Acid Gel Stain (Biotium, Inc., Fremont, CA, USA). Bands of the appropriate size were identified by comparison with a 100 bp DNA ladder (Quick-Load Purple 100 bp DNA Ladder, New England Biolabs). A sample was considered appropriate if a signal band corresponding to 1500 bp was visualized under UV light.
Vials containing the different PCR products were sent for Sanger sequencing to Eton Bioscientific (Eton Bioscience, Inc., San Diego, CA, USA). The sequences obtained in ab1 format were converted to FASTQ, quality controlled with FastQC, and the first 20 bases removed from all sequences by Fastq Trimmer, followed by filtering to remove sequences less than 200 bp using filtlong; these analyses were conducted at usegalaxy.eu [21]. The 16S sequences were exported from Galaxy and analyzed in the Ribosomal Database Project (RDP 11) Classifier to obtain taxonomic assignments to the genus level [22,23]. The sequences identified as belonging to family Enterobacteriaceae were then checked and manually edited using the BioEdit 7.2 Sequence Alignment editor. These sequences were then searched by BLASTn against the nt database at the National Center for Biotechnology Information (NCBI), for identification to species level.
2.7. Experimental Design and Statistical Analysis of Data
Experiments testing the selective/differential recovery of inoculated E. albertii, E. coli, and Salmonella from irradiated ground chicken with the three cocktails (i.e., Cocktails 1, 2, and 3) were completed as a complete block and replicated three times on differing dates. Each replicate possessed three independently completed samples derived from differing 250 g sample packs of irradiated chicken (n = 9). Data were analyzed by the general linear method for a two-way analysis of variance for the main effects of cocktail, medium (MXgMac or XRM-Mac agar), and their interaction for recovery of E. albertii. A similar analysis was completed wherein the counts of E. coli and Salmonella from MXgMac were first summed together for each sample and then compared to the count of E. coli and Salmonella-typical colonies from XRM-Mac, again testing the main effects of medium and cocktail, and their interaction. Means were separated post-ANOVA using Bonferroni’s method with significance set at p < 0.05. Statistical analyses were completed with Prism v9.4.1 (GraphPad Software, LLC, San Diego, CA, USA).
3. Results
3.1. Differential Identification of E. albertii from E. coli and Salmonella enterica on MXgMac Agar Surfaces
As indicated in Table 1, isolates of E. albertii did not effectively hydrolyze the lactose analogue X-gal or melibiose, resulting in colonies displaying colorless growth. Figure 2 depicts the typical appearance of the three inoculated pathogenic organisms when streaked individually onto MXgMac agar surfaces and incubated as described in Section 2.2.
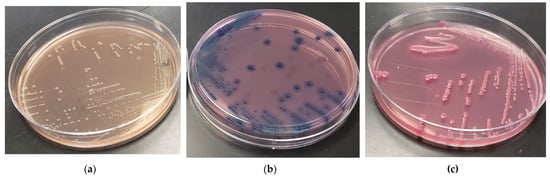
Figure 2.
Typical appearance of (a) E. albertii, (b) E. coli, and (c) Salmonella on surfaces of melibiose–X-gal–MacConkey (MXgMac) agar surfaces following 36 h incubation at 36 ± 1 °C.
Isolates of E. albertii on MXgMac, unable typically to utilize melibiose or lactose, appeared colorless with at times small zones of precipitated bile salts surrounding the colonies. E. coli isolates routinely appeared bluish green from the degradation of the X-gal, whereas Salmonella, negative for lactose use but positive for melibiose usage, took on a reddish/pink tinge in the colony center following plate incubation with colorless edges (Figure 2). Zones of bile precipitation were intermittently observed for all three organisms but were most pronounced for E. coli.
3.2. Comparisons of Recoveries of Inoculated Pathogens on MXgMac and XRM-Mac from Ground Chicken
Mean numbers of E. albertii, E. coli, and Salmonella enterica isolates following mixing together, prior to final cocktail preparation for chicken inoculation, were 7.76 ± 0.18, 7.82 ± 0.12, and 7.82 ± 0.07 log10 CFU/mL. Counts of organisms did not statistically differ from one another (p = 0.498). Figure 3 depicts the recoveries of E. albertii on MXgMac and XRM-Mac agars as a function of the three cocktail setups (1, 2, and 3), wherein the targeted number of the organism was inoculated at 102 CFU/g chicken meat while numbers of E. coli and Salmonella were systematically increased for the three cocktails from 102 to 104 CFU/g chicken meat. Least squares means of E. albertii on MXgMac ranged from 1.8 to 2.5 log10 CFU/g chicken meat across the three cocktail setups, and from 2.2–2.5 log10 CFU/g chicken meat on XRM-Mac, but did not statistically differ as a function of the medium (MXgMac vs. XRM-Mac), cocktail setup, or their interaction (p = 0.600).
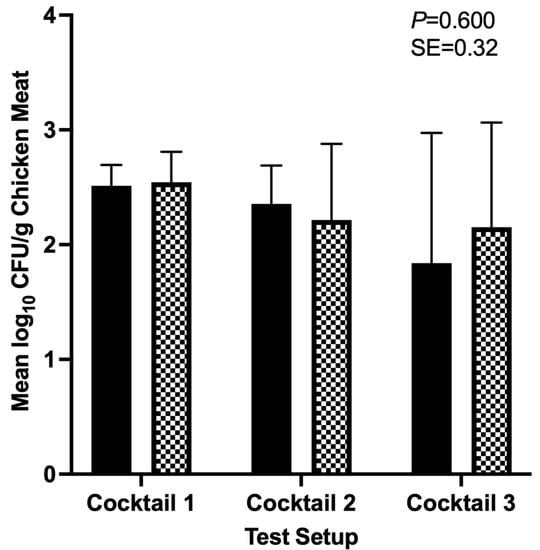
Figure 3.
Least squares means of Escherichia albertii on MXgMac (solid black bars) and XRM-Mac (checkered bars) recovered from inoculated ground chicken meat, following incubation of inoculated plates for 24–36 h at 36 ± 1 °C. Bars indicate the mean of triplicate identically completed replicates, with each replicate possessing three independently completed samples (n = 9). Error bars depict one sample standard deviation. SE: pooled standard error.
Counts of E. coli and Salmonella individually and summed together on MXgMac, as well as collectively on XRM-Mac, are presented in Table 2. Means of these pathogens did not statistically differ by medium, when E. coli and Salmonella counts were summed together, allowing comparison of E. coli and Salmonella counts on MXgMac with the total count of these organisms on XRM-Mac (p = 0.0595). The summed counts of these pathogens must be compared to recovered counts from XRM-Mac, given the latter’s lack of differentiation of E. coli from Salmonella [7]. Similar to results for E. albertii, the interaction of cocktail setup x medium did not result in one main effect significantly influencing the other main effect with respect to resulting mean counts (p = 0.511).

Table 2.
Least squares means ± one standard deviation (log10 CFU/g chicken meat) of E. coli, Salmonella from MXgMac agar, summed counts of Escherichia coli + Salmonella enterica cocktails on MXgMac and XRM-Mac agar.
3.3. 16S rRNA Identification of E. albertii-Typical Colonies from Chicken Feces/Litter Samples
Following completion of colony isolation, DNA extraction, and 16S rRNA gene sequencing by described procedures, no isolated organisms displaying non-fermentation on MXgMac from chicken feces/litter samples were identified as E. albertii. Of 22 NCBI-submitted isolate sequences, five isolates were identified by the RDP Classifier as belonging to the Escherichia/Shigella genus grouping, with one being identified by BLAST alignment as most closely related to S. sonnei, with the remaining four most closely related to E. coli (Table A1). Figure 4 below depicts phylogenetic relatedness of isolates from chicken litter/feces samples. This analysis illustrates that it was difficult to unambiguously assign strains to the species level based on available partial 16S rDNA sequences, with the possible exceptions of strain 22 (E. cloacae), strain 16 (E. hormaechei), and strain 28 (C. sakazakii). The necessity for improved techniques for E. albertii isolation is clearly apparent, such as the pre-application of rigorous selective enrichment to prohibit growth of undesirable microbes at the expense of E. albertii isolation.
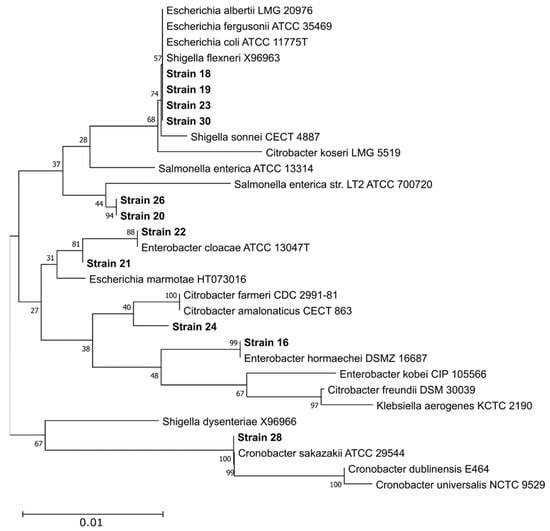
Figure 4.
Phylogenetic relatedness of enterobacterial chicken litter/feces-recovered isolates from 16S rRNA sequence analysis. A reference set of 20 16S rDNA sequences from species most related to the enterobacterial isolate sequences (based on BLASTn results vs. the NT database) were retrieved from the RDP and aligned with 11 partial 16S rDNA sequences using the ClustalW algorithm at default parameters. Sample 27 was not included in this alignment due to excessive gaps. A neighbor-joining tree was generated from this alignment using the Jukes–Cantor method and omitting all positions containing gaps or ambiguous bases, leaving 521 positions in the final dataset. Numbers at each branch indicate the percentage score from a 500-replicate bootstrap analysis. All analyses were conducted in MEGA11 v11.0.13 [24].
4. Discussion
The detection of human enteric pathogens from foods, despite the advent of modern genomic analyses, still frequently employs the use of selective enrichment and/or plating media for purposes of isolating the pathogen from background microorganisms. These procedures facilitate researchers and regulatory technicians’ efforts to confirm the identity of the presumptively detected pathogen, allowing for execution of regulatory food safety requirements and/or pathogen surveillance from foods [25,26]. The emerging pathogen E. albertii has become of increased interest due to its being identified as the causative agent of at least one human foodborne disease outbreak and its global dissemination [4].
While mEA and XRM-Mac agars have been described as useful for the differentiation of E. albertii from other enteric bacteria from clinical samples, to date no medium other than the MXgMac described herein is known to the authors allowing for differential identification of E. albertii from other pathogens from a food sample [7,18]. Our group focused on the differentiation of E. albertii from Salmonella enterica and E. coli, including both non-human-pathogenic and human-pathogenic O157 and non-O157 Shiga-toxin producing E. coli (STEC), given E. albertii’s previous recovery from US poultry production and commercial poultry meat samples [11,12]. In the first set of experiments, the use of melibiose and X-gal demonstrated useful differentiation of the typically lactose-negative and melibiose-negative E. albertii from the lactose-positive/melibiose-negative E. coli and lactose-negative/melibiose-positive Salmonella (Section 2.2). Nevertheless, subsequent testing of the medium should incorporate a far broader range of isolates than those we were able to access during the project, to further screen its differential capabilities.
In experiments testing selective/differential enumeration of E. albertii from E. coli and Salmonella in irradiated ground chicken meat, MXgMac and XRM-Mac agars did not differ in their ability to support the presumptive identification of E. albertii across the three cocktail setups. Even as numbers of inoculated E. coli and Salmonella were systematically increased over the three cocktail setups, E. albertii numbers did not differ between the two selective/differential media, demonstrating both were sufficiently useful for E. albertii recovery and enumeration from poultry meat. Nonetheless, the MXgMac demonstrated additional utility versus XRM-Mac due to its ability to allow presumptive discrimination of other possible human pathogens in addition to E. albertii, facilitating those organisms’ subsequent identification (Table 2). The USDA-FSIS implements mandatory performance standards for young chicken carcasses and fabricated chicken parts testing the prevalence of Salmonella, and has proposed new standards that will include a quantitative maximum allowable Salmonella count on certain not ready-to-eat poultry products [27,28]. MXgMac is expected to be of enhanced utility versus other E. albertii-differentiating media when seeking to simultaneously identify other poultry-borne human pathogens in addition to E. albertii, or when used for enumerating the pathogen from other poultry-borne pathogens.
Testing of feces/litter samples from chickens located at the Texas A&M University Department of Poultry Science’s Teaching, Research, and Extension Center did not yield any confirmed E. albertii. This was likely a result of having only a small number of chickens for which testing could be completed, as compared to commercial establishments that may house several thousand chickens together, such as that reported by other researchers [11]. The presence of multiple non-lactose- or non-melibiose-using organisms from feces leading to needs for genetic identification of the organism is not surprising, and multiple culture media integrated into routine testing procedures are known to support the growth of multiple organisms other than the targeted microorganism(s). The use of antibiotics to suppress other Gram-negatives in addition to the presence of bile salts in MacConkey agar could improve the selectivity of E. albertii; the base of knowledge regarding the organism’s resistance to various antibiotics is growing [2,6,29]. Likewise, the use of E. albertii-specific selective enrichment would improve opportunity for pathogen recovery on culture media or by molecular testing, with E. albertii-specific selective enrichment formulae only very recently being reported in the literature [7,30]. In combination with continued optimization of E. albertii-specific multiplex PCR and/or MLST analyses, such efforts should improve efforts for E. albertii surveillance and improved food safety protection.
Author Contributions
Conceptualization, S.D.A. and T.M.T.; methodology, S.D.A., S.D.P., J.J.G. and T.M.T.; formal analysis, C.R.K. and J.J.G.; data curation, T.M.T.; writing—original draft preparation, T.M.T.; writing—review and editing, S.D.A., K.S.S., S.D.P., C.R.K., J.J.G. and T.M.T.; supervision, J.J.G., C.R.K. and T.M.T.; project administration, T.M.T.; funding acquisition, T.M.T. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the USDA National Institute of Food and Agriculture (Agriculture and Food Research Initiative Award No. 2017-67017-26227).
Data Availability Statement
Original data may be provided upon reasonable request. CDC- and ATCC-acquired bacterial isolates were collected subject to materials transfer agreement (MTA) or purchase and may be obtained directly from those entities.
Acknowledgments
The authors thank Sapna C. Dass, Department of Animal Science, Texas A&M AgriLife Research, College Station, TX, for technical advice related to genetic screening of presumptive E. albertii isolates. The authors acknowledge Morgan Farnell, Department of Poultry Science, Texas A&M AgriLife Research, for provision of poultry animal fecal/litter samples.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A

Table A1.
Taxonomic identification and National Center for Biotechnology Information (NCBI) accession numbers for 16s rRNA sequences of poultry litter/feces-recovered bacterial isolates on MXgMac agar surfaces.
Table A1.
Taxonomic identification and National Center for Biotechnology Information (NCBI) accession numbers for 16s rRNA sequences of poultry litter/feces-recovered bacterial isolates on MXgMac agar surfaces.
| Taxonomic ID (RDP 11 > 70% Confidence | Sequence ID | NCBI Accession |
|---|---|---|
| Cronobacter sp. | 27 | OQ283624 |
| Escherichia sp. | 30 | OQ283625 |
| Cronobacter sp. | 28 | OQ283626 |
| Salmonella sp. | 26 | OQ283627 |
| Citrobacter sp. | 24 | OQ283628 |
| Escherichia sp. | 23 | OQ283629 |
| Enterobacter sp. | 22 | OQ283630 |
| Escherichia sp. | 21 | OQ283631 |
| Salmonella sp. | 20 | OQ283632 |
| Escherichia sp. | 19 | OQ283633 |
| Escherichia sp. | 18 | OQ283634 |
| Enterobacter sp. | 16 | OQ283635 |
| Empedobacter sp. | 15 | OQ283636 |
| Myroides sp. | 14 | OQ283637 |
| Acinetobacter sp. | 13 | OQ283638 |
| Myroides sp. | 7 | OQ283639 |
| Enterococcus sp. | 5 | OQ283640 |
| Acinetobacter sp. | 4 | OQ283641 |
| Gammaproteobacteria bacterium | 3r | OQ283642 |
| Gammaproteobacteria bacterium | 3f | OQ283643 |
| Acinetobacter sp. | 2r | OQ283644 |
| Acinetobacter sp. | 2f | OQ283645 |
References
- Konno, T.; Yatsuyanagi, J.; Takahashi, S.; Kumagai, Y.; Wada, E.; Chiba, M.; Saito, S. Isolation and identification of Escherichia albertii from a patient in an outbreak of gastroenteritis. Jpn. J. Infect. Dis. 2012, 65, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Ooka, T.; Seto, K.; Kawano, K.; Kobayashi, H.; Etoh, Y.; Ichihara, S.; Kaneko, A.; Isobe, J.; Yamaguchi, K.; Horiwaka, K.; et al. Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 2012, 18, 488–592. [Google Scholar] [CrossRef] [PubMed]
- Asoshima, N.; Matsuda, M.; Shigemura, K.; Honda, M.; Yoshida, H.; Hiwaki, H.; Ogata, K.; Oda, T. Identification of Escherichia albertii as a causative agent of a food-borne outbreak occurred in 2003. Jpn. J. Infect. Dis. 2014, 67, 139–140. [Google Scholar] [CrossRef]
- Muchaamba, F.; Barmettler, K.; Treier, A.; Houf, K.; Stephan, R. Microbiology and epidemiology of Escherichia albertii-an emerging elusive foodborne pathogen. Microorganisms 2022, 10, 875. [Google Scholar] [CrossRef] [PubMed]
- Abbott, S.L.; O’Connor, J.; Robin, T.; Zimmer, B.L.; Janda, J.M. Biochemical properties of a newly described Escherichia species, Escherichia albertii. J. Clin. Microbiol. 2003, 41, 4852–4854. [Google Scholar] [CrossRef] [PubMed]
- Huys, G.; Cnockaert, M.; Janda, J.M.; Swings, J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 2003, 53, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Hinenoya, A.; Nagano, K.; Okuno, K.; Nagita, A.; Hatanaka, N.; Awasthi, S.P.; Yamasaki, S. Development of XRM-MacConkey agar selective medium for the isolation of Escherichia albertii. Diagn. Microbiol. Infect. Dis. 2020, 7, 115006. [Google Scholar] [CrossRef]
- Inglis, T.J.J.; Merritt, A.J.; Bzdyl, N.; Lansley, S.; Urosevic, M.N. First bacteraemic human infection with Escherichia albertii. New Microbe New Infect. 2015, 8, 171–173. [Google Scholar] [CrossRef]
- Maheux, A.F.; Boudreau, D.K.; Bergeron, M.G.; Rodriguez, M.J. Characterization of Escherichia fergusonii and Escherichia albertii isolated from water. J. Appl. Microbiol. 2014, 117, 597–609. [Google Scholar] [CrossRef]
- Oaks, J.L.; Besser, T.E.; Walk, S.T.; Gordon, D.M.; Beckmen, K.B.; Burek, K.A.; Haldorson, G.J.; Bradway, D.S.; Ouellette, L.; Rurangirwa, F.R.; et al. Escherichia albertii in wild and domestic birds. Emerg. Infect. Dis. 2010, 16, 638–646. [Google Scholar] [CrossRef]
- Hinenoya, A.; Li, X.-P.; Zeng, X.; Sahin, O.; Moxley, R.A.; Logue, C.M.; Gillespie, B.; Yamasaki, S.; Lin, J. Isolation and characterization of Escherichia albertii in poultry at the pre-harvest level. Zoonoses Public Hlth. 2021, 68, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, R.L.; Fedorka-Cray, P.J.; Abley, M.; Turpin, J.B.; Meinersmann, R.J. Evaluating the occurrence of Escherichia albertii in chicken carcass rinses by PCR, Vitek analysis, and sequencing of the rpoB gene. Appl. Environ. Microbiol. 2015, 81, 1727–1734. [Google Scholar] [CrossRef]
- Lacher, D.W.; Steinsland, H.; Whittam, T.S. Allelic subtyping of the intimin locus (eae) of pathogenic Escherichia coli by fluorescent RFLP. FEMS Microbiol. Lett. 2006, 261, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Brandal, L.T.; Tunsjø, H.S.; Ranheim, T.E.; Løbersli, I.; Lange, H.; Wester, A.L. Shiga toxin 2a in Escherichia albertii. J. Clin. Microbiol. 2015, 53, 1454–1455. [Google Scholar] [CrossRef] [PubMed]
- Hyma, K.E.; Lacher, D.W.; Nelson, A.M.; Bumbaugh, A.C.; Janda, J.M.; Strockbine, N.A.; Young, V.B.; Whittam, T.S. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 2005, 187, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Etoh, Y.; Tanaka, E.; Ichihara, S.; Horikawa, K.; Kawano, K.; Ooka, T.; Kawamura, Y.; Ito, K. Shiga toxin 2f-producing Escherichia albertii from a symptomatic human. Jpn. J. Infect. Dis. 2014, 67, 204–208. [Google Scholar] [CrossRef]
- Wang, H.; Li, Q.; Bai, X.; Xu, Y.; Zhao, A.; Sun, H.; Deng, J.; Xiao, B.; Liu, X.; Sun, S.; et al. Prevalence of eae-positive, lactose non-fermenting Escherichia albertii from retail raw meat in China. Epidemiol. Infect. 2016, 144, 45–52. [Google Scholar] [CrossRef]
- Maheux, A.F.; Brodeur, S.; Bérubé, È.; Boudreau, D.K.; Abed, J.Y.; Boissinot, M.; Bissonnette, L.; Bergeron, M.G. Method for isolation of both lactose-fermenting and -non-fermenting Escherichia albertii strains from stool samples. J. Microbiol. Methods 2018, 154, 134–140. [Google Scholar] [CrossRef]
- USDA-FSIS. Food Safety Research Priorities & Studies. Available online: https://www.fsis.usda.gov/science-data/research-priorities (accessed on 19 January 2023).
- Arai, S.; Ohtsuka, K.; Konishi, N.; Ohya, K.; Konno, T.; Tokoi, Y.; Nagaoka, H.; Asano, Y.; Maruyama, H.; Uchiyama, H.; et al. Evaluating methods for detecting Escherichia albertii in chicken meat. J. Food Protect. 2021, 84, 553–562. [Google Scholar] [CrossRef]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res. 2020, 48, W395–W402. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA 11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- USDA-FSIS. Raw Pork Products Exploratory Sampling Program. Available online: https://www.fsis.usda.gov/science-data/sampling-program/raw-pork-products-exploratory-sampling-program (accessed on 19 January 2023).
- USDA-FSIS. Serotypes Profile of Salmonella isolates from Meat and Poultry Products January 1998 through December 2012. Available online: http://www.fsis.usda.gov/wps/portal/fsis/topics/data-collection-and-reports/microbiology/annual-serotyping-reports (accessed on 26 June 2015).
- USDA-FSIS. Proposed Regulatory Framework to Reduce Salmonella Illnesses Attributable to Poultry. Available online: https://www.fsis.usda.gov/sites/default/files/media_file/documents/FINAL-Salmonella-Framework-10112022-508-edited.pdf (accessed on 12 November 2022).
- U.S. Department of Agriculture-Food Safety and Inspection Service. New Performance Standards for Salmonella and Campylobacter in Not-Ready-to-Eat Comminuted Chicken and Turkey Products and Raw Chicken Parts and Changes to Related Agency Verification Procedures: Response to Comments and Announcement of Implementation Schedule. Fed. Reg. 2016, 81, 7285–7300. [Google Scholar]
- Ooka, T.; Ogura, Y.; Katsura, K.; Seto, K.; Kobayashi, H.; Kawano, K.; Tokuoka, E.; Furukawa, M.; Harada, S.; Yoshino, S.; et al. Defining the genome features of Escherichia albertii, an emerging enteropathogen closely related to Escherichia coli. Genome Biol. Evol. 2015, 7, 3170–3179. [Google Scholar]
- Hirose, S.; Nakamura, Y.; Arai, S.; Hara-Kudo, Y. The development and evaluation of a selective enrichment for the detection of Escherichia albertii in food. Foodborne Pathog. Dis. 2022, 19, 704–712. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).