Complete Genome Sequence and Benzophenone-3 Mineralisation Potential of Rhodococcus sp. USK10, A Bacterium Isolated from Riverbank Sediment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Rhodococcus sp. USK10
2.2. BP3 Biodegradation Experiment
2.3. DNA Extraction and Library Preparation
2.4. Bioinformatics Analyses
3. Results and Discussion
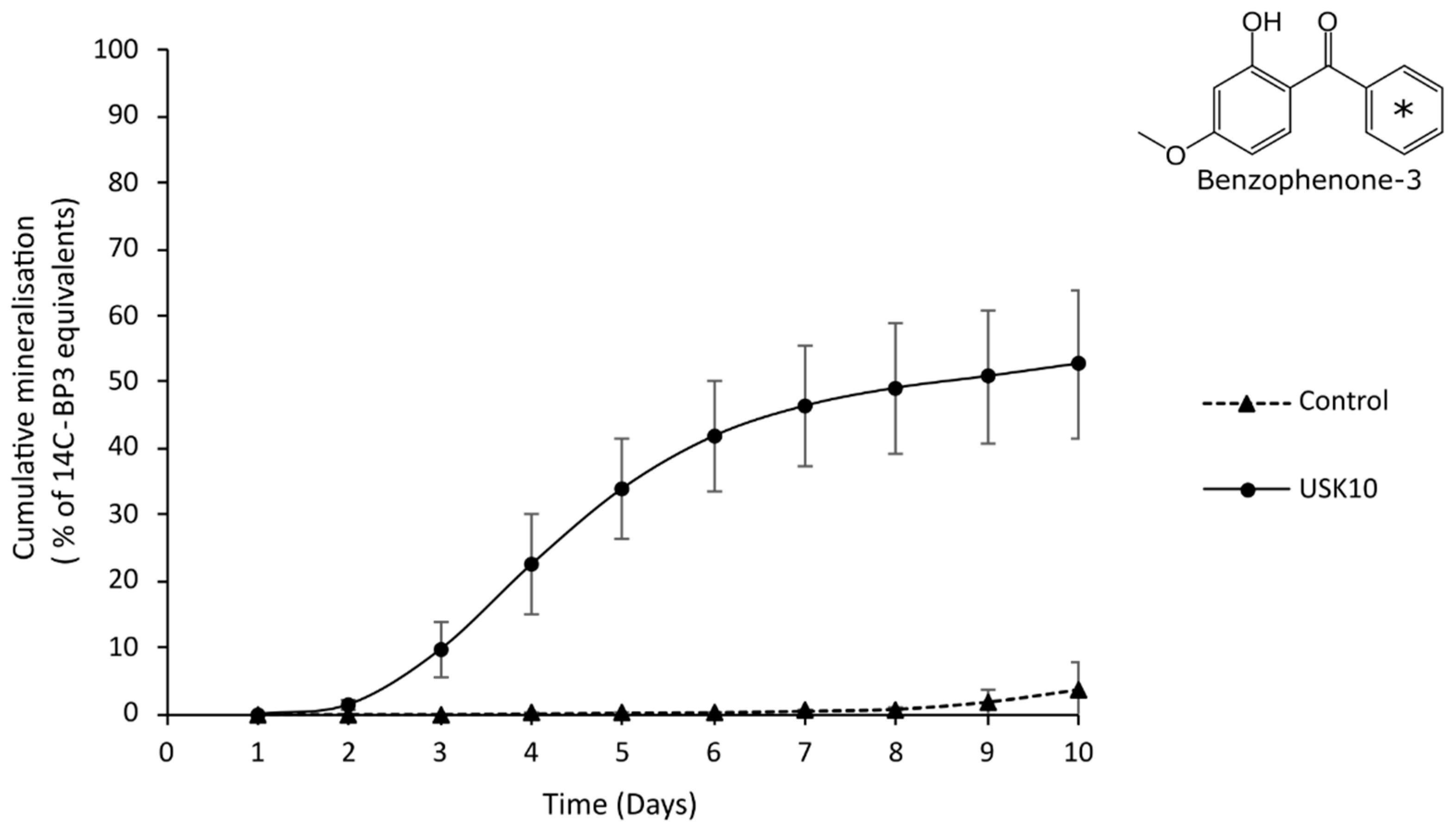
3.1. BP3 Degradation Potential of Rhodococcus sp. USK10
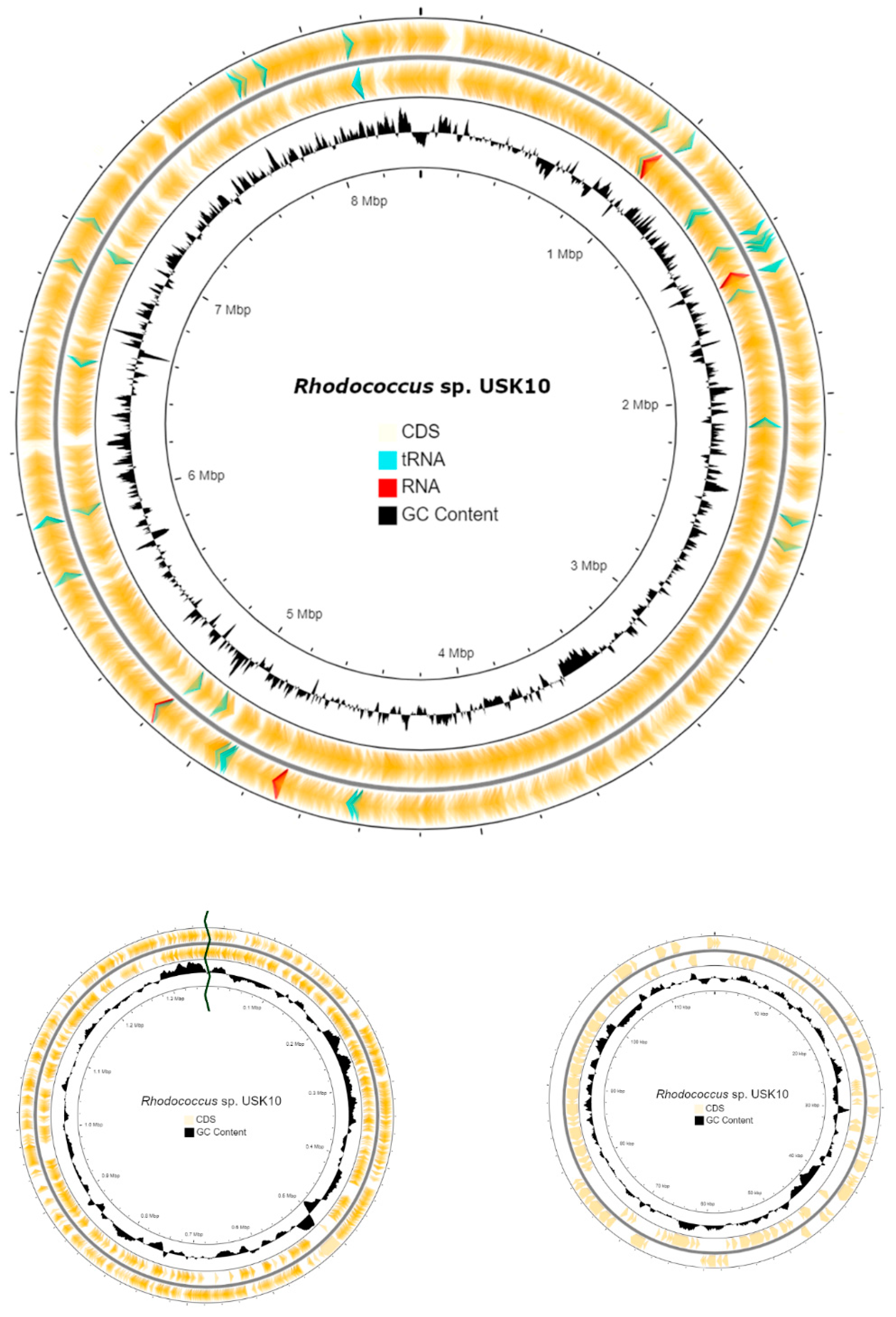
3.2. Genome Analysis
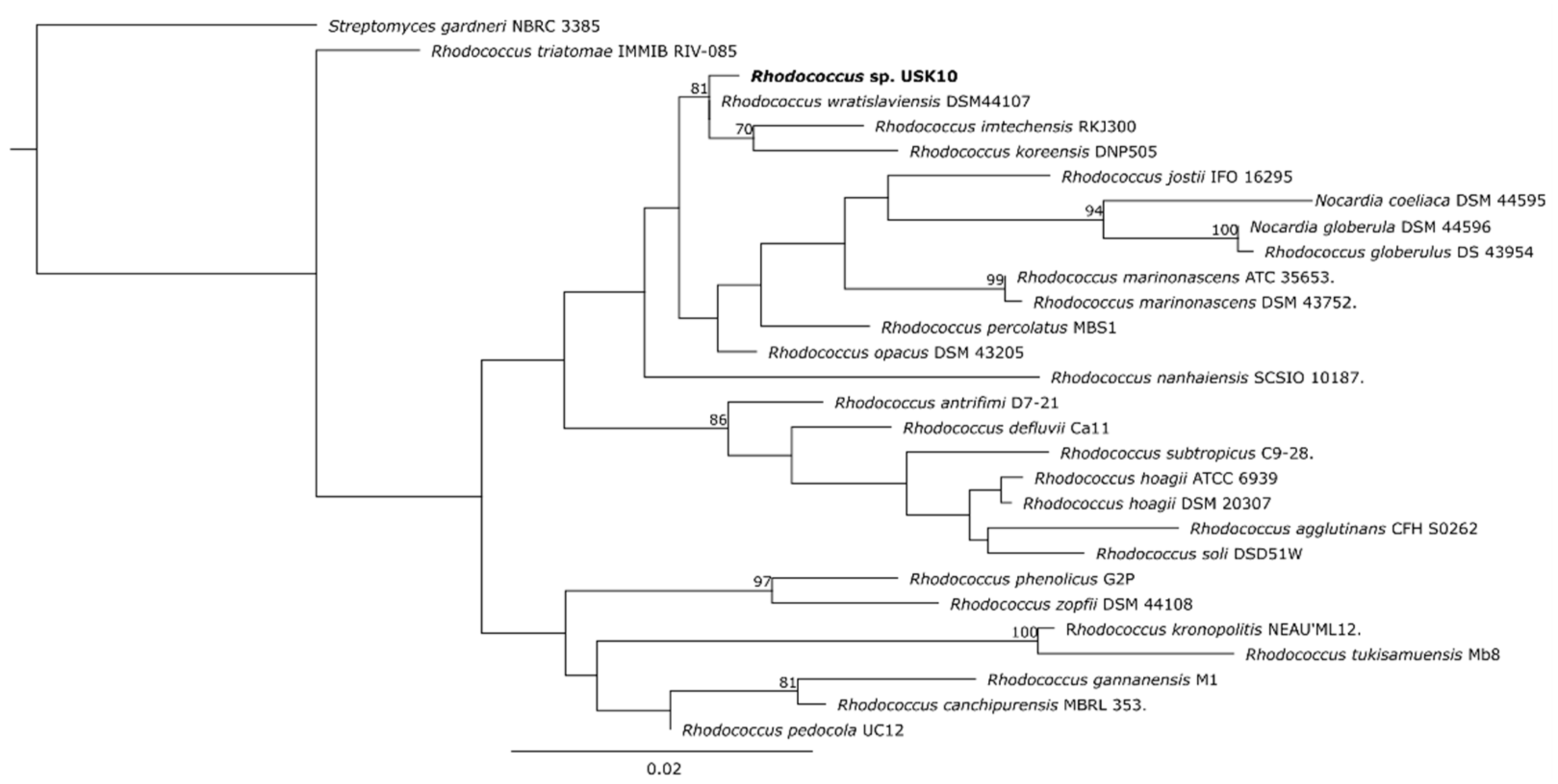
3.3. Phylogenetic Placement of Rhodococcus sp. USK10
3.4. Genome Annotation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, T.; Tsui, M.M.P.; Tan, C.J.; Ng, K.Y.; Guo, F.W.; Wang, L.H.; Chen, T.H.; Fan, T.Y.; Lam, P.K.S.; Murphy, M.B. Comparative Toxicities of Four Benzophenone Ultraviolet Filters to Two Life Stages of Two Coral Species. Sci. Total Environ. 2019, 651, 2391–2399. [Google Scholar] [CrossRef]
- Tsui, M.M.P.; Leung, H.W.; Wai, T.-C.; Yamashita, N.; Taniyasu, S.; Liu, W.; Lam, P.K.S.; Murphy, M.B. Occurrence, Distribution and Ecological Risk Assessment of Multiple Classes of UV Filters in Surface Waters from Different Countries. Water Res. 2014, 67, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Emnet, P.; Gaw, S.; Northcott, G.; Storey, B.; Graham, L. Personal Care Products and Steroid Hormones in the Antarctic Coastal Environment Associated with Two Antarctic Research Stations, McMurdo Station and Scott Base. Environ. Res. 2015, 136, 331–342. [Google Scholar] [CrossRef]
- Downs, C.A.; Kramarsky-Winter, E.; Segal, R.; Fauth, J.; Knutson, S.; Bronstein, O.; Ciner, F.R.; Jeger, R.; Lichtenfeld, Y.; Woodley, C.M.; et al. Toxicopathological Effects of the Sunscreen UV Filter, Oxybenzone (Benzophenone-3), on Coral Planulae and Cultured Primary Cells and Its Environmental Contamination in Hawaii and the US Virgin Islands. Arch. Environ. Contam. Toxicol. 2016, 70, 265–288. [Google Scholar] [CrossRef]
- Balázs, A.; Krifaton, C.; Orosz, I.; Szoboszlay, S.; Kovács, R.; Csenki, Z.; Urbányi, B.; Kriszt, B. Hormonal Activity, Cytotoxicity and Developmental Toxicity of UV Filters. Ecotoxicol. Environ. Saf. 2016, 131, 45–53. [Google Scholar] [CrossRef]
- DiNardo, J.C.; Downs, C.A. Dermatological and Environmental Toxicological Impact of the Sunscreen Ingredient Oxybenzone/Benzophenone-3. J. Cosmet. Dermatol. 2018, 17, 15–19. [Google Scholar] [CrossRef]
- Fitt, W.K.; Hofmann, D.K. The Effects of the UV-Blocker Oxybenzone (Benzophenone-3) on Planulae Swimming and Metamorphosis of the Scyphozoans Cassiopea xamachana and Cassiopea frondosa. Oceans 2020, 1, 174–180. [Google Scholar] [CrossRef]
- Kim, S.; Choi, K. Occurrences, Toxicities, and Ecological Risks of Benzophenone-3, a Common Component of Organic Sunscreen Products: A Mini-Review. Environ. Int. 2014, 70, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Ghazipura, M.; McGowan, R.; Arslan, A.; Hossain, T. Exposure to Benzophenone-3 and Reproductive Toxicity: A Systematic Review of Human and Animal Studies. Reprod. Toxicol. 2017, 73, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Geng, Z.; Pang, X.; Zhang, Y.; Wang, G.; Ji, J.; Li, X.; Guan, C. Isolation and Characterization of a Novel Benzophenone-3-Degrading Bacterium Methylophilus sp. Strain FP-6. Ecotoxicol. Environ. Saf. 2019, 186, 109780. [Google Scholar] [CrossRef]
- Fagervold, S.K.; Rohée, C.; Rodrigues, A.M.S.; Stien, D.; Lebaron, P. Efficient Degradation of the Organic UV Filter Benzophenone-3 by Sphingomonas wittichii Strain BP14P Isolated from WWTP Sludge. Sci. Total Environ. 2021, 758, 143674. [Google Scholar] [CrossRef]
- Vaser, R.; Šikić, M. Raven: A de Novo Genome Assembler for Long Reads. BioRxiv 2021. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaser, R.; Sović, I.; Nagarajan, N.; Šikić, M. Fast and Accurate de Novo Genome Assembly from Long Uncorrected Reads. Genome Res. 2017, 27, 737–746. [Google Scholar] [CrossRef] [Green Version]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate Paired Shotgun Read Merging via Overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-Suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum Likelihood-Based Phylogenetic Analyses with Thousands of Taxa and Mixed Models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A Standardized Bacterial Taxonomy Based on Genome Phylogeny Substantially Revises the Tree of Life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A Complete Domain-to-Species Taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A Toolkit to Classify Genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Resour. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchez, M.; Blanchet, D.; Vandecasteele, J.-P. The Microbiological Fate of Polycyclic Aromatic Hydrocarbons: Carbon and Oxygen Balances for Bacterial Degradation of Model Compounds. Appl. Microbiol. Biotechnol. 1996, 45, 556–561. [Google Scholar] [CrossRef]
- Liu, Y.-S.; Ying, G.-G.; Shareef, A.; Kookana, R.S. Biodegradation of the Ultraviolet Filter Benzophenone-3 under Different Redox Conditions. Environ. Toxicol. Chem. 2012, 31, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-M.; Lee, G.; Zoh, K.-D. Benzophenone-3 Degradation via UV/H2O2 and UV/Persulfate Reactions. Journal of Hazardous Materials 2021, 403, 123591. [Google Scholar] [CrossRef]
- Pan, X.; Yan, L.; Qu, R.; Wang, Z. Degradation of the UV-Filter Benzophenone-3 in Aqueous Solution Using Persulfate Activated by Heat, Metal Ions and Light. Chemosphere 2018, 196, 95–104. [Google Scholar] [CrossRef]
- Wang, Z.; Deb, A.; Srivastava, V.; Iftekhar, S.; Ambat, I.; Sillanpää, M. Investigation of Textural Properties and Photocatalytic Activity of PbO/TiO2 and Sb2O3/TiO2 towards the Photocatalytic Degradation Benzophenone-3 UV Filter. Sep. Purif. Technol. 2019, 228, 115763. [Google Scholar] [CrossRef]
- Kundu, D.; Hazra, C.; Chaudhari, A. Bioremediation Potential of Rhodococcus pyridinivorans NT2 in Nitrotoluene-Contaminated Soils: The Effectiveness of Natural Attenuation, Biostimulation and Bioaugmentation Approaches. Soil Sediment Contam. Int. J. 2016, 25, 637–651. [Google Scholar] [CrossRef]
- Martínková, L.; Uhnáková, B.; Pátek, M.; Nešvera, J.; Křen, V. Biodegradation Potential of the Genus rhodococcus. Environ. Int. 2009, 35, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Gűrtler, V.; Seviour, R.J. Systematics of Members of the Genus Rhodococcus (Zopf 1891) Emend Goodfellow et al. 1998. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Microbiology Monographs; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–28. ISBN 978-3-642-12937-7. [Google Scholar]
- Dib, J.R.; Wagenknecht, M.; Hill, R.T.; Farías, M.E.; Meinhardt, F. First Report of Linear Megaplasmids in the Genus Micrococcus. Plasmid 2010, 63, 40–45. [Google Scholar] [CrossRef]
- König, C.; Eulberg, D.; Gröning, J.; Lakner, S.; Seibert, V.; Kaschabek, S.R.; Schlömann, M. A Linear Megaplasmid, P1CP, Carrying the Genes for Chlorocatechol Catabolism of Rhodococcus opacus 1CP. Microbiology 2004, 150, 3075–3087. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Wishart, D.S. Circular Genome Visualization and Exploration Using CGView. Bioinformatics 2005, 21, 537–539. [Google Scholar] [CrossRef] [Green Version]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [Green Version]
- Zervas, A.; Aggerbeck, M.R.; Allaga, H.; Güzel, M.; Hendriks, M.; Jonuškienė, I.; Kedves, O.; Kupeli, A.; Lamovšek, J.; Mülner, P.; et al. Identification and Characterization of 33 Bacillus cereus sensu lato Isolates from Agricultural Fields from Eleven Widely Distributed Countries by Whole Genome Sequencing. Microorganisms 2020, 8, 2028. [Google Scholar] [CrossRef]
- Hennessy, R.C.; Dichmann, S.I.; Martens, H.J.; Zervas, A.; Stougaard, P. Serratia inhibens sp. nov., a New Antifungal Species Isolated from Potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2020, 70, 4204–4211. [Google Scholar] [CrossRef] [PubMed]
- Tropel, D.; van der Meer, J.R. Bacterial Transcriptional Regulators for Degradation Pathways of Aromatic Compounds. Microbiol. Mol. Biol. Rev. 2004, 68, 474–500. [Google Scholar] [CrossRef] [Green Version]
- Miller, T.R.; Delcher, A.L.; Salzberg, S.L.; Saunders, E.; Detter, J.C.; Halden, R.U. Genome sequence of the dioxin-mineralizing bacterium Sphingomonas wittichii RW1. J. Bacteriol. 2010, 192, 6101–6102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, M.; Yan, X.; He, J.; Qiu, J.; Chen, Q. Comparative genome analysis reveals 611 the evolution of chloroacetanilide herbicide mineralization in Sphingomonas wittichii DC-6. Arch. Microbiol. 2019, 201, 907–918. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, J.D.; Krüger, U.S.; Zervas, A.; Schostag, M.D.; Nielsen, T.K.; Aamand, J.; Hansen, L.H.; Ellegaard-Jensen, L. Complete Genome Sequence and Benzophenone-3 Mineralisation Potential of Rhodococcus sp. USK10, A Bacterium Isolated from Riverbank Sediment. Appl. Microbiol. 2022, 2, 104-112. https://doi.org/10.3390/applmicrobiol2010006
Martin JD, Krüger US, Zervas A, Schostag MD, Nielsen TK, Aamand J, Hansen LH, Ellegaard-Jensen L. Complete Genome Sequence and Benzophenone-3 Mineralisation Potential of Rhodococcus sp. USK10, A Bacterium Isolated from Riverbank Sediment. Applied Microbiology. 2022; 2(1):104-112. https://doi.org/10.3390/applmicrobiol2010006
Chicago/Turabian StyleMartin, Joseph Donald, Urse Scheel Krüger, Athanasios Zervas, Morten Dencker Schostag, Tue Kjærgaard Nielsen, Jens Aamand, Lars Hestbjerg Hansen, and Lea Ellegaard-Jensen. 2022. "Complete Genome Sequence and Benzophenone-3 Mineralisation Potential of Rhodococcus sp. USK10, A Bacterium Isolated from Riverbank Sediment" Applied Microbiology 2, no. 1: 104-112. https://doi.org/10.3390/applmicrobiol2010006
APA StyleMartin, J. D., Krüger, U. S., Zervas, A., Schostag, M. D., Nielsen, T. K., Aamand, J., Hansen, L. H., & Ellegaard-Jensen, L. (2022). Complete Genome Sequence and Benzophenone-3 Mineralisation Potential of Rhodococcus sp. USK10, A Bacterium Isolated from Riverbank Sediment. Applied Microbiology, 2(1), 104-112. https://doi.org/10.3390/applmicrobiol2010006





