Characterization of Gentisate 1,2-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and Its Stabilization by Immobilization on Nickel-Functionalized Magnetic Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Bacterial Strains and Media
2.3. Preparation of Sphe3 Cell-Free Extracts
2.4. Cloning and Heterologous Expression of the Gentisate 1,2-Dioxygenase Gene
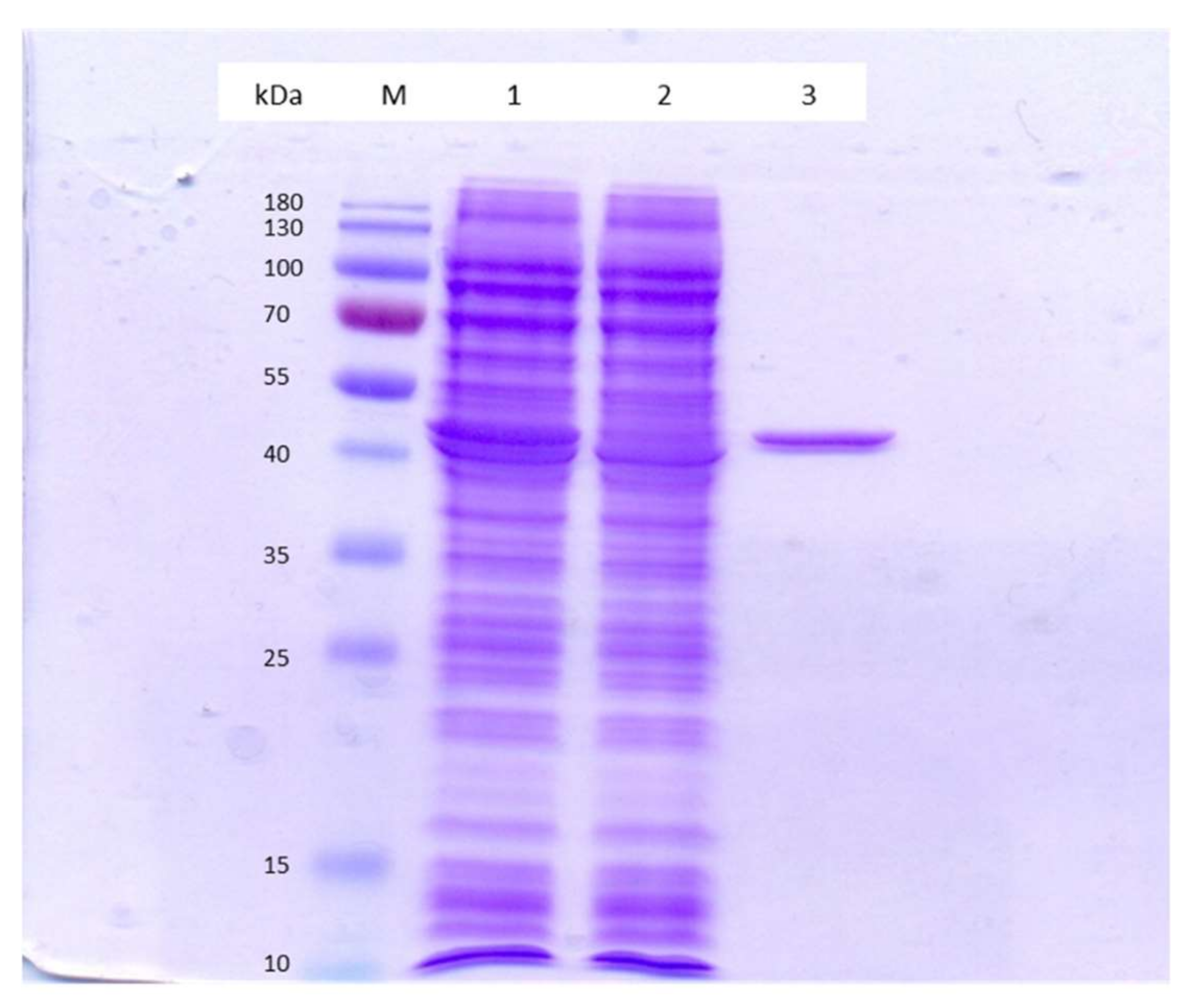
2.5. Crude Lysate Preparation and GDO Purification
2.6. Enzyme Assays, Kinetic Measurements
2.7. Synthesis of Ni2+-Polydopamine Magnetic Nanoparticles
2.8. Selective Immobilization of the Recombinant GDO on Ni2+-PDA-MNPs
2.9. Characterization of the Nanobiocatalyst
2.9.1. Fourier Transform Infrared Spectroscopy
2.9.2. Identification of Nanobiocatalyst Reaction Product
2.9.3. Determination of Apparent Kinetic Parameters
2.9.4. Thermal Stability of Free and Immobilized Biocatalyst
2.9.5. Storage Stability Studies
2.9.6. Reusability Studies of Immobilized Enzyme
3. Results and Discussion
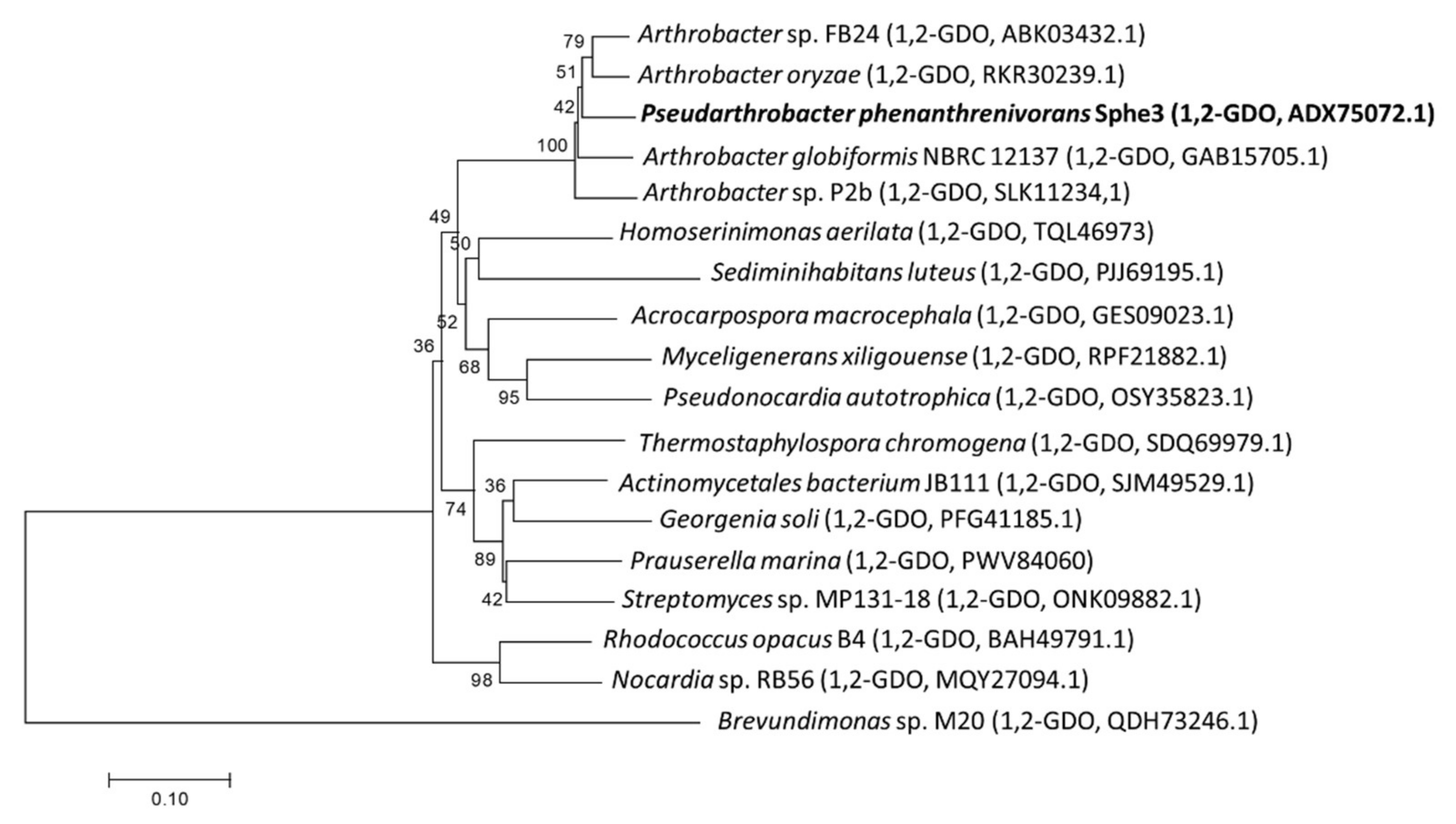
3.1. Phylogenetic Analysis, Cloning, and Overexpression
3.2. Enzyme Activity Assay and Characteristics of Purified GDO
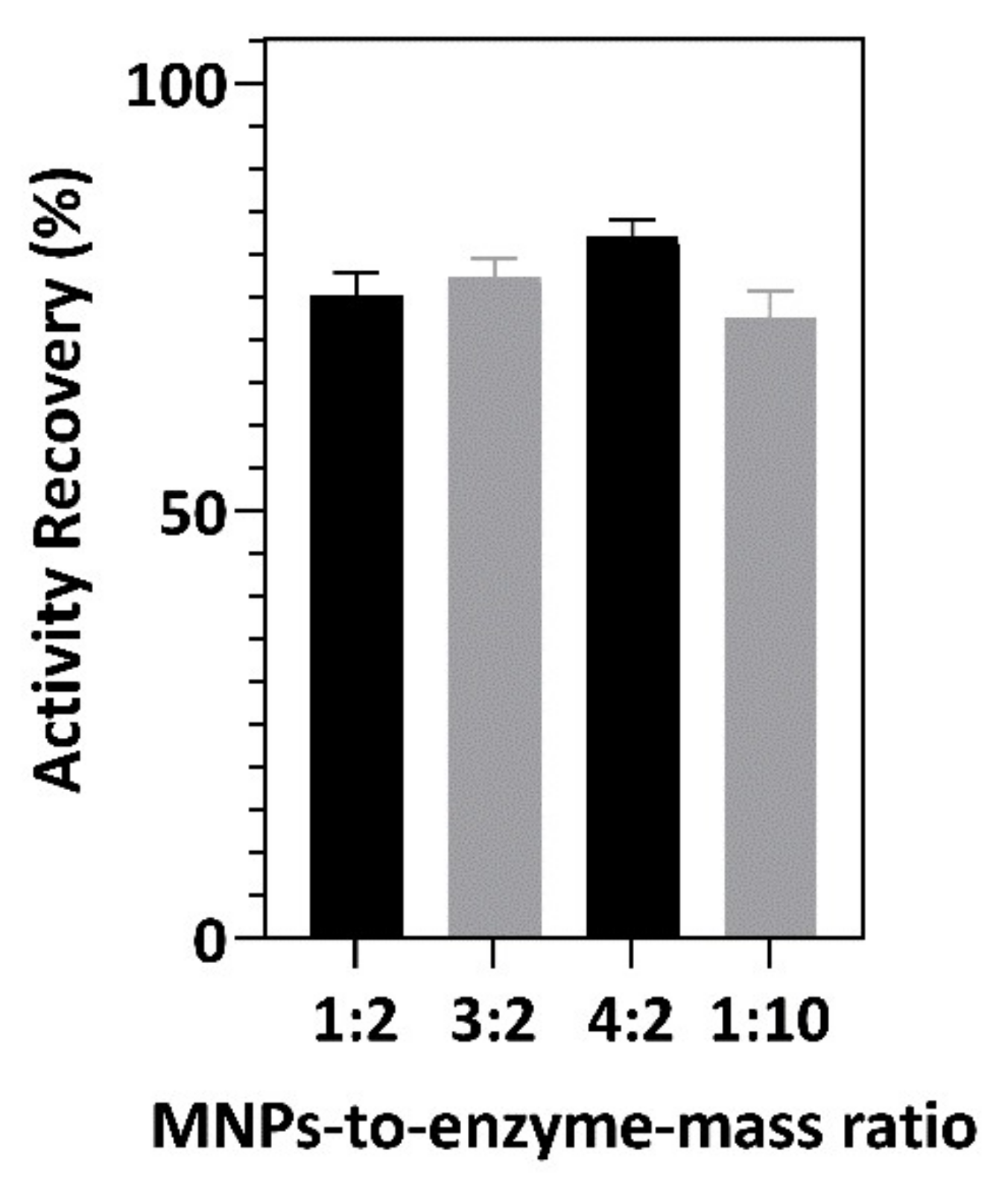
3.3. Immobilization of Recombinant Gentisate 1,2-Dioxygenase
3.4. Characterization of the Nanobiocatalyst
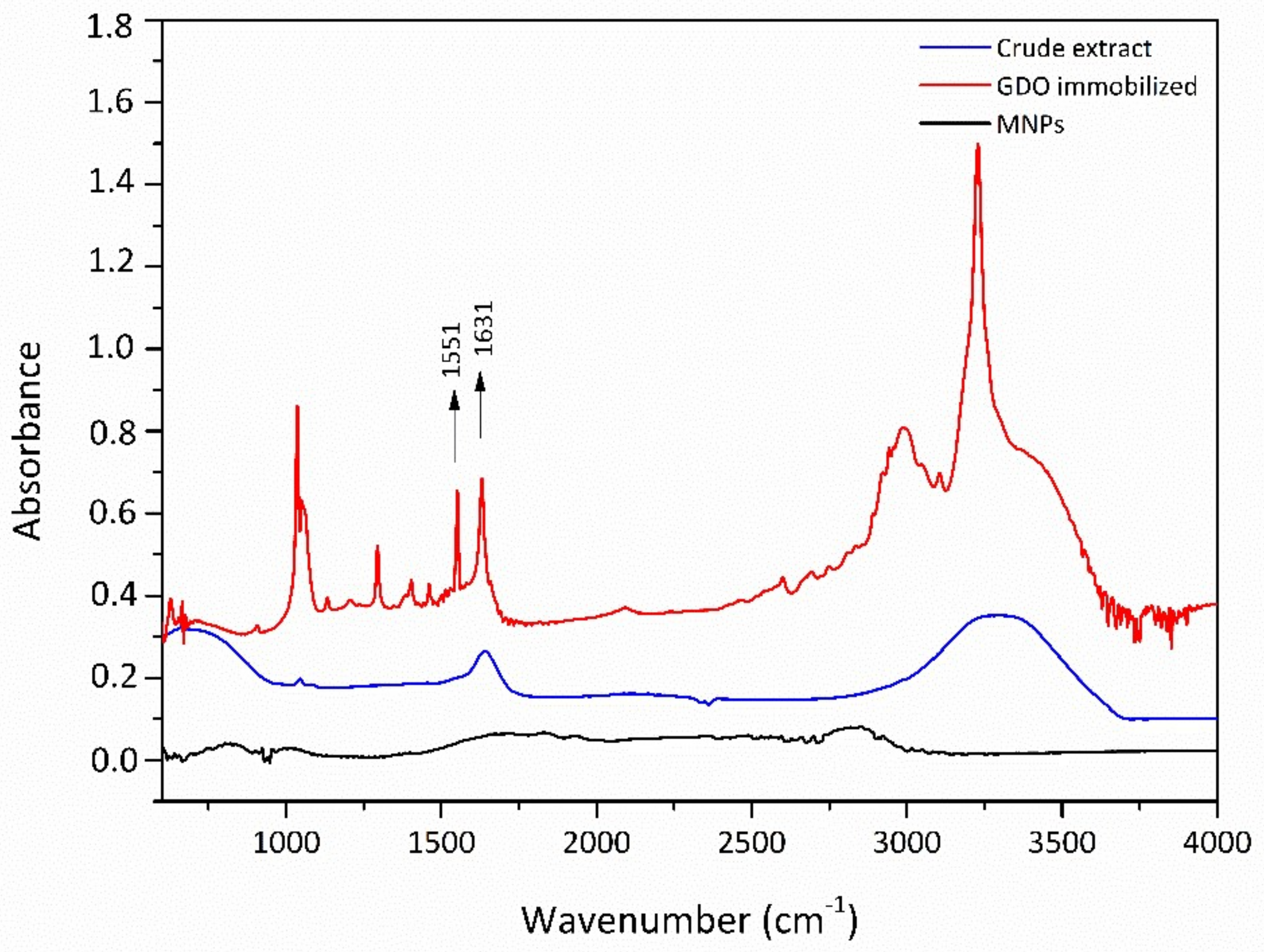
3.4.1. Fourier Transform Infrared Spectroscopy
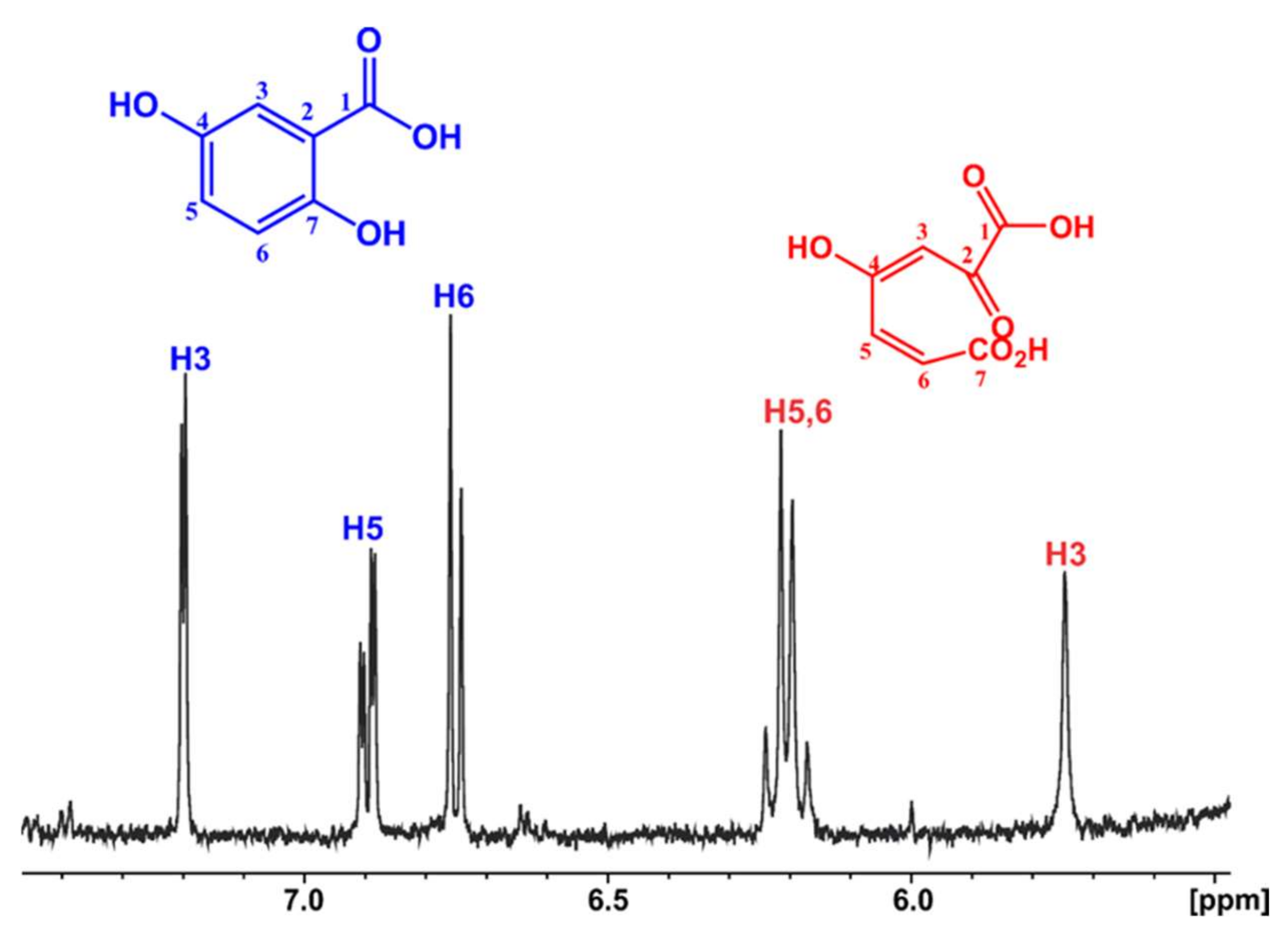
3.4.2. Identification of the Nanobiocatalyst Reaction Product
3.4.3. Comparison of Free and Immobilized GDO Kinetic Parameters
3.5. Stability of Free and Immobilized Biocatalyst
3.5.1. pH and Temperature Stability Studies
3.5.2. Thermal Stability Studies
3.5.3. Storage Stability Studies
3.5.4. Reusability Studies of Immobilized Enzyme
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vaillancourt, F.H.; Bolin, J.T.; Eltis, L.D. The Ins and Outs of Ring-Cleaving Dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006, 41, 241–267. [Google Scholar] [CrossRef] [PubMed]
- Lack, L. The Enzymic Oxidation of Gentisic Acid. Biochim. Biophys. Acta 1959, 34, 117–123. [Google Scholar] [CrossRef]
- Fetzner, S. Ring-Cleaving Dioxygenases with a Cupin Fold. Appl. Environ. Microbiol. 2012, 78, 2505–2514. [Google Scholar] [CrossRef] [PubMed]
- Subbotina, N.M.; Chernykh, A.M.; Taranov, A.I.; Shebanova, A.D.; Moiseeva, O.V.; Ferraroni, M.; Kolomytseva, M.P. Gentisate 1,2-Dioxygenase from the Gram-Positive Bacteria Rhodococcus opacus 1CP: Identical Active Sites vs. Different Substrate Selectivities. Biochimie 2021, 180, 90–103. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, S.; Guo, L. Novel Gene Encoding 5-Aminosalicylate 1,2-Dioxygenase from Comamonas sp. Strain QT12 and Catalytic Properties of the Purified Enzyme. J. Bacteriol. 2018, 200, e00395-17. [Google Scholar] [CrossRef]
- Adams, M.A.; Singh, V.K.; Keller, B.O.; Jia, Z. Structural and Biochemical Characterization of Gentisate 1,2-Dioxygenase from Escherichia coli O157:H7. Mol. Microbiol. 2006, 61, 1469–1484. [Google Scholar] [CrossRef]
- Feng, J.; Che, Y.; Milse, J.; Yin, Y.-J.; Liu, L.; Rückert, C.; Shen, X.-H.; Qi, S.-W.; Kalinowski, J.; Liu, S.-J. The Gene Ncgl2918 Encodes a Novel Maleylpyruvate Isomerase That Needs Mycothiol as Cofactor and Links Mycothiol Biosynthesis and Gentisate Assimilation in Corynebacterium glutamicum. J. Biol. Chem. 2006, 281, 10778–10785. [Google Scholar] [CrossRef]
- Di Gennaro, P.; Terreni, P.; Masi, G.; Botti, S.; de Ferra, F.; Bestetti, G. Identification and Characterization of Genes Involved in Naphthalene Degradation in Rhodococcus opacus R7. Appl. Microbiol. Biotechnol. 2010, 87, 297–308. [Google Scholar] [CrossRef]
- Grund, E.; Denecke, B.; Eichenlaub, R. Naphthalene Degradation via Salicylate and Gentisate by Rhodococcus sp. Strain B4. Appl. Environ. Microbiol. 1992, 58, 1874–1877. [Google Scholar] [CrossRef]
- Jones, D.C.N.; Cooper, R.A. Catabolism of 3-Hydroxybenzoate by the Gentisate Pathway in Klebsiella pneumoniae M5a1. Arch. Microbiol. 1990, 154, 489–495. [Google Scholar] [CrossRef]
- Ladd, J.N. Oxidation of Anthranilic Acid by a Species of Achromobacter Isolated from Soil. Nature 1962, 194, 1099–1100. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-T.; Xu, Y.; Liu, H.; Luo, S.; Yin, Y.-J.; Liu, S.-J.; Zhou, N.-Y. Functional Characterization of a Gene Cluster Involved in Gentisate Catabolism in Rhodococcus sp. Strain NCIMB 12038. Appl. Microbiol. Biotechnol. 2011, 90, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.T.; Zhou, N.Y. Novel L-Cysteine-Dependent Maleylpyruvate Isomerase in the Gentisate Pathway of Paenibacillus sp. Strain NyZ101. J. Bacteriol. 2012, 194, 3987–3994. [Google Scholar] [CrossRef] [PubMed]
- Ohmoto, T.; Sakai, K.; Hamada, N.; Ohe, T. Salicylic Acid Metabolism through a Gentisate Pathway by Pseudomonas sp. TA-2. Agric. Biol. Chem. 1991, 55, 1733–1737. [Google Scholar] [CrossRef][Green Version]
- Feng, Y.; Khoo, H.E.; Laa Poh, C.; Poh, C.L. Purification and Characterization of Gentisate 1,2-Dioxygenases from Pseudomonas alcaligenes NCIB 9867 and Pseudomonas putida NCIB 9869. Appl. Environ. Microbiol. 1999, 65, 946–950. [Google Scholar] [CrossRef]
- Harpel, M.R.; Lipscomb, J.D. Gentisate 1,2-Dioxygenase from Pseudomonas. Purification, Characterization, and Comparison of the Enzymes from Pseudomonas testosteroni and Pseudomonas acidovorans. J. Biol. Chem. 1990, 265, 6301–6311. [Google Scholar] [CrossRef]
- Hirano, S.; Morikawa, M.; Takano, K.; Imanaka, T.; Kanaya, S. Gentisate 1,2-Dioxygenase from Xanthobacter polyaromaticivorans 127W. Biosci. Biotechnol. Biochem. 2007, 71, 192–199. [Google Scholar] [CrossRef]
- Werwath, J.; Arfmann, H.A.; Pieper, D.H.; Timmis, K.N.; Wittich, R.M. Biochemical and Genetic Characterization of a Gentisate 1,2-Dioxygenase from Sphingomonas sp. Strain RW5. J. Bacteriol. 1998, 180, 4171–4176. [Google Scholar] [CrossRef]
- Liu, D.; Zhu, T.; Fan, L.; Quan, J.; Guo, H.; Ni, J. Identification of a Novel Gentisate 1,2-Dioxygenase from Silicibacter pomeroyi. Biotechnol. Lett. 2007, 29, 1529–1535. [Google Scholar] [CrossRef]
- Lee, H.J.; Kim, J.M.; Lee, S.H.; Park, M.; Lee, K.; Madsen, E.L.; Jeon, C.O. Gentisate 1,2-Dioxygenase, in the Third Naphthalene Catabolic Gene Cluster of Polaromonas naphthalenivorans CJ2, Has a Role in Naphthalene Degradation. Microbiology 2011, 157, 2891–2903. [Google Scholar] [CrossRef]
- Chen, J.; Li, W.; Wang, M.; Zhu, G.; Liu, D.; Sun, F.; Hao, N.; Li, X.; Rao, Z.; Zhang, X.C. Crystal Structure and Mutagenic Analysis of GDOsp, a Gentisate 1,2-Dioxygenase from Silicibacter pomeroyi. Protein Sci. 2008, 17, 1362–1373. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhu, S.; Ni, J. Purification and Characterisation of a Gentisate 1,2-Dioxygenase from Ralstonia solanacearum GMI 1000. Ann. Microbiol. 2007, 57, 307–312. [Google Scholar] [CrossRef]
- Eppinger, E.; Stolz, A. Expansion of the Substrate Range of the Gentisate 1,2-Dioxygenase from Corynebacterium glutamicum for the Conversion of Monohydroxylated Benzoates. Protein Eng. Des. Sel. 2017, 30, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Suemori, A.; Kurane, R.; Tomizuka, N. Purification and Properties of Gentisate 1,2-Dioxygenase from Rhodococcus erythropolis S-1. Biosci. Biotechnol. Biochem. 1993, 57, 1781–1783. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Wojcieszynska, D. Immobilization as a Strategy for Improving Enzyme Properties—Application to Oxidoreductases. Molecules 2014, 19, 8995–9018. [Google Scholar] [CrossRef]
- Rodrigues, R.C.; Berenguer-Murcia, Á.; Carballares, D.; Morellon-Sterling, R.; Fernandez-Lafuente, R. Stabilization of Enzymes via Immobilization: Multipoint Covalent Attachment and Other Stabilization Strategies. Biotechnol. Adv. 2021, 52, 107821. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R. Stabilization of Multimeric Enzymes: Strategies to Prevent Subunit Dissociation. Enzym. Microb. Technol. 2009, 45, 405–418. [Google Scholar] [CrossRef]
- Bilal, M.; Asgher, M.; Cheng, H.; Yan, Y.; Iqbal, H.M.N. Multi-Point Enzyme Immobilization, Surface Chemistry, and Novel Platforms: A Paradigm Shift in Biocatalyst Design. Crit. Rev. Biotechnol. 2019, 39, 202–219. [Google Scholar] [CrossRef]
- Pessela, B.C.C.; Mateo, C.; Fuentes, M.; Vian, A.; García, J.L.; Carrascosa, A.V.; Guisán, J.M.; Fernández-Lafuente, R. Stabilization of a Multimeric Beta-Galactosidase from Thermus sp. Strain T2 by Immobilization on Novel Heterofunctional Epoxy Supports plus Aldehyde-Dextran Cross-Linking. Biotechnol. Prog. 2004, 20, 388–392. [Google Scholar] [CrossRef]
- Arana-Peña, S.; Rios, N.S.; Carballares, D.; Mendez-Sanchez, C.; Lokha, Y.; Gonçalves, L.R.B.; Fernandez-Lafuente, R. Effects of Enzyme Loading and Immobilization Conditions on the Catalytic Features of Lipase From Pseudomonas fluorescens Immobilized on Octyl-Agarose Beads. Front. Bioeng. Biotechnol. 2020, 8, 36. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of Enzyme Activity, Stability and Selectivity via Immobilization Techniques. Enzym. Microb. Technol. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Silva, A.S.; Jacques, R.J.S.; Andreazza, R.; Bento, F.M.; Roesch, L.F.W.; Camargo, F.A.O. Properties of Catechol 1, 2-Dioxygenase in the Cell Free Extract and Immobilized Extract of Mycobacterium fortuitum. Braz. J. Microbiol. 2013, 44, 291–297. [Google Scholar] [CrossRef] [PubMed]
- Suma, Y.; Kang, C.S.; Kim, H.S. Noncovalent and Covalent Immobilization of Oxygenase on Single-Walled Carbon Nanotube for Enzymatic Decomposition of Aromatic Hydrocarbon Intermediates. Environ. Sci. Pollut. Res. 2016, 23, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Suma, Y.; Lim, H.; Kwean, O.S.; Cho, S.; Yang, J.; Kim, Y.; Kang, C.S.; Kim, H.S. Enzymatic Degradation of Aromatic Hydrocarbon Intermediates Using a Recombinant Dioxygenase Immobilized onto Surfactant-Activated Carbon Nanotube. Bioresour. Technol. 2016, 210, 117–122. [Google Scholar] [CrossRef]
- Das, R.; Hamid, S.B.A.; Annuar, M.S.M. Highly Efficient and Stable Novel NanoBiohybrid Catalyst to Avert 3,4-Dihydroxybenzoic Acid Pollutant in Water. Sci. Rep. 2016, 6, 33572. [Google Scholar] [CrossRef]
- Zhang, L.-S.; Fang, Y.; Zhou, Y.; Ye, B.-C. Improvement of the Stabilization and Activity of Protocatechuate 3,4-Dioxygenase Isolated from Rhizobium sp. LMB-1 and Immobilized on Fe3O4 Nanoparticles. Appl. Biochem. Biotechnol. 2017, 183, 1035–1048. [Google Scholar] [CrossRef]
- Kwean, O.S.; Cho, S.Y.; Yang, J.W.; Cho, W.; Park, S.; Lim, Y.; Shin, M.C.; Kim, H.S.; Park, J.; Kim, H.S. 4-Chlorophenol Biodegradation Facilitator Composed of Recombinant Multi-Biocatalysts Immobilized onto Montmorillonite. Bioresour. Technol. 2018, 259, 268–275. [Google Scholar] [CrossRef]
- Gkantzou, E.; Chatzikonstantinou, A.V.; Fotiadou, R.; Giannakopoulou, A.; Patila, M.; Stamatis, H. Trends in the Development of Innovative Nanobiocatalysts and Their Application in Biocatalytic Transformations. Biotechnol. Adv. 2021, 51, 107738. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of Different Enzyme Immobilization Strategies to Improve Enzyme Performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Giannakopoulou, A.; Patila, M.; Spyrou, K.; Chalmpes, N.; Zarafeta, D.; Skretas, G.; Gournis, D.; Stamatis, H. Development of a Four-Enzyme Magnetic Nanobiocatalyst for Multi-Step Cascade Reactions. Catalysts 2019, 9, 995. [Google Scholar] [CrossRef]
- Yang, J.; Ni, K.; Wei, D.; Ren, Y. One-Step Purification and Immobilization of His-Tagged Protein via Ni2+-Functionalized Fe3O4@polydopamine Magnetic Nanoparticles. Biotechnol. Bioprocess Eng. 2015, 20, 901–907. [Google Scholar] [CrossRef]
- Porath, J. Immobilized Metal Ion Affinity Chromatography. Protein Expr. Purif. 1992, 3, 263–281. [Google Scholar] [CrossRef]
- Mateo, C.; Fernández-Lorente, G.; Cortés, E.; Garcia, J.L.; Fernández-Lafuente, R.; Guisan, J.M. One-Step Purification, Covalent Immobilization, and Additional Stabilization of Poly-His-Tagged Proteins Using Novel Heterofunctional Chelate-Epoxy Supports. Biotechnol. Bioeng. 2001, 76, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Chang, J.H. Facile and High-Efficient Immobilization of Histidine-Tagged Multimeric Protein G on Magnetic Nanoparticles. Nanoscale Res. Lett. 2014, 9, 664. [Google Scholar] [CrossRef]
- Ball, V. Polydopamine Nanomaterials: Recent Advances in Synthesis Methods and Applications. Front. Bioeng. Biotechnol. 2018, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Han, S.; Li, X.; He, J.; Secundo, F.; Liang, H. Co-Immobilization of Two-Component Hydroxylase Monooxygenase by Functionalized Magnetic Nanoparticles for Preserving High Catalytic Activity and Enhancing Enzyme Stabilty. Int. J. Biol. Macromol. 2020, 164, 3163–3170. [Google Scholar] [CrossRef]
- Wang, W.; Wang, D.I.C.; Li, Z. Facile Fabrication of Recyclable and Active Nanobiocatalyst: Purification and Immobilization of Enzyme in One Pot with Ni-NTA Functionalized Magnetic Nanoparticle. Chem. Commun. 2011, 47, 8115–8117. [Google Scholar] [CrossRef]
- Sung, H.K.; Jeyakumar, M.; Katzenellenbogen, J.A. Dual-Mode Fluorophore-Doped Nickel Nitrilotriacetic Acid-Modified Silica Nanoparticles Combine Histidine-Tagged Protein Purification with Site-Specific Fluorophore Labeling. J. Am. Chem. Soc. 2007, 129, 13254–13264. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, Z.; Deng, C.; Zhang, X.; Yang, P. Facile Synthesis of Ti4+-Immobilized Fe3O4@polydopamine Core–Shell Microspheres for Highly Selective Enrichment of Phosphopeptides. Chem. Commun. 2013, 49, 5055. [Google Scholar] [CrossRef]
- Kallimanis, A.; Kavakiotis, K.; Perisynakis, A.; Spröer, C.; Pukall, R.; Drainas, C.; Koukkou, A.I. Arthrobacter phenanthrenivorans Sp. Nov., to Accommodate the Phenanthrene-Degrading Bacterium Arthrobacter sp. Strain Sphe3. Int. J. Syst. Evol. Microbiol. 2009, 59, 275–279. [Google Scholar] [CrossRef]
- Vandera, E.; Kavakiotis, K.; Kallimanis, A.; Kyrpides, N.C.; Drainas, C.; Koukkou, A. Heterologous Expression and Characterization of Two 1-Hydroxy-2-Naphthoic Acid Dioxygenases from Arthrobacter phenanthrenivorans. Appl. Environ. Microbiol. 2012, 78, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Tsagogiannis, E.; Vandera, E.; Primikyri, A.; Asimakoula, S.; Tzakos, A.G.; Gerothanassis, I.P.; Koukkou, A.-I. Characterization of Protocatechuate 4,5-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and In Situ Reaction Monitoring in the NMR Tube. Int. J. Mol. Sci. 2021, 22, 9647. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- William, S.; Feil, H.; Copeland, A. Bacterial Genomic DNA Isolation Using CTAB. Sigma 2012, 50, 6876. [Google Scholar]
- Hanahan, D. Studies on Transformation of Escherichia coli with Plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Chung, C.T.; Miller, R.H. A Rapid and Convenient Method for the Preparation and Storage of Competent Bacterial Cells. Nucleic Acids Res. 1988, 16, 3580. [Google Scholar] [CrossRef]
- Altschul, S. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Sambrook, J.F.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2003; Volumes 1–3. [Google Scholar]
- Boudrant, J.; Woodley, J.M.; Fernandez-Lafuente, R. Parameters Necessary to Define an Immobilized Enzyme Preparation. Process Biochem. 2020, 90, 66–80. [Google Scholar] [CrossRef]
- Dias Gomes, M.; Woodley, J.M. Considerations When Measuring Biocatalyst Performance. Molecules 2019, 24, 3573. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Suárez, M.; Ferrer, E.; Martín, M. Purification and Biochemical Characterization of Gentisate 1,2-Dioxygenase from Klebsiella pneumoniae M5a1. FEMS Microbiol. Lett. 1996, 143, 89–95. [Google Scholar] [CrossRef]
- Kiemer, P.; Tshisuaka, B.; Fetzner, S.; Lingens, F. Degradation of Benzoate via Benzoyl-Coenzyme A and Gentisate by Bacillus stearothermophilus PK1, and Purification of Gentisate 1,2-Dioxygenase. Biol. Fertil. Soils 1996, 23, 307–313. [Google Scholar] [CrossRef]
- Kotake, T.; Matsuzawa, J.; Suzuki-Minakuchi, C.; Okada, K.; Nojiri, H.; Iwata, K. Purification and Partial Characterization of the Extradiol Dioxygenase, 2′-Carboxy-2,3-Dihydroxybiphenyl 1,2-Dioxygenase, in the Fluorene Degradation Pathway from Rhodococcus sp. Strain DFA3. Biosci. Biotechnol. Biochem. 2016, 80, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Harpel, M.R.; Lipscomb, J.D. Gentisate 1,2-Dioxygenase from Pseudomonas. Substrate Coordination to Active Site Fe2+ and Mechanism of Turnover. J. Biol. Chem. 1990, 265, 22187–22196. [Google Scholar] [CrossRef]
- Yeo, C.C.; Tan, C.L.; Gao, X.; Zhao, B.; Poh, C.L. Characterization of HbzE-Encoded Gentisate 1,2-Dioxygenase from Pseudomonas alcaligenes NCIMB 9867. Res. Microbiol. 2007, 158, 608–616. [Google Scholar] [CrossRef]
- Huang, L.; Hu, H.; Tang, H.; Liu, Y.; Xu, P.; Shi, J.; Lin, K.; Luo, Q.; Cui, C.; Luo, S.; et al. Identification and Characterization of a Novel Gentisate 1,2-Dioxygenase Gene from a Halophilic Martelella Strain. Sci. Rep. 2015, 5, 14307. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Krysiak, M.; Wojcieszyńska, D. Degradation Potential of Protocatechuate 3,4-Dioxygenase from Crude Extract of Stenotrophomonas maltophilia Strain KB2 Immobilized in Calcium Alginate Hydrogels and on Glyoxyl Agarose. Biomed. Res. Int. 2014, 2014, 138768. [Google Scholar] [CrossRef]
- Barry, K.P.; Taylor, E.A. Characterizing the Promiscuity of LigAB, a Lignin Catabolite Degrading Extradiol Dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry 2013, 52, 6724–6736. [Google Scholar] [CrossRef]
- Ha, E.J.; Kim, B.S.; Park, E.K.; Song, K.W.; Lee, S.G.; An, S.S.A.; Paik, H.-j. Site-Specific Reversible Immobilization and Purification of His-Tagged Protein on Poly(2-Acetamidoacrylic Acid) Hydrogel Beads. Polym. Adv. Technol. 2013, 24, 75–80. [Google Scholar] [CrossRef]
- Vahidi, A.K.; Yang, Y.; Ngo, T.P.N.; Li, Z. Simple and Efficient Immobilization of Extracellular His-Tagged Enzyme Directly from Cell Culture Supernatant as Active and Recyclable Nanobiocatalyst: High-Performance Production of Biodiesel from Waste Grease. ACS Catal. 2015, 5, 3157–3161. [Google Scholar] [CrossRef]
- Zhou, L.J.; Li, R.F.; Li, X.Y.; Zhang, Y.W. One-step Selective Affinity Purification and Immobilization of His-tagged Enzyme by Recyclable Magnetic Nanoparticles. Eng. Life Sci. 2021, 21, 364. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Zhao, Y.; Rasheed, T.; Iqbal, H.M.N. Magnetic Nanoparticles as Versatile Carriers for Enzymes Immobilization: A Review. Int. J. Biol. Macromol. 2018, 120, 2530–2544. [Google Scholar] [CrossRef] [PubMed]
- Giannakopoulou, A.; Chatzikonstantinou, A.V.; Chalmpes, N.; Tsapara, G.; Gournis, D.; Polydera, A.C.; Stamatis, H. Development of a Novel Bi-Enzymatic Nanobiocatalyst for the Efficient Bioconversion of Oleuropein to Hydroxytyrosol. Catalysts 2021, 11, 749. [Google Scholar] [CrossRef]
- Pathmamanoharan, C.; Wijkens, P.; Grove, D.M.; Philipse, A.P. Paramagnetic Silica Particles: Synthesis and Grafting of a Silane Coupling Agent Containing Nickel Ions onto Colloidal Silica Particles. Langmuir 1996, 12, 4372–4377. [Google Scholar] [CrossRef]
- Ji, Y.; Yang, X.; Ji, Z.; Zhu, L.; Ma, N.; Chen, D.; Jia, X.; Tang, J.; Cao, Y. DFT-Calculated IR Spectrum Amide I, II, and III Band Contributions of N -Methylacetamide Fine Components. ACS Omega 2020, 5, 8572–8578. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A Knowledgebase for the Human Metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef]
- Jiao, M.; He, J.; Sun, S.; Vriesekoop, F.; Yuan, Q.; Liu, Y.; Liang, H. Fast Immobilization of Human Carbonic Anhydrase II on Ni-Based Metal-Organic Framework Nanorods with High Catalytic Performance. Catalysts 2020, 10, 401. [Google Scholar] [CrossRef]
- Zhang, C.; Luo, S.; Chen, W. Activity of Catalase Adsorbed to Carbon Nanotubes: Effects of Carbon Nanotube Surface Properties. Talanta 2013, 113, 142–147. [Google Scholar] [CrossRef]
- Di Nardo, G.; Roggero, C.; Campolongo, S.; Valetti, F.; Trotta, F.; Gilardi, G. Catalytic Properties of Catechol 1,2-Dioxygenase from Acinetobacter radioresistens S13 Immobilized on Nanosponges. Dalt. Trans. 2009, 2, 6507. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.L.; Chaudhary, R.; Tsuzuki, T.; Barrow, C.J.; Puri, M. Immobilization of β-Glucosidase on a Magnetic Nanoparticle Improves Thermostability: Application in Cellobiose Hydrolysis. Bioresour. Technol. 2013, 135, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yuan, S.; Liu, Q.; Yan, D.; Wang, Y.; Gao, L.; Han, J.; Shi, H. Synchronized Purification and Immobilization of His-Tagged β-Glucosidase via Fe3O4/PMG Core/Shell Magnetic Nanoparticles. Sci. Rep. 2017, 7, 41741. [Google Scholar] [CrossRef] [PubMed]
- Alemzadeh, I.; Nejati, S. Phenols Removal by Immobilized Horseradish Peroxidase. J. Hazard. Mater. 2009, 166, 1082–1086. [Google Scholar] [CrossRef] [PubMed]
- Schnell, S.; Hanson, S.M. A Test for Measuring the Effects of Enzyme Inactivation. Biophys. Chem. 2007, 125, 269–274. [Google Scholar] [CrossRef]
- Kalogeris, E.; Sanakis, Y.; Mamma, D.; Christakopoulos, P.; Kekos, D.; Stamatis, H. Properties of Catechol 1,2-Dioxygenase from Pseudomonas putida Immobilized in Calcium Alginate Hydrogels. Enzyme Microb. Technol. 2006, 39, 1113–1121. [Google Scholar] [CrossRef]
- Wojcieszyńska, D.; Hupert-Kocurek, K.; Jankowska, A.; Guzik, U. Properties of Catechol 2,3-Dioxygenase from Crude Extract of Stenotrophomonas maltophilia Strain KB2 Immobilized in Calcium Alginate Hydrogels. Biochem. Eng. J. 2012, 66, 1–7. [Google Scholar] [CrossRef]
- Verma, M.L.; Barrow, C.J.; Kennedy, J.F.; Puri, M. Immobilization of β-d-Galactosidase from Kluyveromyces lactis on Functionalized Silicon Dioxide Nanoparticles: Characterization and Lactose Hydrolysis. Int. J. Biol. Macromol. 2012, 50, 432–437. [Google Scholar] [CrossRef]
- Darwesh, O.M.; Matter, I.A.; Eida, M.F. Development of Peroxidase Enzyme Immobilized Magnetic Nanoparticles for Bioremediation of Textile Wastewater Dye. J. Environ. Chem. Eng. 2019, 7, 102805. [Google Scholar] [CrossRef]





| Concentration (μM) | Relative Activity (%) | |
|---|---|---|
| EDTA | o-Phenanthroline | |
| 0 | 100 | 100 |
| 5 | 95 | 41 |
| 10 | 50 | 10 |
| 15 | 48 | 4 |
| 20 | 46 | 0 |
| 25 | 44 | N/A |
| 30 | 40 | N/A |
| Enzyme | Km (μM) | Vmax (mM·s−1) |
|---|---|---|
| GDO free form | 25.9 ± 4.4 | 1.2 ± 0.1 |
| GDO immobilized | 82.5 ± 14.2 | 0.03 ± 0.002 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asimakoula, S.; Giannakopoulou, A.; Lappa, E.; Tsagogiannis, E.; Primikyri, A.; Stamatis, H.; Koukkou, A.-I. Characterization of Gentisate 1,2-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and Its Stabilization by Immobilization on Nickel-Functionalized Magnetic Nanoparticles. Appl. Microbiol. 2022, 2, 113-132. https://doi.org/10.3390/applmicrobiol2010007
Asimakoula S, Giannakopoulou A, Lappa E, Tsagogiannis E, Primikyri A, Stamatis H, Koukkou A-I. Characterization of Gentisate 1,2-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and Its Stabilization by Immobilization on Nickel-Functionalized Magnetic Nanoparticles. Applied Microbiology. 2022; 2(1):113-132. https://doi.org/10.3390/applmicrobiol2010007
Chicago/Turabian StyleAsimakoula, Stamatia, Archontoula Giannakopoulou, Eirini Lappa, Epameinondas Tsagogiannis, Alexandra Primikyri, Haralambos Stamatis, and Anna-Irini Koukkou. 2022. "Characterization of Gentisate 1,2-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and Its Stabilization by Immobilization on Nickel-Functionalized Magnetic Nanoparticles" Applied Microbiology 2, no. 1: 113-132. https://doi.org/10.3390/applmicrobiol2010007
APA StyleAsimakoula, S., Giannakopoulou, A., Lappa, E., Tsagogiannis, E., Primikyri, A., Stamatis, H., & Koukkou, A.-I. (2022). Characterization of Gentisate 1,2-Dioxygenase from Pseudarthrobacter phenanthrenivorans Sphe3 and Its Stabilization by Immobilization on Nickel-Functionalized Magnetic Nanoparticles. Applied Microbiology, 2(1), 113-132. https://doi.org/10.3390/applmicrobiol2010007






_Stamatis.png)


