Synthetic Biology of Thermophiles: Taking Bioengineering to the Extremes?
Abstract
1. Introduction—Life at the Extremes, Thermophilic Microorganisms
2. Thermophilic Biotechnology and Biocatalysis
2.1. Physiology and Metabolism
2.1.1. Molecular Mechanisms
2.1.2. Metabolism
2.2. Thermophiles in Industry
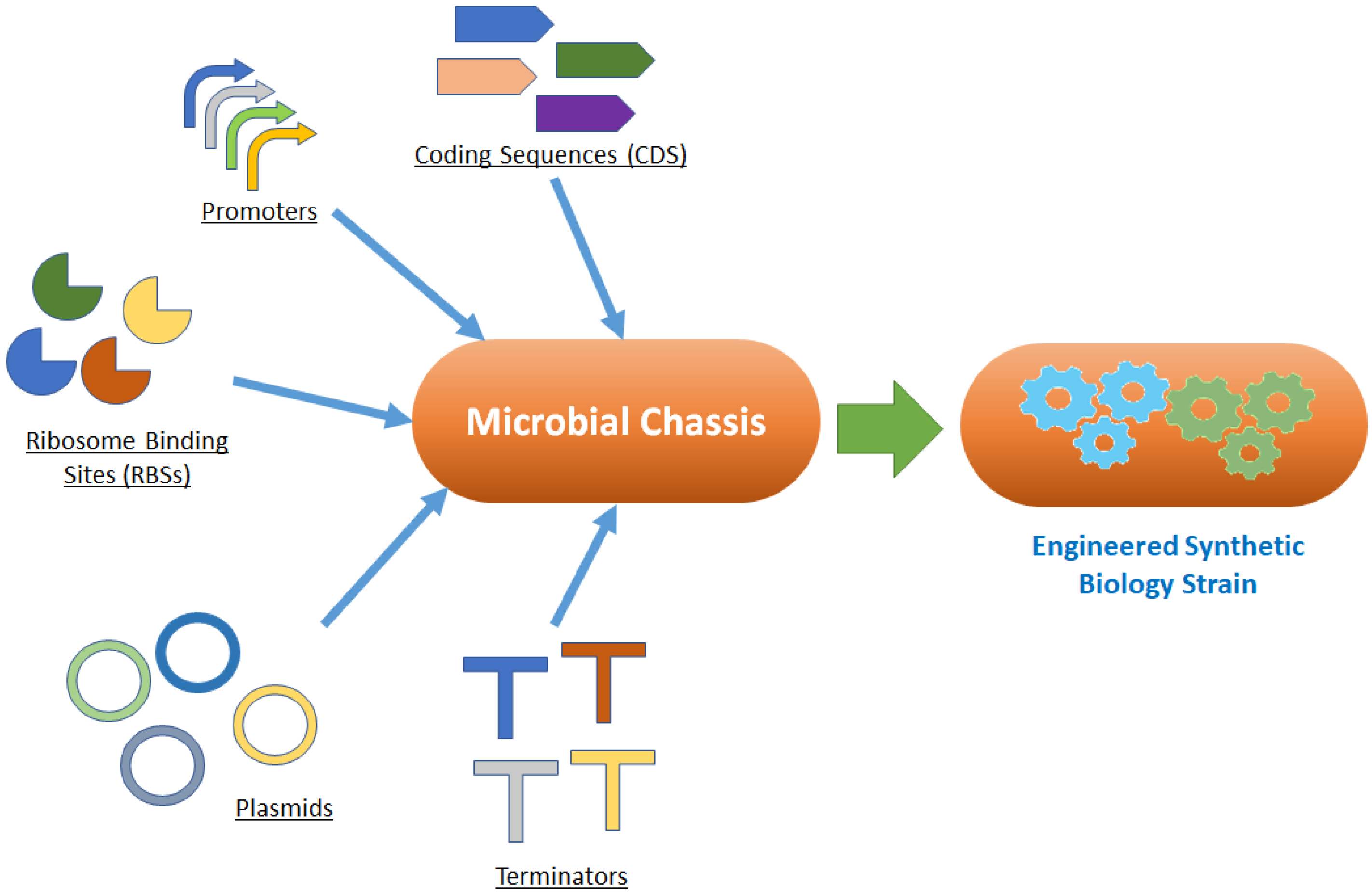
3. Synthetic Biology Chassis and the Need for a “Thermochassis”
- Ease of genetic manipulation;
- Sequenced genome;
- Availability of standardized genetic parts, with predictable and reported characteristics.
4. Future Perspectives in the Synthetic Biology of Thermophiles
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atalah, J.; Cáceres-Moreno, P.; Espina, G.; Blamey, J.M. Thermophiles and the applications of their enzymes as new biocatalysts. Bioresour. Technol. 2019, 280, 478–488. [Google Scholar] [CrossRef]
- Urbieta, M.S.; Donati, E.R.; Chan, K.-G.; Shahar, S.; Sin, L.L.; Goh, K.M. Thermophiles in the genomic era: Biodiversity, science, and applications. Biotechnol. Adv. 2015, 33, 633–647. [Google Scholar] [CrossRef]
- Acharya, S.; Chaudhary, A. Bioprospecting thermophiles for cellulase production: A review. Braz. J. Microbiol. 2012, 43, 844–856. [Google Scholar] [CrossRef]
- Satyanarayana, T.; Littlechild, J.; Kawarabayasi, Y. (Eds.) Thermophilic Microbes in Environmental and Industrial Biotechnology: Biotechnology of Thermophiles; Springer: Dordrecht, The Netherlands, 2013; ISBN 9789400758988. [Google Scholar]
- Zeldes, B.M.; Keller, M.W.; Loder, A.J.; Straub, C.T.; Adams, M.W.W.; Kelly, R.M. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front. Microbiol. 2015, 6, 1209. [Google Scholar] [CrossRef]
- Hussein, A.H.; Lisowska, B.K.; Leak, D.J. Chapter One—The Genus Geobacillus and Their Biotechnological Potential. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2015; Volume 92, pp. 1–48. [Google Scholar]
- Stathopoulou, P.M.; Galanopoulou, A.P.; Anasontzis, G.E.; Karagouni, A.D.; Hatzinikolaou, D.G. Assessment of the biomass hydrolysis potential in bacterial isolates from a volcanic environment: Biosynthesis of the corresponding activities. World J. Microbiol. Biotechnol. 2012, 28, 2889–2902. [Google Scholar] [CrossRef]
- Brumm, P.J.; De Maayer, P.; Mead, D.A.; Cowan, D.A. Genomic analysis of six new Geobacillus strains reveals highly conserved carbohydrate degradation architectures and strategies. Front. Microbiol. 2015, 6, 430. [Google Scholar] [CrossRef]
- Cripps, R.E.; Eley, K.; Leak, D.J.; Rudd, B.; Taylor, M.; Todd, M.; Boakes, S.; Martin, S.; Atkinson, T. Metabolic engineering of Geobacillus thermoglucosidasius for high yield ethanol production. Metab. Eng. 2009, 11, 398–408. [Google Scholar] [CrossRef]
- Talarico, L.A.; Gil, M.A.; Yomano, L.P.; Ingram, L.O.; Maupin-Furlow, J.A. Construction and expression of an ethanol production operon in Gram-positive bacteria. Microbiology 2005, 151, 4023–4031. [Google Scholar] [CrossRef][Green Version]
- Adams, B.L. The Next Generation of Synthetic Biology Chassis: Moving Synthetic Biology from the Laboratory to the Field. ACS Synth. Biol. 2016, 5, 1328–1330. [Google Scholar] [CrossRef]
- Abram, K.Z.; Udaondo, Z. Towards a better metabolic engineering reference: The microbial chassis. Microb. Biotechnol. 2020, 13, 17–18. [Google Scholar] [CrossRef]
- Danchin, A. Scaling up synthetic biology: Do not forget the chassis. FEBS Lett. 2012, 586, 2129–2137. [Google Scholar] [CrossRef]
- Stetter, K.O. Hyperthermophiles in the history of life. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 1837–1842. [Google Scholar] [CrossRef]
- Han, H.; Ling, Z.; Khan, A.; Virk, A.K.; Kulshrestha, S.; Li, X. Improvements of thermophilic enzymes: From genetic modifications to applications. Bioresour. Technol. 2019, 279, 350–361. [Google Scholar] [CrossRef]
- Steiner, K.; Schwab, H. Recent advances in rational approaches for enzyme engineering. Comput. Struct. Biotechnol. J. 2012, 2, e201209010. [Google Scholar] [CrossRef]
- Kumwenda, B.; Litthauer, D.; Bishop, O.T.; Reva, O. Analysis of protein thermostability enhancing factors in industrially important thermus bacteria species. Evol. Bioinform. Online 2013, 9, 327–342. [Google Scholar] [CrossRef]
- Suleiman, M.; Krüger, A.; Antranikian, G. Biomass-degrading glycoside hydrolases of archaeal origin. Biotechnol. Biofuels 2020, 13, 153. [Google Scholar] [CrossRef]
- Fiala, G.; Stetter, K.O. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 °C. Arch. Microbiol. 1986, 145, 56–61. [Google Scholar] [CrossRef]
- Konings, W.N.; Albers, S.-V.; Koning, S.; Driessen, A.J.M. The cell membrane plays a crucial role in survival of bacteria and archaea in extreme environments. Antonie Van Leeuwenhoek 2002, 81, 61–72. [Google Scholar] [CrossRef]
- Patel, A.; Matsakas, L.; Rova, U.; Christakopoulos, P. A perspective on biotechnological applications of thermophilic microalgae and cyanobacteria. Bioresour. Technol. 2019, 278, 424–434. [Google Scholar] [CrossRef]
- Wang, Q.; Cen, Z.; Zhao, J. The survival mechanisms of thermophiles at high temperatures: An angle of omics. Physiology 2015, 30, 97–106. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1014. [Google Scholar] [CrossRef]
- Lusk, B.G. Thermophiles; or, the Modern Prometheus: The Importance of Extreme Microorganisms for Understanding and Applying Extracellular Electron Transfer. Front. Microbiol. 2019, 10, 818. [Google Scholar] [CrossRef]
- Amend, J.P.; Shock, E.L. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and bacteria. FEMS Microbiol. Rev. 2001, 25, 175–243. [Google Scholar] [CrossRef]
- Blumer-Schuette, S.E.; Kataeva, I.; Westpheling, J.; Adams, M.W.; Kelly, R.M. Extremely thermophilic microorganisms for biomass conversion: Status and prospects. Curr. Opin. Biotechnol. 2008, 19, 210–217. [Google Scholar] [CrossRef]
- Rakotoarivonina, H.; Revol, P.-V.; Aubry, N.; Rémond, C. The use of thermostable bacterial hemicellulases improves the conversion of lignocellulosic biomass to valuable molecules. Appl. Microbiol. Biotechnol. 2016, 100, 7577–7590. [Google Scholar] [CrossRef]
- Guerriero, G.; Hausman, J.-F.; Strauss, J.; Ertan, H.; Siddiqui, K.S. Destructuring plant biomass: Focus on fungal and extremophilic cell wall hydrolases. Plant Sci. 2015, 234, 180–193. [Google Scholar] [CrossRef]
- Basit, A.; Liu, J.; Rahim, K.; Jiang, W.; Lou, H. Thermophilic xylanases: From bench to bottle. Crit. Rev. Biotechnol. 2018, 38, 989–1002. [Google Scholar] [CrossRef]
- Collins, T.; Gerday, C.; Feller, G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol. Rev. 2005, 29, 3–23. [Google Scholar] [CrossRef]
- Sheng, L.; Kovács, K.; Winzer, K.; Zhang, Y.; Minton, N.P. Development and implementation of rapid metabolic engineering tools for chemical and fuel production in Geobacillus thermoglucosidasius NCIMB 11955. Biotechnol. Biofuels 2017, 10, 5. [Google Scholar] [CrossRef]
- Keasling, J.D. Synthetic biology for synthetic chemistry. ACS Chem. Biol. 2008, 3, 64–76. [Google Scholar] [CrossRef]
- Nikel, P.I.; de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab. Eng. 2018, 50, 142–155. [Google Scholar] [CrossRef]
- Fredens, J.; Wang, K.; de la Torre, D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beránek, V.; et al. Total synthesis of Escherichia coli with a recoded genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef]
- Yu, J.; Liberton, M.; Cliften, P.F.; Head, R.D.; Jacobs, J.M.; Smith, R.D.; Koppenaal, D.W.; Brand, J.J.; Pakrasi, H.B. Synechococcus elongatus UTEX 2973, a fast growing cyanobacterial chassis for biosynthesis using light and CO₂. Sci. Rep. 2015, 5, 8132. [Google Scholar] [CrossRef]
- Larroude, M.; Celinska, E.; Back, A.; Thomas, S.; Nicaud, J.-M.; Ledesma-Amaro, R. A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol. Bioeng. 2018, 115, 464–472. [Google Scholar] [CrossRef]
- Weinstock, M.T.; Hesek, E.D.; Wilson, C.M.; Gibson, D.G. Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 2016, 13, 849–851. [Google Scholar] [CrossRef]
- Vavitsas, K.; Crozet, P.; Vinde, M.H.; Davies, F.; Lemaire, S.D.; Vickers, C.E. The Synthetic Biology Toolkit for Photosynthetic Microorganisms. Plant Physiol. 2019, 181, 14–27. [Google Scholar] [CrossRef]
- Vavitsas, K.; Kugler, A.; Satta, A.; Hatzinikolaou, D.G.; Lindblad, P.; Fewer, D.P.; Lindberg, P.; Toivari, M.; Stensjö, K. Doing synthetic biology with photosynthetic microorganisms. Physiol. Plant 2021, 173, 624–638. [Google Scholar] [CrossRef]
- Lin, L.; Xu, J. Dissecting and engineering metabolic and regulatory networks of thermophilic bacteria for biofuel production. Biotechnol. Adv. 2013, 31, 827–837. [Google Scholar] [CrossRef]
- Tominaga, Y.; Ohshiro, T.; Suzuki, H. Conjugative plasmid transfer from Escherichia coli is a versatile approach for genetic transformation of thermophilic Bacillus and Geobacillus species. Extremophiles 2016, 20, 375–381. [Google Scholar] [CrossRef]
- Reeve, B.; Martinez-Klimova, E.; de Jonghe, J.; Leak, D.J.; Ellis, T. The Geobacillus Plasmid Set: A Modular Toolkit for Thermophile Engineering. ACS Synth. Biol. 2016, 5, 1342–1347. [Google Scholar] [CrossRef]
- Pogrebnyakov, I.; Jendresen, C.B.; Nielsen, A.T. Genetic toolbox for controlled expression of functional proteins in Geobacillus spp. PLoS ONE 2017, 12, e0171313. [Google Scholar] [CrossRef]
- Marcano-Velazquez, J.G.; Lo, J.; Nag, A.; Maness, P.-C.; Chou, K.J. Developing Riboswitch-Mediated Gene Regulatory Controls in Thermophilic Bacteria. ACS Synth. Biol. 2019, 8, 633–640. [Google Scholar] [CrossRef]
- Bashir, Z.; Sheng, L.; Anil, A.; Lali, A.; Minton, N.P.; Zhang, Y. Engineering Geobacillus thermoglucosidasius for direct utilisation of holocellulose from wheat straw. Biotechnol. Biofuels 2019, 12, 199. [Google Scholar] [CrossRef]
- Carr, J.F.; Danziger, M.E.; Huang, A.L.; Dahlberg, A.E.; Gregory, S.T. Engineering the genome of Thermus thermophilus using a counterselectable marker. J. Bacteriol. 2015, 197, 1135–1144. [Google Scholar] [CrossRef]
- Verdú, C.; Sanchez, E.; Ortega, C.; Hidalgo, A.; Berenguer, J.; Mencía, M. A Modular Vector Toolkit with a Tailored Set of Thermosensors To Regulate Gene Expression in Thermus thermophilus. ACS Omega 2019, 4, 14626–14632. [Google Scholar] [CrossRef]
- Gallo, G.; Mougiakos, I.; Bianco, M.; Carbonaro, M.; Carpentieri, A.; Illiano, A.; Pucci, P.; Bartolucci, S.; van der Oost, J.; Fiorentino, G. A Hyperthermoactive-Cas9 Editing Tool Reveals the Role of a Unique Arsenite Methyltransferase in the Arsenic Resistance System of Thermus thermophilus HB27. mBio 2021, 12, e0281321. [Google Scholar] [CrossRef]
- Adalsteinsson, B.T.; Kristjansdottir, T.; Merre, W.; Helleux, A.; Dusaucy, J.; Tourigny, M.; Fridjonsson, O.; Hreggvidsson, G.O. Efficient genome editing of an extreme thermophile, Thermus thermophilus, using a thermostable Cas9 variant. Sci. Rep. 2021, 11, 9586. [Google Scholar] [CrossRef]
- Li, Y.; Pan, S.; Zhang, Y.; Ren, M.; Feng, M.; Peng, N.; Chen, L.; Liang, Y.X.; She, Q. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016, 44, e34. [Google Scholar] [CrossRef]
- Straub, C.T.; Counts, J.A.; Nguyen, D.M.N.; Wu, C.-H.; Zeldes, B.M.; Crosby, J.R.; Conway, J.M.; Otten, J.K.; Lipscomb, G.L.; Schut, G.J.; et al. Biotechnology of extremely thermophilic archaea. FEMS Microbiol. Rev. 2018, 42, 543–578. [Google Scholar] [CrossRef]
- Wagner, M.; Wagner, A.; Ma, X.; Kort, J.C.; Ghosh, A.; Rauch, B.; Siebers, B.; Albers, S.-V. Investigation of the malE promoter and MalR, a positive regulator of the maltose regulon, for an improved expression system in Sulfolobus acidocaldarius. Appl. Environ. Microbiol. 2014, 80, 1072–1081. [Google Scholar] [CrossRef]
- Jiang, Y.; Jiang, W.; Xin, F.; Zhang, W.; Jiang, M. Thermophiles: Potential chassis for lignocellulosic biorefinery. Trends Biotechnol. 2022, in press. [CrossRef]

| Advantages | Disadvantages |
|---|---|
| Reduced need for cooling | Poorly understood genetics and metabolism |
| Fewer contaminations | Lack of bioengineering tools |
| Thermostable enzymes | Infrastructure and fermentation expertise built around mesophiles |
| High growth rates | Not suitable for all heterologous protein expression (denaturation risks) |
| Description | Organism | References |
|---|---|---|
| Plasmid vectors for transformation; reporter genes; origins of replication | G. thermoglucosidasius | [42] |
| Transformation vectors, knock-in/knockout system | G. thermoglucosidasius | [31] |
| Library of semi-synthetic constitutive promoters | G. thermoglucosidasius | [43] |
| Riboswitches that work at high temperatures | G. thermoglucosidasius, Clostridium thermocellum | [44] |
| Heterologous expression of glycoside hydrolases to degrade lignocellulosic biomass | G. thermoglucosidasius | [45] |
| Counterselection system for introduction of genomic point mutations or deletions | T. thermophilus | [46] |
| Modular vector toolkit for gene expression | T. thermophilus | [47] |
| ThermoCas9-based genome-editing tool | T. thermophilus | [48] |
| Thermostable Cas9 | T. thermophilus | [49] |
| CRISPR-mediated genetic transformation | Sulfolobus islandicus | [50] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vavitsas, K.; Glekas, P.D.; Hatzinikolaou, D.G. Synthetic Biology of Thermophiles: Taking Bioengineering to the Extremes? Appl. Microbiol. 2022, 2, 165-174. https://doi.org/10.3390/applmicrobiol2010011
Vavitsas K, Glekas PD, Hatzinikolaou DG. Synthetic Biology of Thermophiles: Taking Bioengineering to the Extremes? Applied Microbiology. 2022; 2(1):165-174. https://doi.org/10.3390/applmicrobiol2010011
Chicago/Turabian StyleVavitsas, Konstantinos, Panayiotis D. Glekas, and Dimitris G. Hatzinikolaou. 2022. "Synthetic Biology of Thermophiles: Taking Bioengineering to the Extremes?" Applied Microbiology 2, no. 1: 165-174. https://doi.org/10.3390/applmicrobiol2010011
APA StyleVavitsas, K., Glekas, P. D., & Hatzinikolaou, D. G. (2022). Synthetic Biology of Thermophiles: Taking Bioengineering to the Extremes? Applied Microbiology, 2(1), 165-174. https://doi.org/10.3390/applmicrobiol2010011







