A Bioinformatics Approach to Mine the Microbial Proteomic Profile of COVID-19 Mass Spectrometry Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Bacterial and Viral Protein Sequence Compilation
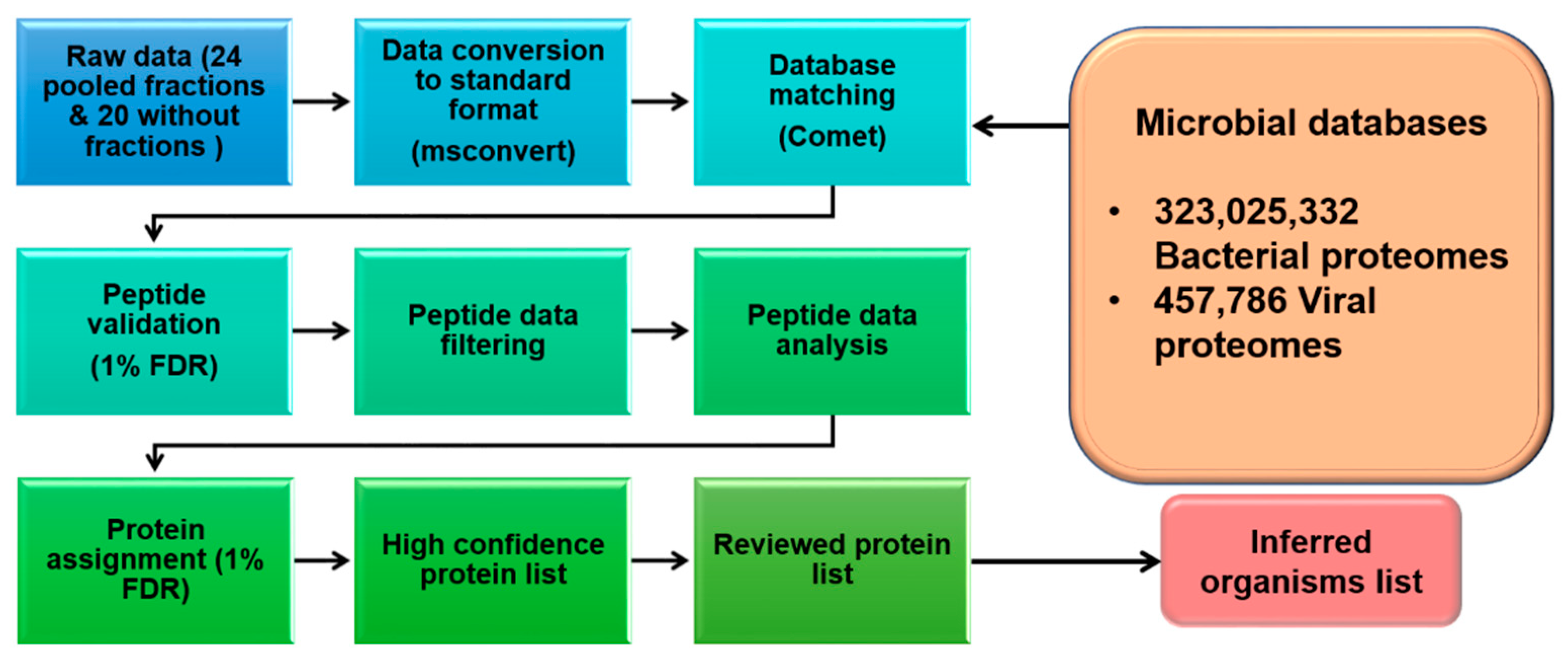
2.3. Data Processing through Trans Proteomic Pipeline (TPP)
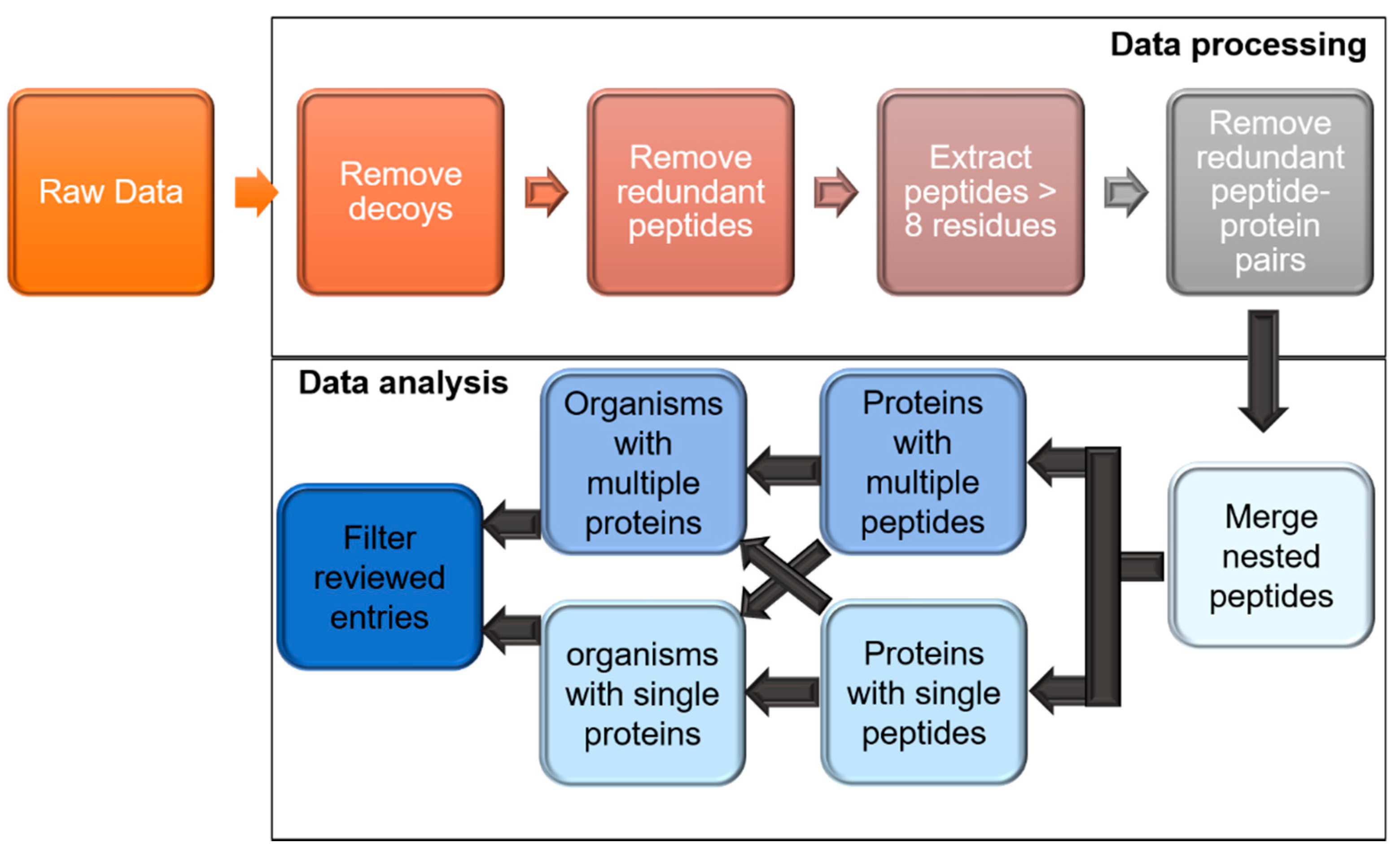
2.4. Analyzing the TPP Processed Data for High-Stringency Microbial Protein Identification
2.5. Compilation of Bacterial and Viral Organisms
3. Results
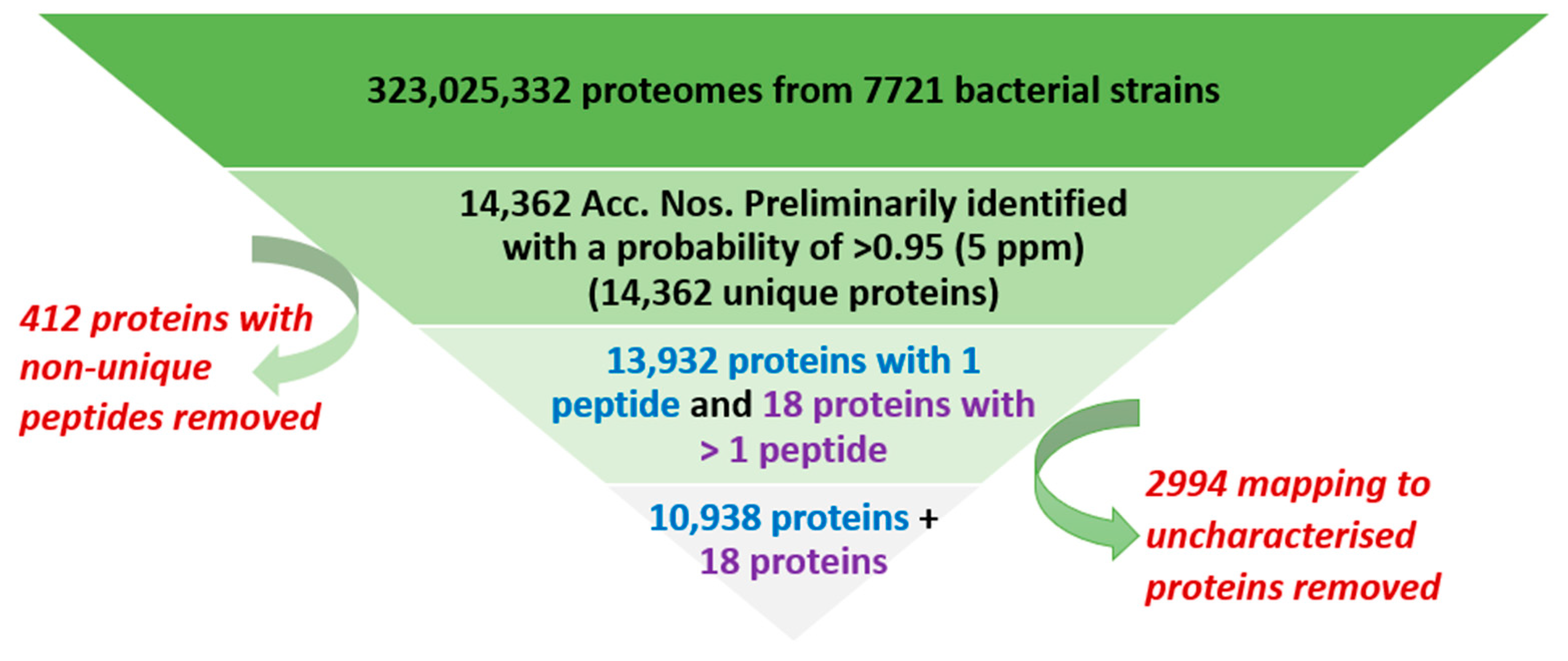
3.1. Overall Process of Bacterial Protein Filtration
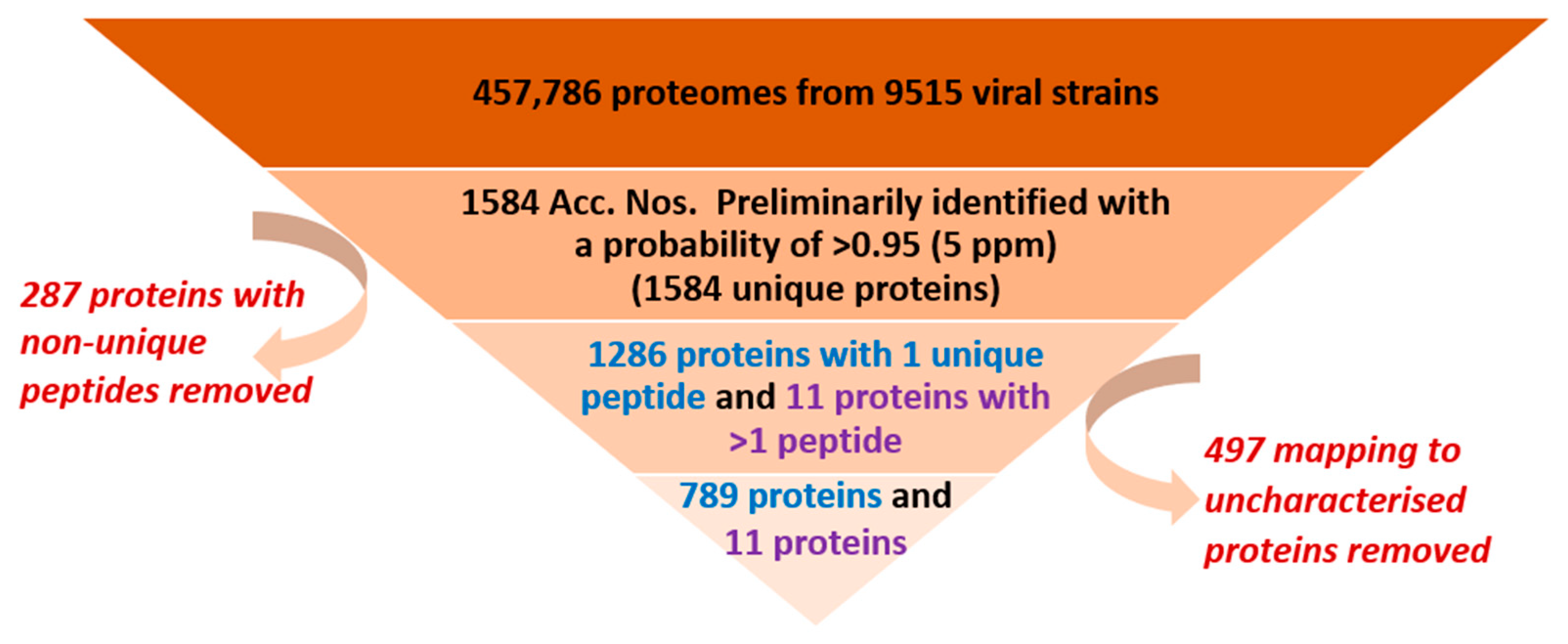
3.2. Overall Process of Viral Protein Filtration in Healthy Human Plasma (HHP)
3.3. SARS-CoV-2 Plasma Proteomics
4. Discussion
4.1. Confidence and Stringency of Identification
4.2. Bacterial and Viral Proteins Identified in Healthy Human Plasma
4.3. Uncharacterized Microbial Proteins in Healthy Human Plasma
4.4. Microbial Organisms Identified in Healthy Human Plasma
4.5. Case Study on SARS-CoV-2
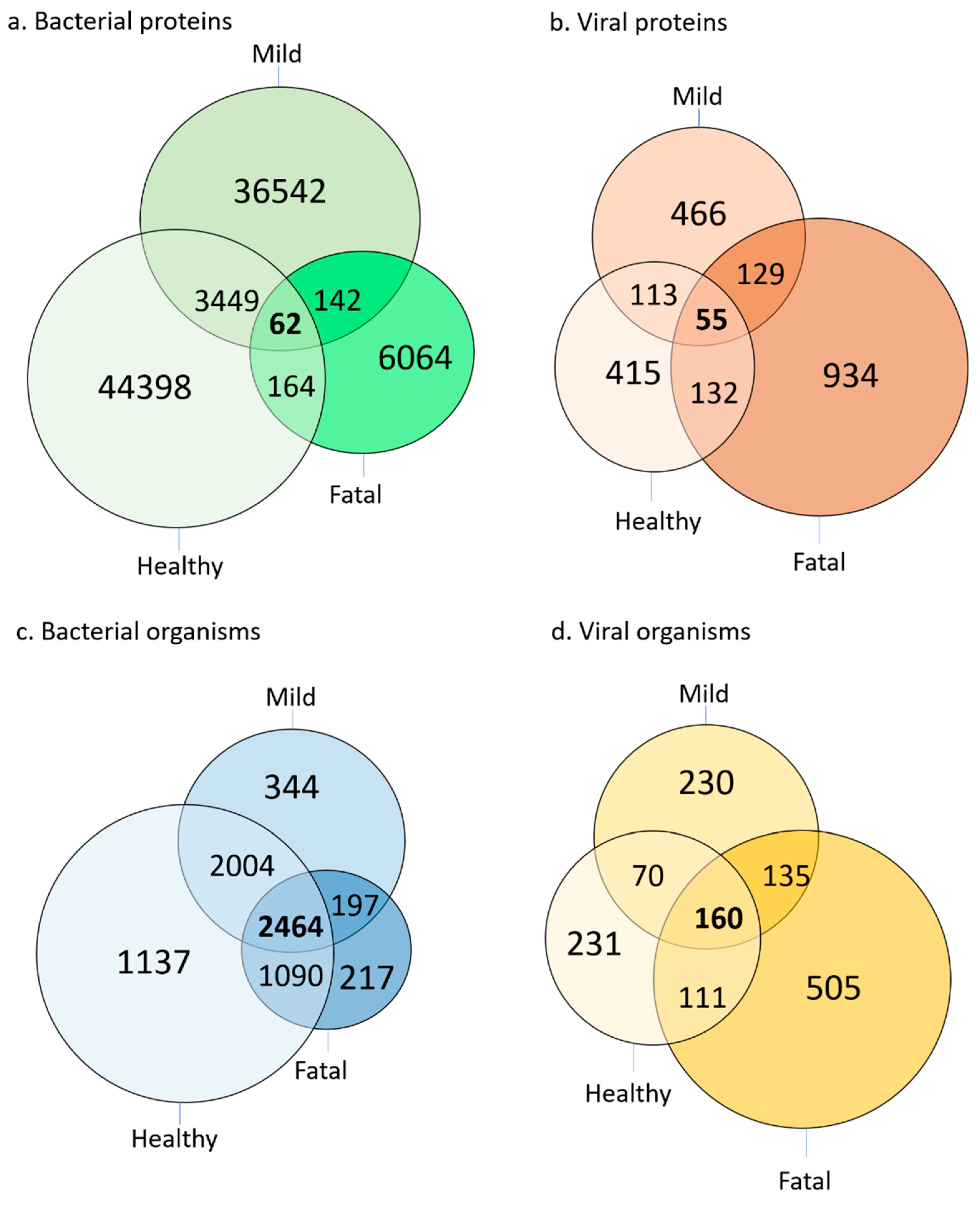
4.6. Bacterial and Viral Proteins Identified in SARS-CoV-2 Patient and Healthy Control Plasma
4.7. The Microbial Proteome Changes in COVID-19 Samples Compared to Healthy
4.8. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burke, W.; Brown Trinidad, S.; Press, N.A. Essential elements of personalized medicine. Urol. Oncol. 2014, 32, 193–197. [Google Scholar] [CrossRef][Green Version]
- Noor, Z.; Ahn, S.B.; Baker, M.S.; Ranganathan, S.; Mohamedali, A. Mass spectrometry-based protein identification in proteomics—A review. Brief. Bioinform. 2021, 22, 1620–1638. [Google Scholar] [CrossRef]
- Chambers, G.; Lawrie, L.; Cash, P.; Murray, G.I. Proteomics: A new approach to the study of disease. J. Pathol. 2000, 192, 280–288. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Foster, J.M.; Martens, L. Proteomics data repositories: Providing a safe haven for your data and acting as a springboard for further research. J. Proteom. 2010, 73, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, R.; Guedes, S.; Trindade, F.; Correia, I.; Moura, G.; Carvalho, P.; Santos, M.A.S.; Amado, F. De novo sequencing of proteins by mass spectrometry. Expert Rev. Proteom. 2020, 17, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Mohamedali, A.; Fernandes, C.S.; Baker, M.S.; Ranganathan, S. De Novo Peptide Sequencing: Deep Mining of High-Resolution Mass Spectrometry Data. Methods Mol. Biol. 2017, 1549, 119–134. [Google Scholar]
- Savage, D.C. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977, 31, 107–133. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Brüls, T.; Weissenbach, J. The human metagenome: Our other genome? Hum. Mol. Genet. 2011, 20, R142–R148. [Google Scholar] [CrossRef]
- Molina, D.K.; DiMaio, V.J. Normal organ weights in men: Part I-the heart. Am. J. Forensic Med. Pathol. 2012, 33, 362–367. [Google Scholar] [CrossRef]
- Moeller, A.H.; Caro-Quintero, A.; Mjungu, D.; Georgiev, A.V.; Lonsdorf, E.V.; Muller, M.N.; Pusey, A.E.; Peeters, M.; Hahn, B.H.; Ochman, H. Cospeciation of gut microbiota with hominids. Science 2016, 353, 380–382. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, G.M. Genomic approaches to studying the human microbiota. Nature 2012, 489, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Ravel, J. The vocabulary of microbiome research: A proposal. Microbiome 2015, 3, 31. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Jeggo, M. Reservoirs and vectors of emerging viruses. Curr. Opin. Virol. 2013, 3, 170–179. [Google Scholar] [CrossRef]
- Vilcek, S. SARS-CoV-2: Zoonotic origin of pandemic coronavirus. Acta Virol. 2020, 64, 281–287. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Smith, G.J.; Zhang, J.X.; Peiris, J.S.; Chen, H.; Guan, Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007, 81, 4012–4020. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Huebner, R.E.; Schein, M.F.; Bass, J.B., Jr. The tuberculin skin test. Clin. Infect. Dis. 1993, 17, 968–975. [Google Scholar] [CrossRef]
- Streeton, J.A.; Desem, N.; Jones, S.L. Sensitivity and specificity of a gamma interferon blood test for tuberculosis infection. Int. J. Tuberc. Lung Dis. 1998, 2, 443–450. [Google Scholar] [PubMed]
- Meyerowitz, E.A.; Richterman, A.; Bogoch, I.I.; Low, N.; Cevik, M. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect. Dis. 2020, 21, e163–e169. [Google Scholar] [CrossRef]
- Dinnes, J.; Deeks, J.J.; Berhane, S.; Taylor, M.; Adriano, A.; Davenport, C.; Dittrich, S.; Emperador, D.; Takwoingi, Y.; Cunningham, J.; et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021, 3, CD013705. [Google Scholar]
- zur Hausen, H. Viruses in human cancers. Science 1991, 254, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Hu, H.; Darido, C.; Vickery, K.; Ranganathan, S. ML218 HCl Is More Efficient Than Capsaicin in Inhibiting Bacterial Antigen-Induced Cal 27 Oral Cancer Cell Proliferation. Int. J. Mol. Sci. 2021, 22, 12559. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Vickery, K.; Darido, C.; Ranganathan, S.; Hu, H. Bacterial Antigens Reduced the Inhibition Effect of Capsaicin on Cal 27 Oral Cancer Cell Proliferation. Int. J. Mol. Sci. 2021, 22, 8686. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Voytik, E.; Treit, P.V.; Doll, S.; Kleinhempel, A.; Niu, L.; Müller, J.B.; Buchholtz, M.L.; Bader, J.M.; Teupser, D.; et al. Plasma Proteome Profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol. Med. 2019, 11, e10427. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Kulak, N.A.; Pichler, G.; Holdt, L.M.; Teupser, D.; Mann, M. Plasma Proteome Profiling to Assess Human Health and Disease. Cell Syst. 2016, 2, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Wewer Albrechtsen, N.J.; Tyanova, S.; Grassl, N.; Iepsen, E.W.; Lundgren, J.; Madsbad, S.; Holst, J.J.; Torekov, S.S.; Mann, M. Proteomics reveals the effects of sustained weight loss on the human plasma proteome. Mol. Syst. Biol. 2016, 12, 901. [Google Scholar] [CrossRef] [PubMed]
- Kulak, N.A.; Geyer, P.E.; Mann, M. Loss-less Nano-fractionator for High Sensitivity, High Coverage Proteomics. Mol. Cell. Proteom. 2017, 16, 694–705. [Google Scholar] [CrossRef]
- Wewer Albrechtsen, N.J.; Geyer, P.E.; Doll, S.; Treit, P.V.; Bojsen-Møller, K.N.; Martinussen, C.; Jørgensen, N.B.; Torekov, S.S.; Meier, F.; Niu, L.; et al. Plasma Proteome Profiling Reveals Dynamics of Inflammatory and Lipid Homeostasis Markers after Roux-En-Y Gastric Bypass Surgery. Cell Syst. 2018, 7, 601–612.e3. [Google Scholar] [CrossRef]
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122.e5. [Google Scholar] [CrossRef]
- The UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Keller, A.; Eng, J.; Zhang, N.; Li, X.J.; Aebersold, R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005, 1, 2005.0017. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; Lane, L.; Overall, C.M.; Corrales, F.J.; Schwenk, J.M.; Paik, Y.-K.; Van Eyk, J.E.; Liu, S.; Pennington, S.; Snyder, M.P.; et al. Progress on Identifying and Characterizing the Human Proteome: 2019 Metrics from the HUPO Human Proteome Project. J. Proteome Res. 2019, 18, 4098–4107. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Lane, L.; Overall, C.M.; Bandeira, N.; Baker, M.S.; Pineau, C.; Moritz, R.L.; Corrales, F.; Orchard, S.; Van Eyk, J.E.; et al. Human Proteome Project Mass Spectrometry Data Interpretation Guidelines 3.0. J. Proteome Res. 2019, 18, 4108–4116. [Google Scholar] [CrossRef]
- Chaitanya, K.V. Structure and Organization of Virus Genomes. In Genome and Genomics; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Tan, S.H.; Mohamedali, A.; Kapur, A.; Baker, M.S. Ultradepletion of human plasma using chicken antibodies: A proof of concept study. J. Proteome Res. 2013, 12, 2399–2413. [Google Scholar] [CrossRef] [PubMed]
- Fonslow, B.R.; Carvalho, P.C.; Academia, K.; Freeby, S.; Xu, T.; Nakorchevsky, A.; Paulus, A.; Yates, J.R., 3rd. Improvements in proteomic metrics of low abundance proteins through proteome equalization using ProteoMiner prior to MudPIT. J. Proteome Res. 2011, 10, 3690–3700. [Google Scholar] [CrossRef] [PubMed]
- Schwenk, J.M.; Omenn, G.S.; Sun, Z.; Campbell, D.S.; Baker, M.S.; Overall, C.M.; Aebersold, R.; Moritz, R.L.; Deutsch, E.W. The Human Plasma Proteome Draft of 2017: Building on the Human Plasma PeptideAtlas from Mass Spectrometry and Complementary Assays. J. Proteome Res. 2017, 16, 4299–4310. [Google Scholar] [CrossRef]
- Adhikari, S.; Sharma, S.; Ahn, S.B.; Baker, M.S. In Silico Peptide Repertoire of Human Olfactory Receptor Proteomes on High-Stringency Mass Spectrometry. J. Proteome Res. 2019, 18, 4117–4123. [Google Scholar] [CrossRef]
- Ijaq, J.; Chandrasekharan, M.; Poddar, R.; Bethi, N.; Sundararajan, V.S. Annotation and curation of uncharacterized proteins—Challenges. Front. Genet. 2015, 6, 119. [Google Scholar] [CrossRef]
- Desler, C.; Durhuus, J.A.; Rasmussen, L.J. Genome-wide screens for expressed hypothetical proteins. Methods Mol. Biol. 2012, 815, 25–38. [Google Scholar]
- Tanner, S.; Shen, Z.; Ng, J.; Florea, L.; Guigó, R.; Briggs, S.P.; Bafna, V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007, 17, 231–239. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dollman, N.L.; Griffin, J.H.; Downard, K.M. Detection, Mapping, and Proteotyping of SARS-CoV-2 Coronavirus with High Resolution Mass Spectrometry. ACS Infect. Dis. 2020, 6, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, R.W.; Vali, H.; Lau, P.C.; Palfree, R.G.; De Ciccio, A.; Sirois, M.; Ahmad, D.; Villemur, R.; Desrosiers, M.; Chan, E.C. Are there naturally occurring pleomorphic bacteria in the blood of healthy humans? J. Clin. Microbiol. 2002, 40, 4771–4775. [Google Scholar] [CrossRef] [PubMed]
- Olde Loohui, S.L.M.; Mangul, S.; Ori, A.P.S.; Jospin, G.; Koslicki, D.; Yang, H.T.; Wu, T.; Boks, M.P.; Lomen-Hoerth, C.; Wiedau-Pazos, M.; et al. Transcriptome analysis in whole blood reveals increased microbial diversity in schizophrenia. Transl. Psychiatry 2018, 8, 96. [Google Scholar] [CrossRef]
- Païssé, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Pereira Andrade, A.; de Miranda Boratto, P.V.; Rodrigues, R.A.L.; Bastos, T.M.; Azevedo, B.L.; Dornas, F.P.; Oliveira, D.B.; Drumond, B.P.; Kroon, E.G.; Abrahão, J.S. New Isolates of Pandoraviruses: Contribution to the Study of Replication Cycle Steps. J. Virol. 2019, 93, e01942-18. [Google Scholar] [CrossRef]
- Legendre, M.; Fabre, E.; Poirot, O.; Jeudy, S.; Lartigue, A.; Alempic, J.-M.; Beucher, L.; Philippe, N.; Bertaux, L.; Christo-Foroux, E.; et al. Diversity and evolution of the emerging Pandoraviridae family. Nat. Commun. 2018, 9, 2285. [Google Scholar] [CrossRef]
- Kuehnert, M.J.; Roth, V.R.; Haley, N.R.; Gregory, K.R.; Elder, K.V.; Schreiber, G.B.; Arduino, M.J.; Holt, S.C.; Carson, L.A.; Banerjee, S.N.; et al. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 2001, 41, 1493–1499. [Google Scholar] [CrossRef]
- Theakston, E.P.; Morris, A.J.; Streat, S.J.; Baker, B.W.; Woodfield, D.G. Transfusion transmitted Yersinia enterocolitica infection in New Zealand. Aust. N. Z. J. Med. 1997, 27, 62–67. [Google Scholar] [CrossRef]
- Wagner, S.J. Transfusion-transmitted bacterial infection: Risks, sources and interventions. Vox Sang. 2004, 86, 157–163. [Google Scholar] [CrossRef]
- Staheli, J.P.; Dyen, M.R.; Deutsch, G.H.; Basom, R.S.; Fitzgibbon, M.P.; Lewis, P.; Barcy, S. Complete Unique Genome Sequence, Expression Profile, and Salivary Gland Tissue Tropism of the Herpesvirus 7 Homolog in Pigtailed Macaques. J. Virol. 2016, 90, 6657–6674. [Google Scholar] [CrossRef] [PubMed]
- Delhon, G.; Tulman, E.R.; Afonso, C.L.; Lu, Z.; Becnel, J.J.; Moser, B.A.; Kutish, G.F.; Rock, D.L. Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). J. Virol. 2006, 80, 8439–8449. [Google Scholar] [CrossRef] [PubMed]
- Rouse, B.T.; Sehrawat, S. Immunity and immunopathology to viruses: What decides the outcome? Nat. Rev. Immunol. 2010, 10, 514–526. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alnakli, A.A.A.; Jabeen, A.; Chakraborty, R.; Mohamedali, A.; Ranganathan, S. A Bioinformatics Approach to Mine the Microbial Proteomic Profile of COVID-19 Mass Spectrometry Data. Appl. Microbiol. 2022, 2, 150-164. https://doi.org/10.3390/applmicrobiol2010010
Alnakli AAA, Jabeen A, Chakraborty R, Mohamedali A, Ranganathan S. A Bioinformatics Approach to Mine the Microbial Proteomic Profile of COVID-19 Mass Spectrometry Data. Applied Microbiology. 2022; 2(1):150-164. https://doi.org/10.3390/applmicrobiol2010010
Chicago/Turabian StyleAlnakli, Aziz Abdullah A., Amara Jabeen, Rajdeep Chakraborty, Abidali Mohamedali, and Shoba Ranganathan. 2022. "A Bioinformatics Approach to Mine the Microbial Proteomic Profile of COVID-19 Mass Spectrometry Data" Applied Microbiology 2, no. 1: 150-164. https://doi.org/10.3390/applmicrobiol2010010
APA StyleAlnakli, A. A. A., Jabeen, A., Chakraborty, R., Mohamedali, A., & Ranganathan, S. (2022). A Bioinformatics Approach to Mine the Microbial Proteomic Profile of COVID-19 Mass Spectrometry Data. Applied Microbiology, 2(1), 150-164. https://doi.org/10.3390/applmicrobiol2010010







