Transcription at a Distance in the Budding Yeast Saccharomyces cerevisiae
Abstract
1. Overview and Background
2. Overview of Transcriptional Regulation in the Budding Yeast, Saccharomyces cerevisiae
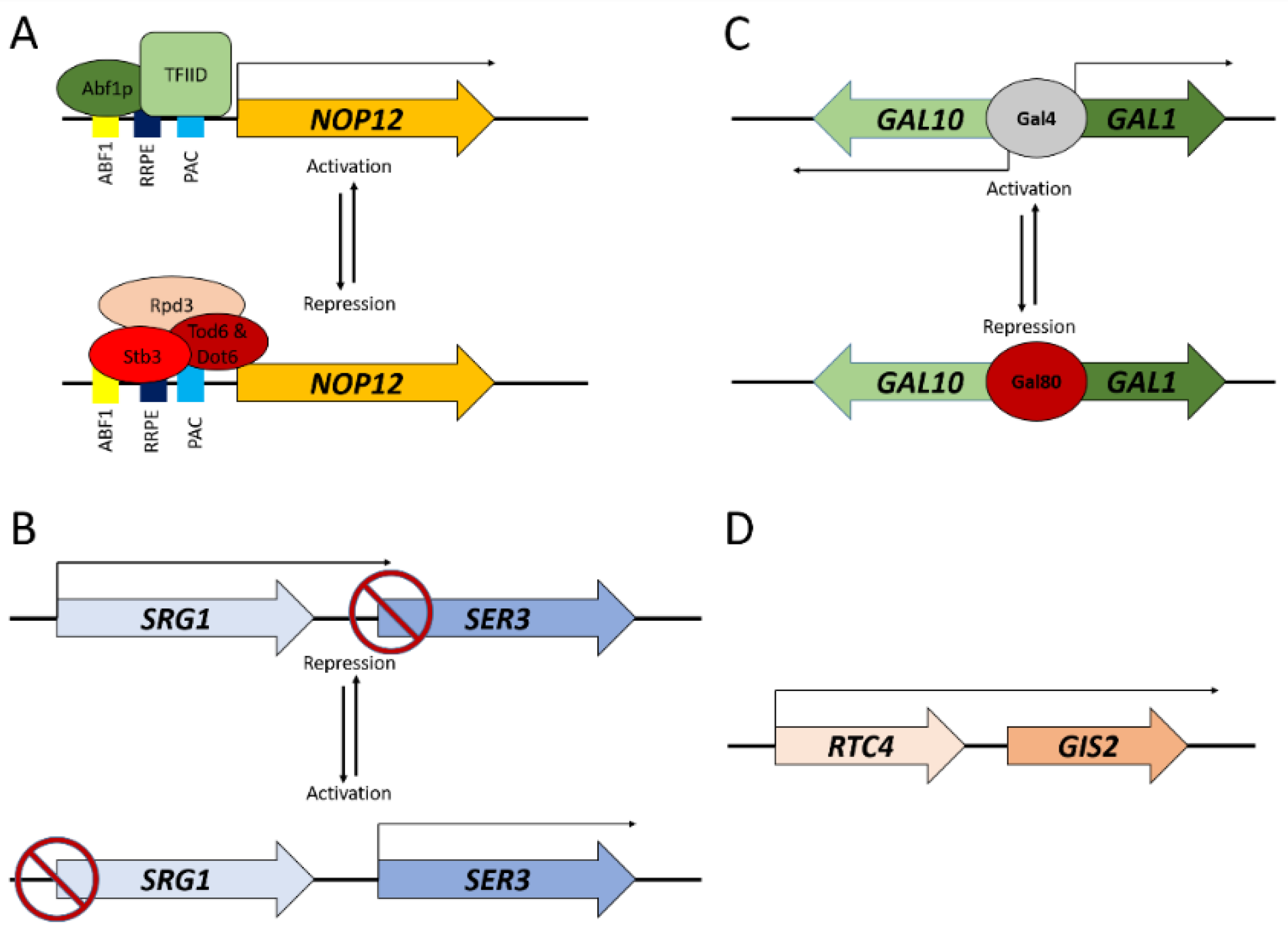
3. Transcriptional Interference and Gene Repression at a Distance
4. Gene Activation at a Distance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958, 12, 138–163. [Google Scholar]
- Crick, F.H. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Gasch, A.P.; Spellman, P.T.; Kao, C.M.; Carmel-Harel, O.; Eisen, M.B.; Storz, G.; Botstein, D.; Brown, P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 2000, 11, 4241–4257. [Google Scholar] [CrossRef]
- Gasch, A.P.; Werner-Washburne, M. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genom. 2002, 2, 181–192. [Google Scholar] [CrossRef]
- Tirosh, I.; Weinberger, A.; Carmi, M.; Barkai, N. A genetic signature of interspecies variations in gene expression. Nat. Genet. 2006, 38, 830–834. [Google Scholar] [CrossRef]
- Ramachandran, S.; Hiratsuka, K.; Chua, N.-H. Transcription factors in plant growth and development. Curr. Opin. Genet. Dev. 1994, 4, 642–646. [Google Scholar] [CrossRef]
- Jennings, R.E.; Scharfmann, R.; Staels, W. Transcription factors that shape the mammalian pancreas. Diabetologia 2020, 63, 1974–1980. [Google Scholar] [CrossRef]
- Lenhard, B.; Sandelin, A.; Carninci, P. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012, 13, 233–245. [Google Scholar] [CrossRef]
- Yusuf, D.; Butland, S.L.; Swanson, M.I.; Bolotin, E.; Ticoll, A.; Cheung, W.A.; Zhang, X.Y.C.; Dickman, C.T.; Fulton, D.L.; Lim, J.S. The transcription factor encyclopedia. Genome Biol. 2012, 13, 1–25. [Google Scholar] [CrossRef]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The human transcription factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Tapias, A.; Wang, Z.-Q. Lysine acetylation and deacetylation in brain development and neuropathies. Genom. Proteom. Bioinform. 2017, 15, 19–36. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, H.; Chen, F.; Liu, Y. The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 2014, 84, 125–133. [Google Scholar] [CrossRef]
- Grunstein, M.; Gasser, S.M. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb. Perspect. Biol. 2013, 5, a017491. [Google Scholar] [CrossRef]
- Gardner, K.E.; Allis, C.D.; Strahl, B.D. Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 2011, 409, 36–46. [Google Scholar] [CrossRef]
- Kaypee, S.; Sudarshan, D.; Shanmugam, M.K.; Mukherjee, D.; Sethi, G.; Kundu, T.K. Aberrant lysine acetylation in tumorigenesis: Implications in the development of therapeutics. Pharmacol. Ther. 2016, 162, 98–119. [Google Scholar] [CrossRef]
- Archer, S.Y.; Hodin, R.A. Histone acetylation and cancer. Curr. Opin. Genet. Dev. 1999, 9, 171–174. [Google Scholar] [CrossRef]
- Orr, J.A.; Hamilton, P.W. Histone acetylation and chromatin pattern in cancer. A review. Anal. Quant. Cytol. Histol. 2007, 29, 17–31. [Google Scholar] [PubMed]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [PubMed]
- Sachs, A.B. Messenger RNA degradation in eukaryotes. Cell 1993, 74, 413–421. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. The many pathways of RNA degradation. Cell 2009, 136, 763–776. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Botstein, D.; Chervitz, S.A.; Cherry, M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Chávez, S.; Pérez-Ortín, J.E. A complete set of nascent transcription rates for yeast genes. PLoS ONE 2010, 5, e15442. [Google Scholar] [CrossRef]
- Li, H.; Johnson, A.D. Evolution of transcription networks—lessons from yeasts. Curr. Biol. 2010, 20, R746–R753. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G. Yeast Systems Biology: The Continuing Challenge of Eukaryotic Complexity. Yeast Syst. Biol. 2019, 2049, 3–13. [Google Scholar]
- Castrillo, J.I.; Oliver, S.G. Yeast systems biology: The challenge of eukaryotic complexity. Yeast Syst. Biol. 2011, 759, 3–28. [Google Scholar]
- Tang, H.; Wu, Y.; Deng, J.; Chen, N.; Zheng, Z.; Wei, Y.; Luo, X.; Keasling, J.D. Promoter architecture and promoter engineering in Saccharomyces cerevisiae. Metabolites 2020, 10, 320. [Google Scholar] [CrossRef] [PubMed]
- Dujon, B. The yeast genome project: What did we learn? Trends Genet. 1996, 12, 263–270. [Google Scholar] [CrossRef]
- Pelechano, V.; Garcıa-Martınez, J.; Pérez-Ortın, J.E. A genomic study of the inter-ORF distances in Saccharomyces cerevisiae. Yeast 2006, 23, 689–699. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Lin, Z. Pervasive and dynamic transcription initiation in Saccharomyces cerevisiae. Genome Res. 2019, 29, 1198–1210. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.H.; Umbarger, M.A.; McAlear, M.A. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast 2006, 23, 293–306. [Google Scholar] [CrossRef]
- Michael Lee, K.; DaSilva, N.A. Evaluation of the Saccharomyces cerevisiae ADH2 promoter for protein synthesis. Yeast 2005, 22, 431–440. [Google Scholar] [CrossRef] [PubMed]
- De Boer, C.G.; Hughes, T.R. YeTFaSCo: A database of evaluated yeast transcription factor sequence specificities. Nucleic Acids Res. 2012, 40, D169–D179. [Google Scholar] [CrossRef]
- Hu, Z.; Killion, P.J.; Iyer, V.R. Genetic reconstruction of a functional transcriptional regulatory network. Nat. Genet. 2007, 39, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Bosio, M.C.; Negri, R.; Dieci, G. Promoter architectures in the yeast ribosomal expression program. Transcription 2011, 2, 71–77. [Google Scholar] [CrossRef]
- Bosio, M.C.; Fermi, B.; Dieci, G. Transcriptional control of yeast ribosome biogenesis: A multifaceted role for general regulatory factors. Transcription 2017, 8, 254–260. [Google Scholar] [CrossRef] [PubMed]
- Bosio, M.C.; Fermi, B.; Spagnoli, G.; Levati, E.; Rubbi, L.; Ferrari, R.; Pellegrini, M.; Dieci, G. Abf1 and other general regulatory factors control ribosome biogenesis gene expression in budding yeast. Nucleic Acids Res. 2017, 45, 4493–4506. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.-C.; Liu, Y.-J.; Dion, M.F.; Slack, M.D.; Wu, L.F.; Altschuler, S.J.; Rando, O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 2005, 309, 626–630. [Google Scholar] [CrossRef]
- Jansen, A.; Verstrepen, K.J. Nucleosome positioning in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2011, 75, 301–320. [Google Scholar] [CrossRef]
- Lee, W.; Tillo, D.; Bray, N.; Morse, R.H.; Davis, R.W.; Hughes, T.R.; Nislow, C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007, 39, 1235–1244. [Google Scholar] [CrossRef]
- Lee, C.-K.; Shibata, Y.; Rao, B.; Strahl, B.D.; Lieb, J.D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 2004, 36, 900–905. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.L.; Kaplan, T.; Kim, M.; Buratowski, S.; Schreiber, S.L.; Friedman, N.; Rando, O.J. Single-nucleosome mapping of histone modifications in S. cerevisiae. PloS Biol. 2005, 3, e328. [Google Scholar] [CrossRef] [PubMed]
- Torrent, M.; Chalancon, G.; de Groot, N.S.; Wuster, A.; Babu, M.M. Cells alter their tRNA abundance to selectively regulate protein synthesis during stress conditions. Sci. Signal. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef]
- Sandell, L.L.; Zakian, V.A. Telomeric position effect in yeast. Trends Cell Biol. 1992, 2, 10–14. [Google Scholar] [CrossRef]
- Arnone, J.T. Genomic considerations for the modification of saccharomyces cerevisiae for biofuel and metabolite biosynthesis. Microorganisms 2020, 8, 321. [Google Scholar] [CrossRef]
- Arnone, J.T.; Arace, J.R.; Soorneedi, A.R.; Citino, T.T.; Kamitaki, T.L.; McAlear, M.A. Dissecting the cis and trans elements that regulate adjacent-gene coregulation in Saccharomyces cerevisiae. Eukaryot. Cell 2014, 13, 738–748. [Google Scholar] [CrossRef] [PubMed]
- Arnone, J.T.; McAlear, M.A. Adjacent gene pairing plays a role in the coordinated expression of ribosome biogenesis genes MPP10 and YJR003C in Saccharomyces cerevisiae. Eukaryot. Cell 2011, 10, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Hagee, D.; Hardan, A.A.; Botero, J.; Arnone, J.T. Genomic Clustering within Functionally Related Gene Families in Ascomycota Fungi. Comput. Struct. Biotechnol. J. 2020, 18, 3267–3277. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.A.; Wu, P.-Y.J.; Winston, F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005, 19, 2695–2704. [Google Scholar] [CrossRef] [PubMed]
- Martens, J.A.; Laprade, L.; Winston, F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 2004, 429, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Steinmetz, L.M. Gene regulation by antisense transcription. Nat. Rev. Genet. 2013, 14, 880–893. [Google Scholar] [CrossRef]
- Dobi, K.C.; Winston, F. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 5575–5586. [Google Scholar] [CrossRef] [PubMed]
- Cera, A.; Holganza, M.K.; Hardan, A.A.; Gamarra, I.; Eldabagh, R.S.; Deschaine, M.; Elkamhawy, S.; Sisso, E.M.; Foley, J.J.; Arnone, J.T. Functionally related genes cluster into genomic regions that coordinate transcription at a distance in saccharomyces cerevisiae. Msphere 2019, 4, e00063-19. [Google Scholar] [CrossRef]
- Uthe, H.; Vanselow, J.T.; Schlosser, A. Proteomic analysis of the mediator complex interactome in Saccharomyces cerevisiae. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Q.; Ding, Z.; Ji, J.; Wang, J.; Kong, X.; Yang, J.; Cai, G. Redefining the modular organization of the core Mediator complex. Cell Res. 2014, 24, 796–808. [Google Scholar] [CrossRef]
- Reavey, C.T.; Hickman, M.J.; Dobi, K.C.; Botstein, D.; Winston, F. Analysis of polygenic mutants suggests a role for mediator in regulating transcriptional activation distance in Saccharomyces cerevisiae. Genetics 2015, 201, 599–612. [Google Scholar] [CrossRef]
- Goffeau, A.; Aert, R.; Agostini-Carbone, M.; Ahmed, A.; Aigle, M.; Alberghina, L.; Albermann, K.; Albers, M.; Aldea, M.; Alexandraki, D. The yeast genome directory. Nature 1997, 387, 5–6. [Google Scholar] [CrossRef]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef]
- Kristiansson, E.; Thorsen, M.; Tamás, M.J.; Nerman, O. Evolutionary forces act on promoter length: Identification of enriched cis-regulatory elements. Mol. Biol. Evol. 2009, 26, 1299–1307. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.A.; Mitra, R.D.; Hughes, J.D.; Church, G.M. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 2000, 26, 183–186. [Google Scholar] [CrossRef]
- Quintero-Cadena, P.; Sternberg, P.W. Enhancer sharing promotes neighborhoods of transcriptional regulation across eukaryotes. G3 Genes Genomes Genet. 2016, 6, 4167–4174. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F.; Ames, G.F.-L.; Barnes, W.M.; Clement, J.M.; Hofnung, M. A novel intercistronic regulatory element of prokaryotic operons. Nature 1982, 298, 760–762. [Google Scholar] [CrossRef]
- Osborn, A.M.; Bruce, K.D.; Strike, P.; Ritchie, D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997, 19, 239–262. [Google Scholar] [CrossRef] [PubMed]
- Blumenthal, T.; Evans, D.; Link, C.D.; Guffanti, A.; Lawson, D.; Thierry-Mieg, J.; Thierry-Mieg, D.; Chiu, W.L.; Duke, K.; Kiraly, M. A global analysis of Caenorhabditis elegans operons. Nature 2002, 417, 851–854. [Google Scholar] [CrossRef] [PubMed]
- Zorio, D.A.; Cheng, N.N.; Blumenthal, T.; Spieth, J. Operons as a common form of chromosomal organization in C. elegans. Nature 1994, 372, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Eldabagh, R.S.; Mejia, N.G.; Barrett, R.L.; Monzo, C.R.; So, M.K.; Foley, J.J.; Arnone, J.T. Systematic identification, characterization, and conservation of adjacent-gene coregulation in the budding yeast Saccharomyces cerevisiae. Msphere 2018, 3, e00220-18. [Google Scholar] [CrossRef]
- Asfare, S.; Eldabagh, R.; Siddiqui, K.; Patel, B.; Kaba, D.; Mullane, J.; Siddiqui, U.; Arnone, J.T. Systematic Analysis of Functionally Related Gene Clusters in the Opportunistic Pathogen, Candida albicans. Microorganisms 2021, 9, 276. [Google Scholar] [CrossRef]
- Arnone, J.T.; Robbins-Pianka, A.; Arace, J.R.; Kass-Gergi, S.; McAlear, M.A. The adjacent positioning of co-regulated gene pairs is widely conserved across eukaryotes. BMC Genom. 2012, 13, 1–15. [Google Scholar] [CrossRef]
- West, R.; Yocum, R.R.; Ptashne, M. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: Location and function of the upstream activating sequence UASG. Mol. Cell. Biol. 1984, 4, 2467–2478. [Google Scholar] [CrossRef]
- Li, Y.-Y.; Yu, H.; Guo, Z.-M.; Guo, T.-Q.; Tu, K.; Li, Y.-X. Systematic analysis of head-to-head gene organization: Evolutionary conservation and potential biological relevance. PLoS Comput. Biol. 2006, 2, e74. [Google Scholar]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Münster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009, 457, 1033–1037. [Google Scholar] [CrossRef]
- Santangelo, G.; Tornow, J.; McLaughlin, C.; Moldave, K. Properties of promoters cloned randomly from the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 1988, 8, 4217–4224. [Google Scholar] [CrossRef]
- Neil, H.; Malabat, C.; d’Aubenton-Carafa, Y.; Xu, Z.; Steinmetz, L.M.; Jacquier, A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 2009, 457, 1038–1042. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.C.; Patel, H.; Chia, M.; Moretto, F.; Frith, D.; Snijders, A.P.; van Werven, F.J. Repression of divergent noncoding transcription by a sequence-specific transcription factor. Mol. Cell 2018, 72, 942–954. e947. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Cox, M.P. Characterization of Bicistronic Transcription in Budding Yeast. Msystems 2021, 6, e01002-20. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shitrit, T.; Yosef, N.; Shemesh, K.; Sharan, R.; Ruppin, E.; Kupiec, M. Systematic identification of gene annotation errors in the widely used yeast mutation collections. Nat. Methods 2012, 9, 373–378. [Google Scholar] [CrossRef]
- Atias, N.; Kupiec, M.; Sharan, R. Systematic identification and correction of annotation errors in the genetic interaction map of Saccharomyces cerevisiae. Nucleic Acids Res. 2016, 44, e50. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spiegel, J.; Arnone, J.T. Transcription at a Distance in the Budding Yeast Saccharomyces cerevisiae. Appl. Microbiol. 2021, 1, 142-149. https://doi.org/10.3390/applmicrobiol1010011
Spiegel J, Arnone JT. Transcription at a Distance in the Budding Yeast Saccharomyces cerevisiae. Applied Microbiology. 2021; 1(1):142-149. https://doi.org/10.3390/applmicrobiol1010011
Chicago/Turabian StyleSpiegel, JerryAnna, and James T. Arnone. 2021. "Transcription at a Distance in the Budding Yeast Saccharomyces cerevisiae" Applied Microbiology 1, no. 1: 142-149. https://doi.org/10.3390/applmicrobiol1010011
APA StyleSpiegel, J., & Arnone, J. T. (2021). Transcription at a Distance in the Budding Yeast Saccharomyces cerevisiae. Applied Microbiology, 1(1), 142-149. https://doi.org/10.3390/applmicrobiol1010011




