Biotechnological Improvement of Nutri-Cereal Finger Millet: Current Status and Future Prospects
Abstract
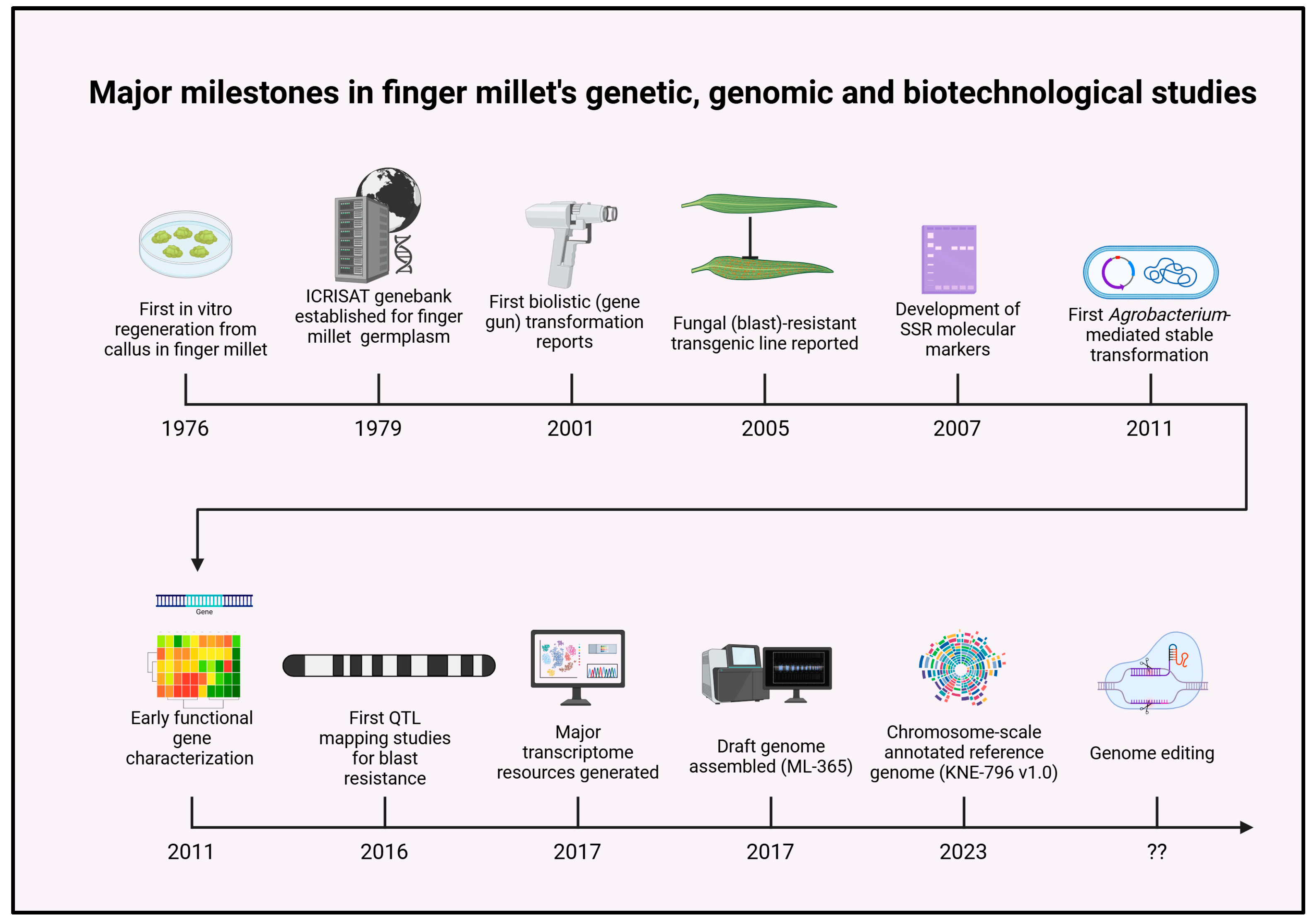
1. Introduction
2. Genetic and Genomic Resources
3. Physiological and Molecular Basis of Stress Resilience
4. Biotechnological Tools for Finger Millet
4.1. In Vitro Regeneration
4.2. Genetic Transformation of Finger Millet
| Year | Method of Transformation | Explant | Strain | Plasmid | Selection Gene | Reporter Gene | Candidate Gene | Reference |
|---|---|---|---|---|---|---|---|---|
| 2001 | Biolistic method | Seed callus | - | - | - | ActI/UqI/RbcS/Ft/uidA | - | [82] |
| 2005 | Biolistic method | Embryogenic calli | - | pPur | bar | uidA | PIN | [78] |
| 2006 | Biolistic method | Shoot tip callus | - | TG0063 of pCAMBIA series | - | - | PcSRP | [83] |
| 2011 | Agrobacterium-mediated | Shoot apex | LBA4404 | pCAMBIA1301 | HptII | uidA | - | [75] |
| 2011 | Agrobacterium-mediated | Seed callus | LBA4404 | pBI-121 | nptII | uidA | - | [77] |
| 2011 | Agrobacterium-mediated | Seed callus | EHA105 | pCNL-56 | nptII | uidA | - | [77] |
| 2012 | Agrobacterium-mediated | Shoot apex | LBA4404 | pHyg-Chi.11 | HptII | - | Chi11 | [80] |
| 2014 | Agrobacterium-mediated | Seed callus | LBA4404 | pCAMBIA1301 | HptII | uidA | SbVPPase | [81] |
| 2014 | Agrobacterium-mediated | Seed callus | EHA105 | pCAMBIA1380 | HptII | uidA | mtlD | [84] |
| 2017 | Agrobacterium-mediated | Shoot apex | EHA105 | pCAMBIA1301 | HptII | uidA | - | [76] |
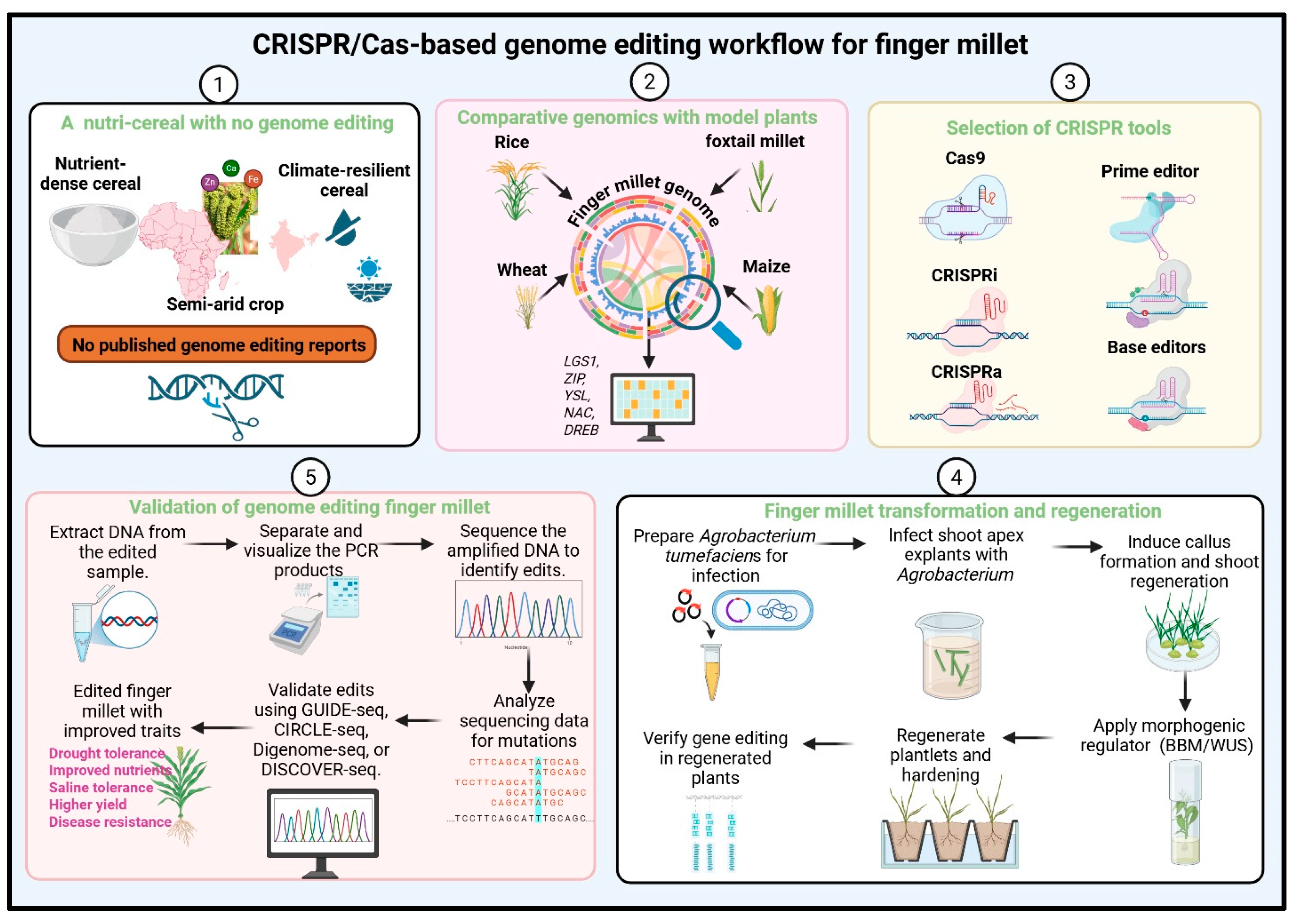
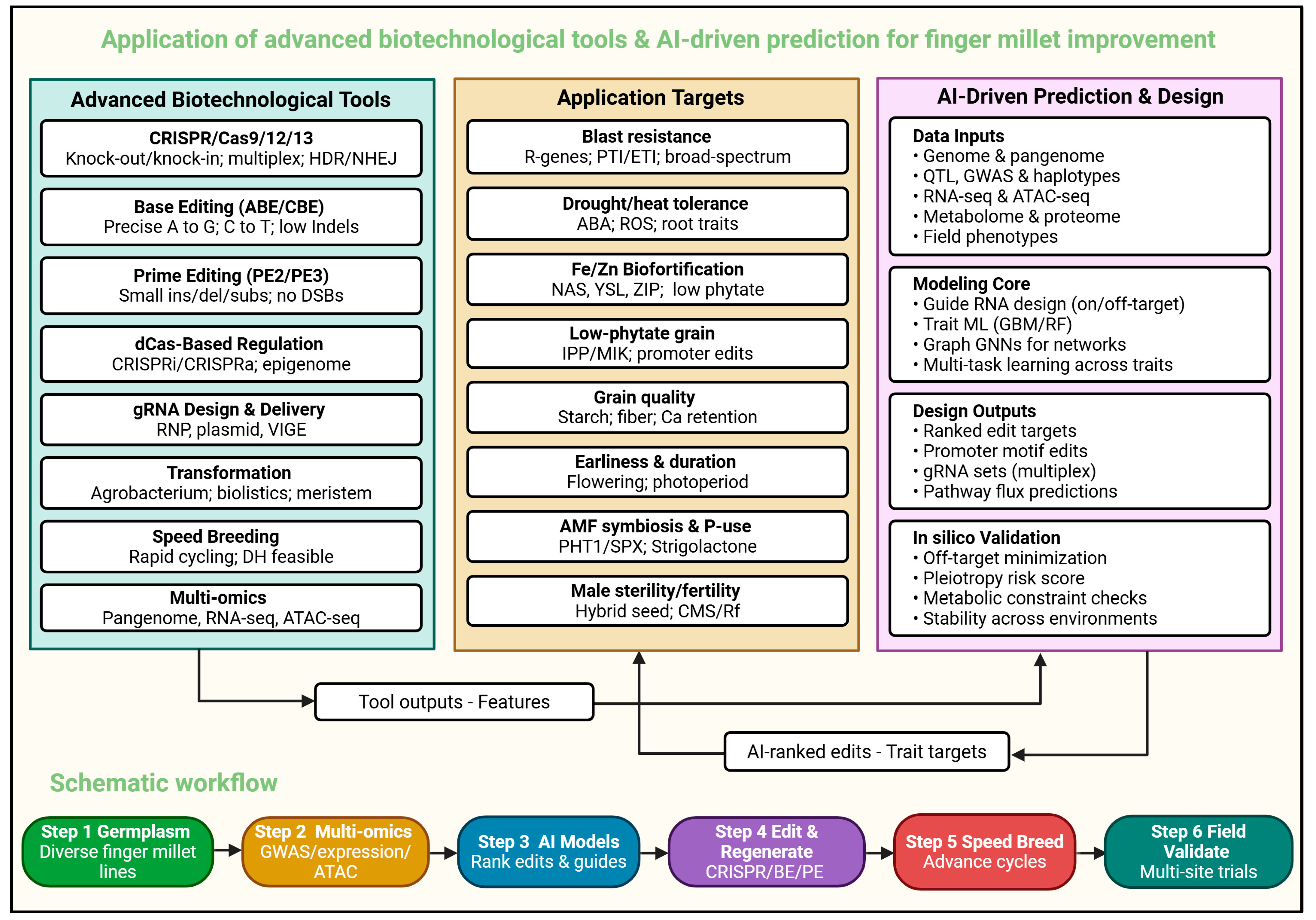
5. Prospectus for CRISPR-Based Genome Editing in Finger Millet
6. Challenges and Future Prospects for Biotechnological Improvement of Finger Millet
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arias, P.; Bellouin, N.; Coppola, E.; Jones, R.; Krinner, G.; Marotzke, J.; Naik, V.; Palmer, M.; Plattner, G.-K.; Rogelj, J. Climate Change 2021: The Physical Science Basis; Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Technical Summary; Intergovernmental Panel on Climate Change (IPCC): Geneva, Switzerland, 2021. [Google Scholar]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of Extreme Weather Disasters on Global Crop Production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y. Temperature Increase Reduces Global Yields of Major Crops in Four Independent Estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef]
- FAO; IFAD; UNICEF; WFP; WHO. The State of Food Security and Nutrition in the World 2020: Transforming Food Systems for Affordable Healthy Diets; Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Muthayya, S.; Rah, J.H.; Sugimoto, J.D.; Roos, F.F.; Kraemer, K.; Black, R.E. The Global Hidden Hunger Indices and Maps: An Advocacy Tool for Action. PLoS ONE 2013, 8, e67860. [Google Scholar] [CrossRef]
- Gupta, S.M.; Arora, S.; Mirza, N.; Pande, A.; Lata, C.; Puranik, S.; Kumar, J.; Kumar, A. Finger Millet: A “Certain” Crop for an “Uncertain” Future and a Solution to Food Insecurity and Hidden Hunger under Stressful Environments. Front. Plant Sci. 2017, 8, 643. [Google Scholar] [CrossRef]
- Goron, T.L.; Raizada, M.N. Genetic Diversity and Physiological Responses of Finger Millet to Abiotic Stress: Opportunities for Crop Improvement. Front. Plant Sci. 2015, 6, 157. [Google Scholar] [CrossRef]
- Devi, P.B.; Vijayabharathi, R.; Sathyabama, S.; Malleshi, N.G.; Priyadarisini, V.B. Health Benefits of Finger Millet (Eleusine coracana L.) Polyphenols and Dietary Fiber: A Review. J. Food Sci. Technol. 2014, 51, 1021–1040. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, N.; Patro, T.S.S.K.; Singamsetti, A.; Sandhya Rani, Y.; Triveni, U.; Nirmala Kumari, A.; Govanakoppa, N.; Lakshmi Pathy, T.; Tonapi, V.A. Comparative Study of AMMI- and BLUP-Based Simultaneous Selection for Grain Yield and Stability of Finger Millet [Eleusine coracana (L.) Gaertn.] Genotypes. Front. Plant Sci. 2022, 12, 786839. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Metwal, M.; Kaur, S.; Gupta, A.K.; Puranik, S.; Singh, S.; Singh, M.; Gupta, S.; Babu, B.K.; Sood, S.; et al. Nutraceutical Value of Finger Millet [Eleusine coracana (L.) Gaertn.], and Their Improvement Using Omics Approaches. Front. Plant Sci. 2016, 7, 934. [Google Scholar] [CrossRef]
- Chandra, D.; Chandra, S.; Pallavi; Sharma, A.K. Review of Finger Millet (Eleusine coracana (L.) Gaertn): A Power House of Health Benefiting Nutrients. Food Sci. Hum. Wellness 2016, 5, 149–155. [Google Scholar] [CrossRef]
- Sharma, A.; Ceasar, S.A.; Pandey, H.; Devadas, V.S.; Kesavan, A.K.; Heisnam, P.; Vashishth, A.; Misra, V.; Mall, A.K. Millets: Nutrient-Rich and Climate-Resilient Crops for Sustainable Agriculture and Diverse Culinary Applications. J. Food Compos. Anal. 2025, 137, 106984. [Google Scholar] [CrossRef]
- Kaur, S.; Kumari, A.; Seem, K.; Kaur, G.; Kumar, D.; Verma, S.; Singh, N.; Kumar, A.; Kumar, M.; Jaiswal, S.; et al. Finger Millet (Eleusine coracana L.): From Staple to Superfood-a Comprehensive Review on Nutritional, Bioactive, Industrial, and Climate Resilience Potential. Planta 2024, 260, 75. [Google Scholar] [CrossRef] [PubMed]
- Anitha, S.; Kane-Potaka, J.; Botha, R.; Givens, D.I.; Sulaiman, N.L.B.; Upadhyay, S.; Vetriventhan, M.; Tsusaka, T.W.; Parasannanavar, D.J.; Longvah, T.; et al. Millets Can Have a Major Impact on Improving Iron Status, Hemoglobin Level, and in Reducing Iron Deficiency Anemia–A Systematic Review and Meta-Analysis. Front. Nutr. 2021, 8, 725529. [Google Scholar] [CrossRef] [PubMed]
- Anitha, S.; Govindaraj, M.; Kane-Potaka, J.; Sulaiman, N.L.B.; Tsusaka, T.W.; Subramaniam, K.; Rajendran, A.; Parasannanavar, D.J.; Bhandari, R.K. Calcium from Finger Millet: A Systematic Review and Meta-Analysis on Calcium Retention, Absorption and Bone Health. Sustainability 2021, 13, 8677. [Google Scholar] [CrossRef]
- Maharajan, T.; Krishna, T.P.A.; Ceasar, S.A. Finger Millet BT-Millets: Crops for Climate Resilience and for Food and Nutritional Security; Ceasar, S.A., Penna, S., Carvalho, C.W.P., Jain, S.M., Eds.; Springer: Singapore, 2025; pp. 1–32. ISBN 978-981-95-1256-0. [Google Scholar]
- Bhatt, A.; Sharma, S.; Joshi, H.C. Finger Millet: A Potential Crop for Climate Resilient Agriculture. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 2190–2200. [Google Scholar]
- Bista, M.K.; Bheemanahalli, R. Finger Millet: A Climate-Resilient and Multi-Nutrient Crop for the Uncertain Future. CSA News 2024, 69, 52–56. [Google Scholar] [CrossRef]
- Maharajan, T.; Antony Ceasar, S.; Ajeesh Krishna, T.P.; Ignacimuthu, S. Finger Millet [Eleusine coracana (L.) Gaertn]: An Orphan Crop with a Potential to Alleviate the Calcium Deficiency in the Semi-Arid Tropics of Asia and Africa. Front. Sustain. Food Syst. 2021, 5, 258. [Google Scholar] [CrossRef]
- Hittalmani, S.; Mahesh, H.B.; Shirke, M.D.; Biradar, H.; Uday, G.; Yadav, M.; Aruna, Y.R.; Lohithaswa, H.C.; Mohanrao, A. Genome and Transcriptome Sequence of Finger Millet (Eleusine coracana) Provides Insights into Drought Tolerance and Nutritional Content. BMC Genom. 2017, 18, 465. [Google Scholar] [CrossRef]
- Ceasar, S.; Maharajan, T.; Ajeesh Krishna, T.P.; Ramakrishnan, M.; Victor Roch, G.; Satish, L.; Ignacimuthu, S. Finger Millet [Eleusine coracana (L.) Gaertn.] Improvement: Current Status and Future Interventions of Whole Genome Sequence. Front. Plant Sci. 2018, 9, 1054. [Google Scholar] [CrossRef]
- Maharajan, T.; Krishna, T.P.A.; Krishnakumar, N.M.; Vetriventhan, M.; Kudapa, H.; Ceasar, S.A. Role of Genome Sequences of Major and Minor Millets in Strengthening Food and Nutritional Security for Future Generations. Agriculture 2024, 14, 670. [Google Scholar] [CrossRef]
- Maharajan, T.; Ceasar, S.A.; Ajeesh Krishna, T.P. Harnessing the Transcriptomic Resources of Millets to Decipher Climate Resilience and Nutrient Enrichment Traits. CRC. Crit. Rev. Plant Sci. 2024, 43, 348–375. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Prabhu, S.; Ebeed, H.T. Protein Research in Millets: Current Status and Way Forward. Planta 2024, 260, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, M.; Aluri, S.; Balachadran, M.T.; Sivarajan, S.R.; Patrignani, A.; Grüter, S.; Poveda, L.; Shimizu-Inatsugi, R.; Baeten, J.; Francoijs, K.-J.; et al. Multiple Hybrid de Novo Genome Assembly of Finger Millet, an Orphan Allotetraploid Crop. DNA Res. 2018, 25, 39–47. [Google Scholar] [CrossRef]
- Devos, K.M.; Qi, P.; Bahri, B.A.; Gimode, D.M.; Jenike, K.; Manthi, S.J.; Lule, D.; Lux, T.; Martinez-Bello, L.; Pendergast, T.H.; et al. Genome Analyses Reveal Population Structure and a Purple Stigma Color Gene Candidate in Finger Millet. Nat. Commun. 2023, 14, 3694. [Google Scholar] [CrossRef] [PubMed]
- Phytozome. Eleusine coracana KNE796-S Reference Genome Assembly (GCA_032690845.1). 2023. Available online: https://phytozome-next.jgi.doe.gov/info/EcoracanaKNE796_S_v1 (accessed on 20 September 2025).
- Upadhyaya, H.D.; Reddy, K.N.; Gowda, C.L.L.; Singh, S. Developing a Mini-Core Collection in Finger Millet Using Multilocation Data. Crop Sci. 2010, 50, 1924–1931. [Google Scholar] [CrossRef]
- International Crops Research Institute for the Semi-Arid Tropics (ICRISAT). ICRISAT Genebank Germplasm Distribution to Global Research Community (as on May 2024). Patancheru, India: ICRISAT Genebank. Available online: https://genebank.icrisat.org/IND/Distributionsummary (accessed on 19 September 2025).
- ICAR NBPGR. Annual Report 2023. New Delhi, India. 2023. Available online: https://nbpgr.org.in/nbpgr2023/wp-content/uploads/2024/07/Annual-report-2023_ICAR-NBPGR.pdf (accessed on 19 September 2025).
- Maharajan, T.; Ceasar, S.A.; Krishna, T.P.A. Finger Millet (Eleusine coracana (L.) Gaertn): Nutritional Importance and Nutrient Transporters. CRC. Crit. Rev. Plant Sci. 2022, 41, 1–31. [Google Scholar] [CrossRef]
- Brhane, H.; Haileselassie, T.; Tesfaye, K.; Ortiz, R.; Hammenhag, C.; Abreha, K.B.; Vetukuri, R.R.; Geleta, M. Finger Millet RNA-Seq Reveals Differential Gene Expression Associated with Tolerance to Aluminum Toxicity and Provides Novel Genomic Resources. Front. Plant Sci. 2022, 13, 1068383. [Google Scholar] [CrossRef]
- Parvathi, M.S.; Nataraja, K.N.; Reddy, Y.A.N.; Naika, M.B.N.; Gowda, M.V.C. Transcriptome Analysis of Finger Millet (Eleusine coracana) Exposed to Progressive Drought Reveals Novel Stress-Responsive Mechanisms. J. Genet. 2019, 98, 46. [Google Scholar] [CrossRef]
- Rahman, H.; Jagadeeshselvam, N.; Valarmathi, R.; Sachin, B.; Sasikala, R.; Senthil, N.; Sudhakar, D.; Robin, S.; Raveendran, M. Transcriptome Analysis of Salinity Responsiveness in Contrasting Genotypes of Finger Millet (Eleusine coracana L.) through RNA-Sequencing. Plant Mol. Biol. 2014, 85, 485–503. [Google Scholar] [CrossRef]
- Ramegowda, V.; Gill, U.S.; Sivalingam, P.N.; Gupta, A.; Gupta, C.; Sharma, A.; Rajamani, V.; Nataraja, K.N.; Pereira, A. GBF3 Transcription Factor Improves Heat and Drought Tolerance in Arabidopsis. Sci. Rep. 2017, 7, 9148. [Google Scholar] [CrossRef] [PubMed]
- Jamra, G.; Agarwal, A.; Singh, N.; Sanyal, S.K.; Kumar, A.; Pandey, G.K. Ectopic Expression of Finger Millet Calmodulin Confers Drought and Salinity Tolerance in Arabidopsis Thaliana. Plant Cell Rep. 2021, 40, 2205–2223. [Google Scholar] [CrossRef]
- Ramegowda, V.; Senthil-Kumar, M.; Nataraja, K.N.; Reddy, M.K.; Mysore, K.S.; Udayakumar, M. Expression of a Finger Millet Transcription Factor, EcNAC1, in Tobacco Confers Abiotic Stress-Tolerance. PLoS ONE 2012, 7, e40397. [Google Scholar] [CrossRef]
- Maharajan, T.; Ajeesh Krishna, T.P.; Rakkammal, K.; Ramakrishnan, M.; Ceasar, S.A.; Ramesh, M.; Ignacimuthu, S. Identification of QTL Associated with Agro-Morphological and Phosphorus Content Traits in Finger Millet under Differential Phosphorus Supply via Linkage Mapping. Agriculture 2023, 13, 262. [Google Scholar] [CrossRef]
- Pendergast, T.H., IV; Qi, P.; Odeny, D.A.; Dida, M.M.; Devos, K.M. A High-Density Linkage Map of Finger Millet Provides QTL for Key Agronomic Traits. Plant Genome 2022, 15, e20175. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Singh, V.K.; Raghavendrarao, S.; Phanindra, M.L.V.; Venkat Raman, K.; Solanke, A.U.; Kumar, P.A.; Sharma, T.R. Expression of Finger Millet EcDehydrin7 in Transgenic Tobacco Confers Tolerance to Drought Stress. Appl. Biochem. Biotechnol. 2015, 177, 207–216. [Google Scholar] [CrossRef]
- Nagarjuna, K.N.; Parvathi, M.S.; Sajeevan, R.S.; Pruthvi, V.; Mamrutha, H.M.; Nataraja, K.N. Full-Length Cloning and Characterization of Abiotic Stress Responsive CIPK31-like Gene from Finger Millet, a Drought-Tolerant Crop. Curr. Sci. 2016, 111, 890–894. [Google Scholar] [CrossRef]
- Rahman, H.; Ramanathan, V.; Nallathambi, J.; Duraialagaraja, S.; Raveendran, M. Over-Expression of a NAC 67 Transcription Factor from Finger Millet (Eleusine coracana L.) Confers Tolerance against Salinity and Drought Stress in Rice. BMC Biotechnol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Parvathi, M.S.; Nataraja, K.N. Discovery of Stress Responsive TATA-Box Binding Protein Associated Factor6 (TAF6) from Finger Millet (Eleusine coracana (L.) Gaertn). J. Plant Biol. 2017, 60, 335–342. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Wang, L.; Zhu, J.; Deng, J.; Tang, R.; Chen, G. Integration of Transcriptomic and Proteomic Analyses for Finger Millet [Eleusine coracana (L.) Gaertn.] in Response to Drought Stress. PLoS ONE 2021, 16, e0247181. [Google Scholar] [CrossRef]
- Singh, S.; Singh, R.K.; Yadav, S.; Tiwari, R. Overexpression of EcDREB2A Transcription Factor from Finger Millet in Tobacco Enhances Tolerance to Heat Stress through ROS Scavenging. J. Biotechnol. 2021, 336, 10–24. [Google Scholar] [CrossRef]
- Ramakrishna, C.; Singh, S.; Raghavendrarao, S.; Padaria, J.C.; Mohanty, S.; Sharma, T.R.; Solanke, A.U. The Membrane Tethered Transcription Factor EcbZIP17 from Finger Millet Promotes Plant Growth and Enhances Tolerance to Abiotic Stresses. Sci. Rep. 2018, 8, 2148. [Google Scholar] [CrossRef]
- Chopperla, R.; Singh, S.; Tomar, R.S.; Mohanty, S.; Khan, S.; Reddy, N.; Padaria, J.C.; Solanke, A.U. Isolation and Allelic Characterization of Finger Millet (Eleusine coracana L.) Small Heat Shock Protein EcHSP17. 8 for Stress Tolerance. Indian J. Genet. Plant Breed. 2018, 78, 95–103. [Google Scholar] [CrossRef]
- Pudake, R.N.; Mehta, C.M.; Mohanta, T.K.; Sharma, S.; Varma, A.; Sharma, A.K. Expression of Four Phosphate Transporter Genes from Finger Millet (Eleusine coracana L.) in Response to Mycorrhizal Colonization and Pi Stress. 3 Biotech 2017, 7, 17. [Google Scholar] [CrossRef]
- Goyal, E.; Singh, A.K.; Mahajan, M.M.; Kanika, K. Comparative Transcriptome Profiling of Contrasting Finger Millet (Eleusine coracana (L.) Gaertn) Genotypes under Heat Stress. Mol. Biol. Rep. 2024, 51, 283. [Google Scholar] [CrossRef] [PubMed]
- Mbinda, W.; Mukami, A. A Review of Recent Advances and Future Directions in the Management of Salinity Stress in Finger Millet. Front. Plant Sci. 2021, 12, 734798. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Tiwari, A.; Sood, S.; Jamra, G.; Singh, N.K.; Meher, P.K.; Kumar, A. Genome Wide Association Mapping of Agro-Morphological Traits among a Diverse Collection of Finger Millet (Eleusine coracana L.) Genotypes Using SNP Markers. PLoS ONE 2018, 13, e0199444. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Ceasar, S.A.; Vinod, K.K.; Duraipandiyan, V.; P, A.K.T.; Upadhyaya, H.D.; Al-Dhabi, N.A.; Ignacimuthu, S. Identification of Putative QTLs for Seedling Stage Phosphorus Starvation Response in Finger Millet (Eleusine coracana L. Gaertn.) by Association Mapping and Cross Species Synteny Analysis. PLoS ONE 2017, 12, e0183261. [Google Scholar] [CrossRef]
- Gaur, V.S.; Tiwari, A.; Kumar, A.; Singh, M. Comparative Proteomic Analysis for Identification and Characterization of Nutritionally and Stress-Responsive Seed Proteins of Finger Millet with Respect to Rice. Plant Cell Biotechnol. Mol. Biol. 2023, 24, 60–73. [Google Scholar] [CrossRef]
- Agrawal, H.; Joshi, R.; Gupta, M. Purification, Identification and Characterization of Two Novel Antioxidant Peptides from Finger Millet (Eleusine coracana) Protein Hydrolysate. Food Res. Int. 2019, 120, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Antony Ceasar, S.; Duraipandiyan, V.; Vinod, K.K.; Kalpana, K.; Al-Dhabi, N.A.; Ignacimuthu, S. Tracing QTLs for Leaf Blast Resistance and Agronomic Performance of Finger Millet (Eleusine coracana (L.) Gaertn.) Genotypes through Association Mapping and in Silico Comparative Genomics Analyses. PLoS ONE 2016, 11, e0159264. [Google Scholar] [CrossRef]
- Chandra, A.K.; Pandey, D.; Sood, S.; Joshi, D.C.; Tiwari, A.; Sharma, D.; Gururani, K.; Kumar, A. Uncovering the Genomic Regions Underlying Grain Iron and Zinc Content Using Genome-Wide Association Mapping in Finger Millet. 3 Biotech 2024, 14, 47. [Google Scholar] [CrossRef]
- Sharma, D.; Tiwari, A.; Sood, S.; Meher, P.K.; Kumar, A. Identification and Validation of Candidate Genes for High Calcium Content in Finger Millet [Eleusine coracana (L.) Gaertn.] through Genome-Wide Association Study. J. Cereal Sci. 2022, 107, 103517. [Google Scholar] [CrossRef]
- Yadav, S.; Kumar, A.; Sood, S. Unraveling the Genetics of Calcium Content in Finger Millet Grains through Association Mapping. Indian J. Genet. Plant Breed. 2020, 80, 432–440. [Google Scholar] [CrossRef]
- Bhatt, D.; Negi, M.; Sharma, P.; Saxena, S.C.; Dobriyal, A.K.; Arora, S. Responses to Drought Induced Oxidative Stress in Five Finger Millet Varieties Differing in Their Geographical Distribution. Physiol. Mol. Biol. Plants 2011, 17, 347–353. [Google Scholar] [CrossRef]
- Bartwal, A.; Pande, A.; Sharma, P.; Arora, S. Intervarietal Variations in Various Oxidative Stress Markers and Antioxidant Potential of Finger Millet (Eleusine coracana) Subjected to Drought Stress. J. Environ. Biol. 2016, 37, 517–522. [Google Scholar]
- Krishnamurthy, L.; Upadhyaya, H.D.; Kashiwagi, J.; Purushothaman, R.; Dwivedi, S.L.; Vadez, V. Variation in Drought-Tolerance Components and Their Interrelationships in the Minicore Collection of Finger Millet Germplasm. Crop Sci. 2016, 56, 1914–1926. [Google Scholar] [CrossRef]
- David, H.; Ramakrishnan, M.; Maharajan, T.; BarathiKannan, K.; Atul Babu, G.; Daniel, M.A.; Agastian, P.; Antony Caesar, S.; Ignacimuthu, S. Mining QTL and Genes for Root Traits and Biochemical Parameters under Vegetative Drought in South Indian Genotypes of Finger Millet (Eleusine coracana (L.) Gaertn) by Association Mapping and in Silico Comparative Genomics. Biocatal. Agric. Biotechnol. 2021, 32, 101935. [Google Scholar] [CrossRef]
- Satish, L.; Rathinapriya, P.; Sagina Rency, A.; Antony Ceasar, S.; Prathibha, M.; Pandian, S.; Rameshkumar, R.; Ramesh, M. Effect of Salinity Stress on Finger Millet (Eleusine coracana (L.) Gaertn): Histochemical and Morphological Analysis of Coleoptile and Coleorhizae. Flora Morphol. Distrib. Funct. Ecol. Plants 2016, 222, 111–120. [Google Scholar] [CrossRef]
- Mukami, A.; Ng’etich, A.; Syombua, E.; Oduor, R.; Mbinda, W. Varietal Differences in Physiological and Biochemical Responses to Salinity Stress in Six Finger Millet Plants. Physiol. Mol. Biol. Plants 2020, 26, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Yogeesh, L.N.; Naryanareddy, A.B.; Nanjareddy, Y.A.; Gowda, M.C.; Channabyre Gowda, M.V. High Temperature Tolerant Genotypes of Finger Millet (Eleusine coracana L.). Nat. Environ. Pollut. Technol. 2016, 15, 1293–1296. [Google Scholar]
- Rakkammal, K.; Maharajan, T.; Shriram, R.N.; Ram, P.S.J.; Ceasar, S.A.; Ramesh, M. Physiological, Biochemical and Molecular Responses of Finger Millet (Eleusine coracana (L.) Gaertn.) Genotypes Exposed to Short-Term Drought Stress Induced by PEG-6000. J. Plant Physiol. 2023, 155, 45–59. [Google Scholar] [CrossRef]
- Sanku, G.; Rajasekaran, R.; Boopathi, N.M.; Krishnamoorthy, I.; Santhanakrishnan, V.P.; Mani, V. Transcriptomic Response of Minor Millets to Abiotic Stresses. Front. Sustain. Food Syst. 2024, 8, 1435437. [Google Scholar] [CrossRef]
- Eapen, S.; George, L. High Frequency Plant Regeneration through Somatic Embryogenesis in Finger Millet (Eleusine coracana Gaertn.). Plant Sci. 1989, 61, 127–130. [Google Scholar] [CrossRef]
- George, L.; Eapen, S. High Frequency Plant Regeneration through Direct Shoot Development and Somatic Embryogenesis from Immature Inflorescence Cultures of Finger Millet (Eleusine coracana Gaertn.). Euphytica 1990, 48, 269–274. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Ignacimuthu, S. Efficient Somatic Embryogenesis and Plant Regeneration from Shoot Apex Explants of Different Indian Genotypes of Finger Millet (Eleusine coracana (L.) Gaertn.). In Vitro Cell. Dev. Biol.-Plant 2008, 44, 427–435. [Google Scholar] [CrossRef]
- Poddar, K.; Vishnoi, R.K.; Kothari, S.L. Plant Regeneration from Embryogenic Callus of Finger Millet (Eleusine coracana (L.) Gaertn.) on Higher Concentrations of NH4NO3 as Replacement of NAA in the Medium. Plant Sci. 1997, 129, 101–106. [Google Scholar] [CrossRef]
- Kothari, S.L.; Agarwal, K.; Kumar, S. Inorganic Nutrient Manipulation for Highly Improved in Vitro Plant Regeneration in Finger Millet (Eleusine coracana (L.) Gaertn.). In Vitro Cell. Dev. Biol.-Plant 2004, 40, 515–519. [Google Scholar] [CrossRef]
- Satish, L.; Ceasar, S.A.; Shilpha, J.; Rency, A.S.; Rathinapriya, P.; Ramesh, M. Direct Plant Regeneration from in Vitro-Derived Shoot Apical Meristems of Finger Millet (Eleusine coracana (L.) Gaertn.). In Vitro Cell. Dev. Biol.-Plant 2015, 51, 192–200. [Google Scholar] [CrossRef]
- Satish, L.; Rency, A.S.; Rathinapriya, P.; Ceasar, S.A.; Pandian, S.; Rameshkumar, R.; Rao, T.B.; Balachandran, S.M.; Ramesh, M. Influence of Plant Growth Regulators and Spermidine on Somatic Embryogenesis and Plant Regeneration in Four Indian Genotypes of Finger Millet (Eleusine coracana (L.) Gaertn). Plant Cell Tissue Organ Cult. 2015, 124, 15–31. [Google Scholar] [CrossRef]
- Ceasar, S.A.; Ignacimuthu, S. Agrobacterium-Mediated Transformation of Finger Millet (Eleusine coracana (L.) Gaertn.) Using Shoot Apex Explants. Plant Cell Rep. 2011, 30, 1759–1770. [Google Scholar] [CrossRef]
- Satish, L.; Ceasar, S.A.; Ramesh, M. Improved Agrobacterium-Mediated Transformation and Direct Plant Regeneration in Four Cultivars of Finger Millet (Eleusine coracana (L.) Gaertn.). Plant Cell Tissue Organ Cult. 2017, 131, 547–565. [Google Scholar] [CrossRef]
- Sharma, M.; Kothari-Chajer, A.; Jagga-Chugh, S.; Kothari, S.L. Factors Influencing Agrobacterium Tumefaciens-Mediated Genetic Transformation of Eleusine coracana (L.) Gaertn. Plant Cell Tissue Organ Cult. 2011, 105, 93–104. [Google Scholar] [CrossRef]
- Latha, A.M.; Rao, K.V.; Reddy, V.D. Production of Transgenic Plants Resistant to Leaf Blast Disease in Finger Millet (Eleusine coracana (L.) Gaertn.). Plant Sci. 2005, 169, 657–667. [Google Scholar] [CrossRef]
- Jagga-Chugh, S.; Kachhwaha, S.; Sharma, M.; Kothari-Chajer, A.; Kothari, S.L. Optimization of Factors Influencing Microprojectile Bombardment-Mediated Genetic Transformation of Seed-Derived Callus and Regeneration of Transgenic Plants in Eleusine coracana (L.) Gaertn. Plant Cell Tissue Organ Cult. 2012, 109, 401–410. [Google Scholar] [CrossRef]
- Ignacimuthu, S.; Ceasar, S.A. Development of Transgenic Finger Millet (Eleusine coracana (L.) Gaertn.) Resistant to Leaf Blast Disease. J. Biosci. 2012, 37, 135–147. [Google Scholar] [CrossRef]
- Anjaneyulu, E.; Reddy, P.S.; Sunita, M.S.; Kishor, P.B.K.; Meriga, B. Salt Tolerance and Activity of Antioxidative Enzymes of Transgenic Finger Millet Overexpressing a Vacuolar H+-Pyrophosphatase Gene (SbVPPase) from Sorghum Bicolor. J. Plant Physiol. 2014, 171, 789–798. [Google Scholar] [CrossRef]
- Gupta, P.; Raghuvanshi, S.; K Tyagi, A. Assessment of the Efficiency of Various Gene Promoters via Biolistics in Leaf and Regenerating Seed Callus of Millets, Eleusine coracana and Echinochloa Crusgalli. Plant Biotechnol. 2001, 18, 275–282. [Google Scholar] [CrossRef]
- Mahalakshmi, S.; Christopher, G.S.B.; Reddy, T.P.; Rao, K.V.; Reddy, V.D. Isolation of a CDNA Clone (PcSrp) Encoding Serine-Rich-Protein from Porteresia Coarctata T. and Its Expression in Yeast and Finger Millet (Eleusine coracana L.) Affording Salt Tolerance. Planta 2006, 224, 347–359. [Google Scholar] [CrossRef]
- Hema, R.; Vemanna, R.S.; Sreeramulu, S.; Reddy, C.P.; Senthil-Kumar, M.; Udayakumar, M. Stable Expression of MtlD Gene Imparts Multiple Stress Tolerance in Finger Millet. PLoS ONE 2014, 9, e99110. [Google Scholar] [CrossRef]
- Sharma, R.; Liang, Y.; Lee, M.Y.; Pidatala, V.R.; Mortimer, J.C.; Scheller, H. V Agrobacterium-Mediated Transient Transformation of Sorghum Leaves for Accelerating Functional Genomics and Genome Editing Studies. BMC Res. Notes 2020, 13, 116. [Google Scholar] [CrossRef]
- Char, S.N.; Wei, J.; Mu, Q.; Li, X.; Zhang, Z.J.; Yu, J.; Yang, B. An Agrobacterium-Delivered CRISPR/Cas9 System for Targeted Mutagenesis in Sorghum. Plant Biotechnol. J. 2020, 18, 319–321. [Google Scholar] [CrossRef]
- Shi, J.; Yu, F. Targeting the Parasite’s Lifeline: Knockout of SL Transporters Confers Durable Striga Resistance in Sorghum. Adv. Biotechnol. 2025, 3, 9. [Google Scholar] [CrossRef]
- Gobena, D.; Shimels, M.; Rich, P.J.; Ruyter-Spira, C.; Bouwmeester, H.; Kanuganti, S.; Mengiste, T.; Ejeta, G. Mutation in Sorghum LOW GERMINATION STIMULANT 1 Reduces Strigolactone Production and Confers Striga Resistance. Proc. Natl. Acad. Sci. USA 2017, 114, 4471–4476. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, Y.; Yang, S.; Zhi, H.; Yin, T.; Ma, X.; Zhang, H.; Diao, X.; Guo, Y.; Li, X.; et al. Establishing in Planta Haploid Inducer Line by Edited SiMTL in Foxtail Millet (Setaria italica). Plant Biotechnol. J. 2021, 19, 1089–1091. [Google Scholar] [CrossRef]
- Zhang, R.; Guo, R.; Zhi, H.; Tang, S.; Wang, L.; Ren, Y.; Ren, G.; Zhang, S.; Feng, J.; Diao, X.; et al. De Novo Creation of Narrowed Plant Architecture via CRISPR/Cas9-Mediated Mutagenesis of SiLGs in Foxtail Millet. Plant Biotechnol. J. 2025, 23, 2400–2402. [Google Scholar] [CrossRef]
- Liang, Z.; Wu, Y.; Ma, L.; Guo, Y.; Ran, Y. Efficient Genome Editing in Setaria italica Using CRISPR/Cas9 and Base Editors. Front. Plant Sci. 2022, 12, 815946. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 Variants Generated by CRISPR-Cas9 Improve Maize Grain Yield under Field Drought Stress Conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Mohr, T.; Horstman, J.; Gu, Y.Q.; Elarabi, N.I.; Abdallah, N.A.; Thilmony, R. CRISPR-Cas9 Gene Editing of the Sal1 Gene Family in Wheat. Plants 2022, 11, 2259. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Zhang, R.; Gao, J.; Qi, Y.; Song, G.; Li, W.; Li, Y.; Li, G. Efficient Multiplex Genome Editing by CRISPR/Cas9 in Common Wheat. Plant Biotechnol. J. 2021, 19, 427–429. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, S.; Jiang, N.; Zhao, X.; Bai, Z.; Liu, J.; Yao, W.; Tang, Q.; Xiao, G.; Lv, C.; et al. Engineering of Rice Varieties with Enhanced Resistances to Both Blast and Bacterial Blight Diseases via CRISPR/Cas9. Plant Biotechnol. J. 2022, 20, 876–885. [Google Scholar] [CrossRef]
- Mikami, M.; Toki, S.; Endo, M. Comparison of CRISPR/Cas9 Expression Constructs for Efficient Targeted Mutagenesis in Rice. Plant Cell Rep. 2015, 88, 561–572. [Google Scholar] [CrossRef]
- Lowder, L.G.; Zhang, D.; Baltes, N.J.; Paul, J.W.; Tang, X.; Zheng, X.; Voytas, D.F.; Hsieh, T.-F.; Zhang, Y.; Qi, Y. A CRISPR/Cas9 Toolbox for Multiplexed Plant Genome Editing and Transcriptional Regulation. Plant Physiol. 2015, 169, 971–985. [Google Scholar] [CrossRef]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.-G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct Design for CRISPR/Cas-Based Genome Editing in Plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef]
- Jung, W.J.; Park, S.-J.; Cha, S.; Kim, K. Factors Affecting the Cleavage Efficiency of the CRISPR-Cas9 System. Animal Cells Syst. 2024, 28, 75–83. [Google Scholar] [CrossRef]
- Kor, S.D.; Chowdhury, N.; Keot, A.K.; Yogendra, K.; Chikkaputtaiah, C.; Sudhakar Reddy, P. RNA Pol III Promoters-Key Players in Precisely Targeted Plant Genome Editing. Front. Genet. 2022, 13, 989199. [Google Scholar] [CrossRef]
- Wang, M.; Mao, Y.; Lu, Y.; Tao, X.; Zhu, J.-K. Multiplex Gene Editing in Rice Using the CRISPR/Cpf1 System. Mol. Plant 2017, 10, 1011–1013. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Self-Processing of Ribozyme-Flanked RNAs into Guide RNAs in Vitro and in Vivo for CRISPR-Mediated Genome Editing. J Integr Plant Biol. 2014, 56, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Atkins, A.; Chung, C.-H.; Allen, A.G.; Dampier, W.; Gurrola, T.E.; Sariyer, I.K.; Nonnemacher, M.R.; Wigdahl, B. Off-Target Analysis in Gene Editing and Applications for Clinical Translation of CRISPR/Cas9 in HIV-1 Therapy. Front. Genome Ed. 2021, 3, 673022. [Google Scholar] [CrossRef] [PubMed]
- Tsakirpaloglou, N.; Septiningsih, E.M.; Thomson, M.J. Guidelines for Performing CRISPR/Cas9 Genome Editing for Gene Validation and Trait Improvement in Crops. Plants 2023, 12, 3564. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Bae, S.; Park, J.; Kim, E.; Kim, S.; Yu, H.R.; Hwang, J.; Kim, J.-I. Digenome-Seq: Genome-Wide Profiling of CRISPR–Cas9 off-Target Effects in Human Cells. Nat. Methods 2015, 12, 237–243. [Google Scholar] [CrossRef]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 Provides Accurate and Rapid Genome Editing Sequence Analysis. Nat. Biotechnol. 2019, 37, 224–226. [Google Scholar] [CrossRef]
- Wienert, B.; Wyman, S.K.; Yeh, C.D.; Conklin, B.R.; Corn, J.E. CRISPR Off-Target Detection with DISCOVER-Seq. Nat. Protoc. 2020, 15, 1775–1799. [Google Scholar] [CrossRef]
- Wienert, B.; Wyman, S.K.; Yeh, C.; Conklin, B.R.; Corn, J.E. Unbiased Detection of CRISPR Off-Targets in Vivo Using DISCOVER-Seq. Science 2019, 364, 286–289. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field High-Throughput Phenotyping: The New Crop Breeding Frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base Editing: Precision Chemistry on the Genome and Transcriptome of Living Cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Jubair, S.; Domaratzki, M. Crop Genomic Selection with Deep Learning and Environmental Data: A Survey. Front. Artif. Intell. 2022, 5, 1040295. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart Breeding Driven by Big Data, Artificial Intelligence, and Integrated Genomic-Enviromic Prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef] [PubMed]
- Lee, M. Deep Learning in CRISPR-Cas Systems: A Review of Recent Studies. Front. Bioeng. Biotechnol. 2023, 11, 1226182. [Google Scholar] [CrossRef]
- Chen, L.; Liu, G.; Zhang, T. Integrating Machine Learning and Genome Editing for Crop Improvement. aBIOTECH 2024, 5, 262–277. [Google Scholar] [CrossRef]
- MacNish, T.R.; Danilevicz, M.F.; Bayer, P.E.; Bestry, M.S.; Edwards, D. Application of Machine Learning and Genomics for Orphan Crop Improvement. Nat. Commun. 2025, 16, 982. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Perez-Rodriguez, P.; Cuevas, J.; Montesinos-Lopez, O.A.; Jarquin, D.; de los Campos, G.; Burgueño, J.; González-Camacho, J.M.; Pérez-Elizalde, S.; Beyene, Y.; et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Theor. Appl. Genet. 2017, 22, 961–975. [Google Scholar] [CrossRef]
- Bellot, P.; de los Campos, G. Can Deep Learning Improve Genomic Prediction of Complex Human Traits? Genetics 2018, 210, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Batley, J.; Edwards, D. Towards Plant Pangenomics. Plant Biotechnol. J. 2016, 14, 1099–1105. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate Solubilizing Bacteria and Their Role in Plant Growth Promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Parniske, M. Arbuscular Mycorrhiza: The Mother of Plant Root Endosymbioses. Nat. Rev. Microbiol. 2008, 6, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Sessitsch, A.; Hardoim, P.; Doring, J.; Weilharter, A.; Krause, A.; Woyke, T.; Mitter, B.; Hauberg-Lotte, L.; Friedrich, F.; Rahalkar, M.; et al. Functional Characteristics of an Endophyte Community Colonizing Rice Roots as Revealed by Metagenomic Analysis. Plant Soil. 2012, 25, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Pretty, J.; Benton, T.G.; Bharucha, Z.P.; Dicks, L.V.; Flora, C.B.; Godfray, H.C.J.; Goulson, D.; Hartley, S.; Lampkin, N.; Morris, C.; et al. Global Assessment of Agricultural System Redesign for Sustainable Intensification. Nat. Sustain. 2018, 1, 441–446. [Google Scholar] [CrossRef]
- Palombi, L.; Sessa, R. Climate Smart Agriculture Sourcebook; FAO: Rome, Italy, 2013. [Google Scholar]
| Trait | Locus/Marker | Type | Evidence Summary | Reference |
|---|---|---|---|---|
| Drought | EcGBF3 (G-box binding factor 3) | Gene (TF) | Overexpression of EcGBF3 improves drought/osmotic tolerance in Arabidopsis | [35] |
| Drought | EcCaM (calmodulin) | Gene | Finger-millet EcCaM enhances drought and salinity tolerance when expressed in Arabidopsis | [36] |
| Drought/salinity/oxidative | EcNAC1 | NAC TF gene | Stress-inducible EcNAC1 from finger millet is strongly up-regulated by stresses. Overexpression in tobacco enhances tolerance to salinity, drought and oxidative stress | [37] |
| Drought | UGEP27–UGEP16 interval (qTPS-1-1; Chr1) | SSR-flanked QTL | Linkage mapping identified QTLs for root & shoot P traits | [38] |
| Heat | UGEP95-candidate HSP70 | SSR -candidate gene | Marker sequence associates with Setaria HSP70 and Ca-binding EGF-domain genes | |
| Cross-trait/diversity resource | DArTseq SNP panel (8778 SNPs) | GBS-SNP markers for GWAS | Genome-enabled DArTseq genotyping of 423 landraces yields 8778 SNPs. Combined with 13 agronomic traits. GWAS and genomic prediction for adaptation and stress-related traits in finger millet. | [39] |
| Drought | EcDehydrin7 | Gene | EcDehydrin7 overexpression in tobacco confers improved drought tolerance | [40] |
| Drought/multiple abiotic stresses | EcCIPK31-like | Gene | EcCIPK31-like in finger millet is induced by dehydration, salinity, heat, cold and oxidative stress. | [41] |
| Drought/salinity (heterologous in rice) | EcNAC67 | NAC TF gene | Salinity-responsive EcNAC67 overexpression in rice improves tolerance to drought and salinity | [42] |
| Drought | EcTAF6 | Gene | EcTAF6 is induced by NaCl, PEG and methyl-viologen | [43] |
| Drought | Genome & transcriptome | Genome &+ RNA-seq | RNA-seq under low-moisture stress reveals numerous drought-induced candidate genes and an expanded repertoire of TFs associated with drought tolerance and nutraceutical traits | [20] |
| Drought-transcriptome & proteome | Drought-responsive DEGs | RNA-seq & label-free proteomics | Time-course drought experiment identifies 80k+ transcripts and 3009 differentially expressed proteins linked to ABA signalling, osmolyte biosynthesis, ROS detoxification and proteostasis modules. | [44] |
| Heat | EcDREB2A | TF gene | EcDREB2A overexpression in tobacco improves tolerance to 42 °C stress via enhanced antioxidant activity, better photosynthetic performance and reduced ROS/membrane damage. | [45] |
| Heat/ER & ROS stress | EcbZIP17 | Gene | EcbZIP17 overexpression in tobacco enhances tolerance to heat and other abiotic stresses, increases growth and yield | [46] |
| Heat | EcHSP17.8 | Gene | Finger-millet small HSP cloned and strong heat-induced upregulation observed across genotypes | [47] |
| Salinity | Salinity-responsive DEGs (transporters, TFs, LEA, etc.) | RNA-seq | Whole-transcriptome profiling of contrasting finger-millet genotypes (Trichy 1 vs. Co-12) under 300 mM NaCl reveals differential induction of ion transporters, LEA proteins and TFs linked to salinity adaptation. | [34] |
| Nutrient stress–phosphorus | EcPT1–EcPT4 (PHT1 family) | Pi transporter genes | Four high-affinity phosphate transporter genes (EcPT1–EcPT4) cloned from finger millet; strongly induced under low-P and AMF colonization in root and shoot tissues. | [48] |
| Heat | HSP/HSF DEGs | RNA-seq candidates | Heat-responsive HSP/HSF transcripts validated by qRT-PCR in contrasting finger millet genotypes. | [49] |
| Salinity | HKT1;5 ortholog(s) | Candidate gene | Na+ retrieval from xylem reduces shoot Na+ and comparative analyses support HKT1;5 as a priority locus in millets | [50] |
| Salinity | NHX1/2 ortholog(s) | Candidate gene | Vacuolar Na+ sequestration via NHX antiporters underpins salt tolerance | [34] |
| Salinity | Salt-responsive DEGs (transporters, LEA) | RNA-seq candidates | Whole-transcriptome profiling of contrasting finger-millet genotypes under salinity identifies ion transporters and LEA-like genes. | |
| Cross-trait | GBS-SNP panels | SNP markers | Genotyping-by-sequencing provides thousands of SNPs enabling GWAS for drought-related and adaptive traits | [32,51] |
| Nutrient stress-low P (association QTL) | SSR markers UGEP19, UGEP68, UGEP13, UGEP90 | QTL (P starvation response) | Association mapping in 128 genotypes identifies four QTL for root and shoot biomass and root hair traits under P starvation at seedling stage. | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceasar, S.A. Biotechnological Improvement of Nutri-Cereal Finger Millet: Current Status and Future Prospects. Crops 2025, 5, 87. https://doi.org/10.3390/crops5060087
Ceasar SA. Biotechnological Improvement of Nutri-Cereal Finger Millet: Current Status and Future Prospects. Crops. 2025; 5(6):87. https://doi.org/10.3390/crops5060087
Chicago/Turabian StyleCeasar, Stanislaus Antony. 2025. "Biotechnological Improvement of Nutri-Cereal Finger Millet: Current Status and Future Prospects" Crops 5, no. 6: 87. https://doi.org/10.3390/crops5060087
APA StyleCeasar, S. A. (2025). Biotechnological Improvement of Nutri-Cereal Finger Millet: Current Status and Future Prospects. Crops, 5(6), 87. https://doi.org/10.3390/crops5060087







