A Genome-Wide Association Study of Seed Morphology-Related Traits in Sorghum Mini-Core and Senegalese Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Phenotypic Evaluations for Seed Morphology-Related Traits
2.2. Statistical Analysis
2.3. GWAS
Genome-Wide Association Study
3. Results
3.1. Seed Morphologies
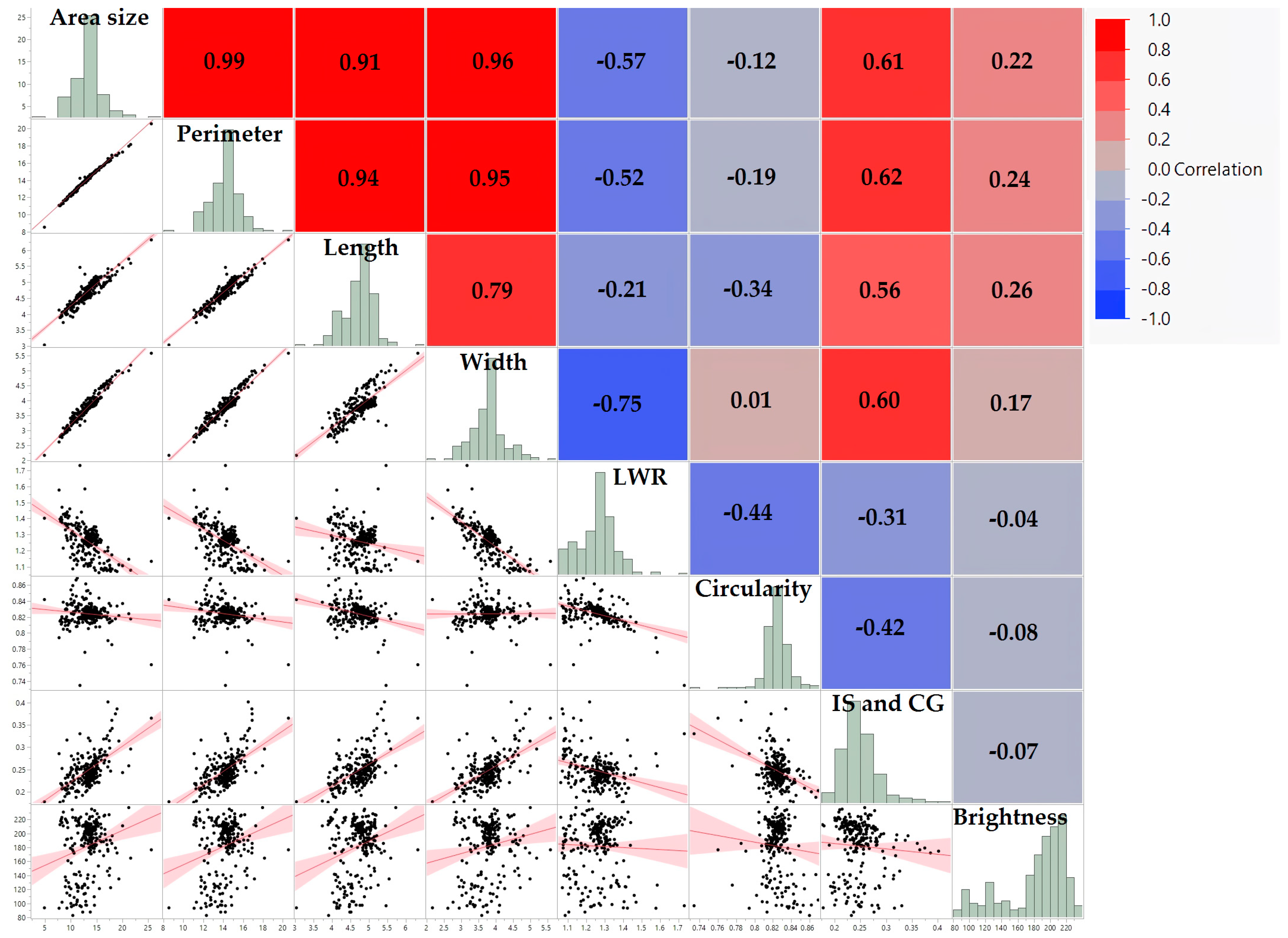
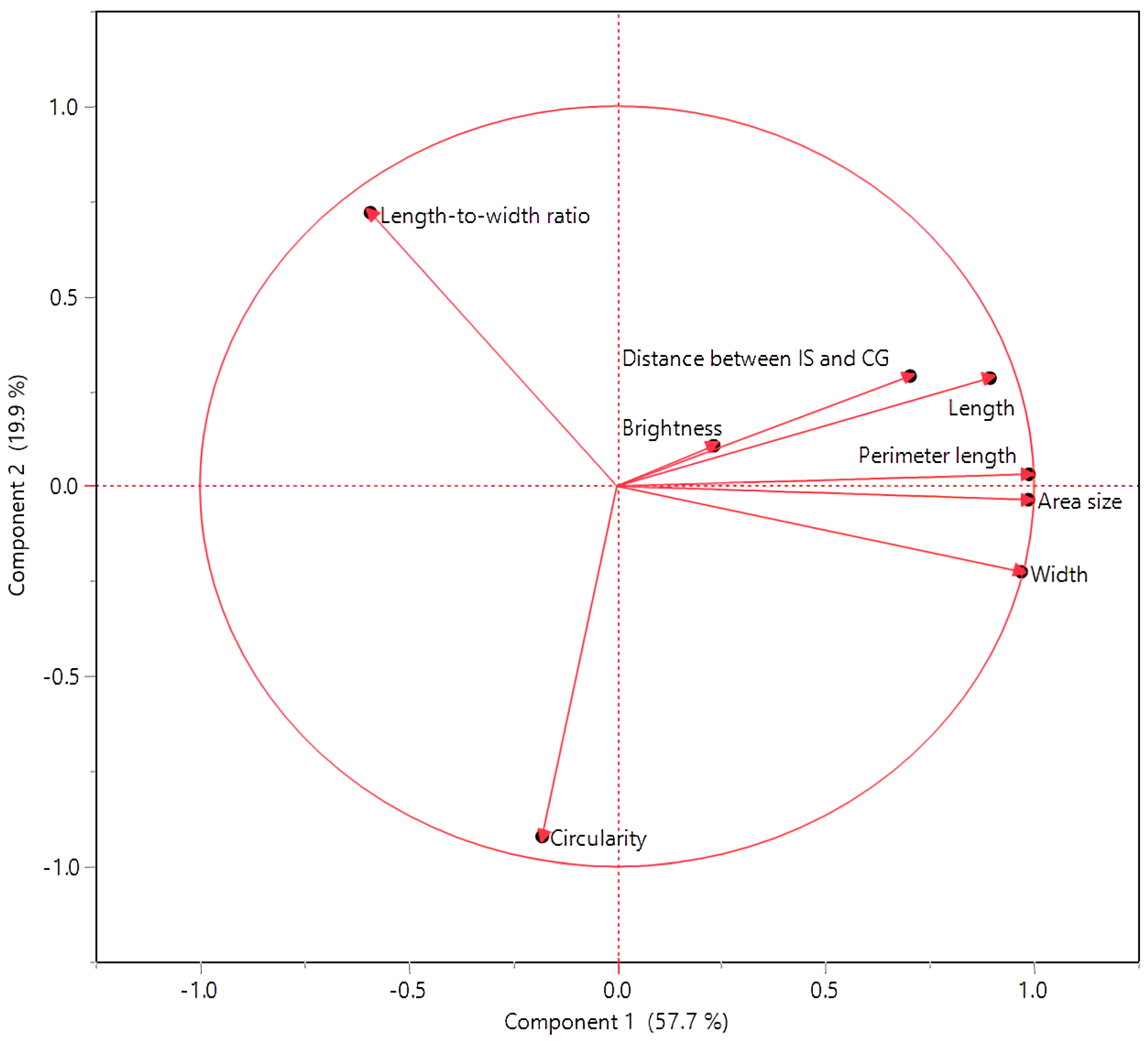
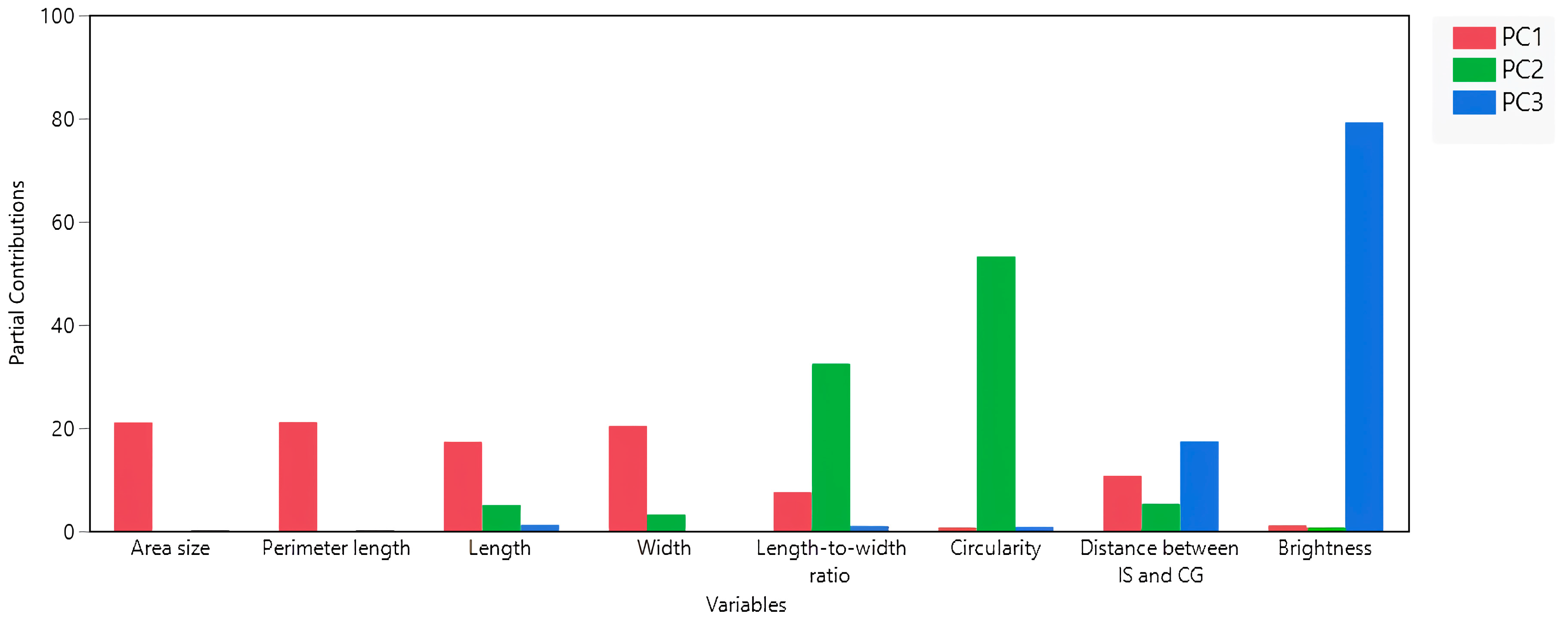
3.2. Interconnections among Seed Morphology Traits
3.3. Associations between Seed Morphology Traits and Resistance to Anthracnose, Head Smut, and Downy Mildew in Mini-Core Lines
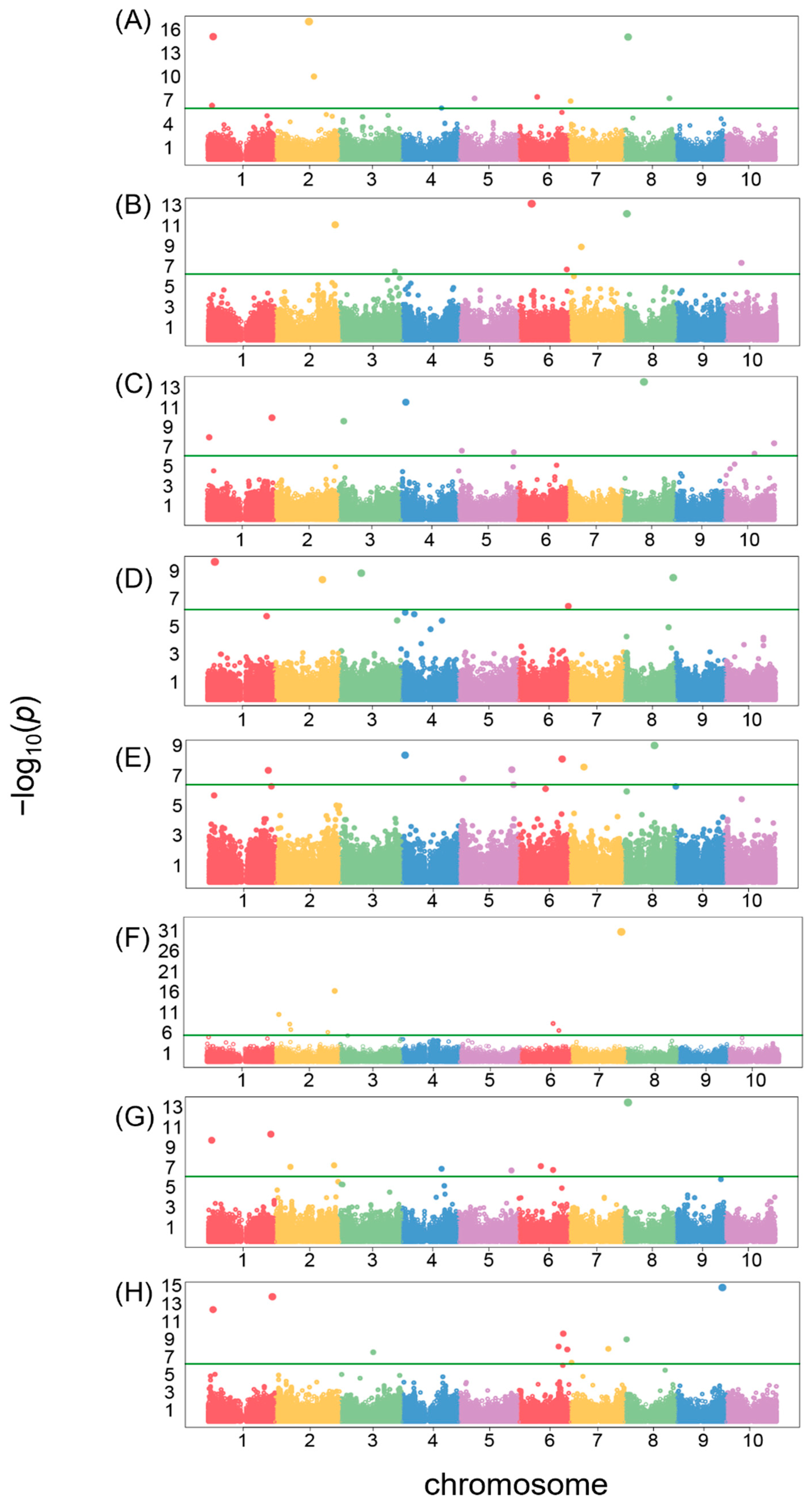
3.4. GWAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Crops and Livestock Products. Available online: http://www.fao.org/faostat/en/#data/QCL (accessed on 16 September 2023).
- Chadalavada, K.; Kumari, B.D.R.; Kumar, T.S. Sorghum mitigates climate variability and change on crop yield and quality. Planta 2021, 253, 113. [Google Scholar] [CrossRef]
- Abreha, K.B.; Enyew, M.; Carlsson, A.S.; Vetukuri, R.R.; Feyissa, T.; Motlhaodi, T.; Ng’uni, D.; Geleta, M. Sorghum in Dryland: Morphological, Physiological, and Molecular Responses of Sorghum under Drought Stress. Planta 2022, 255, 20. [Google Scholar] [CrossRef] [PubMed]
- Enyew, M.; Feyissa, T.; Carlsson, A.S.; Tesfaye, K.; Hammenhag, C.; Seyoum, A.; Geleta, M. Genome-wide analyses using multi-locus models revealed marker-trait associations for major agronomic traits in Sorghum bicolor. Front. Plant Sci. 2022, 13, 999692. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, K.J.; Sokólski, M.M.; Dubis, B.; Załuski, D.; Szempliński, W. Sweet sorghum—Biomass production and energy balance at different levels of agricultural inputs. A six-year field experiment in north-eastern Poland. Eur. J. Agron. 2020, 119, 126119. [Google Scholar] [CrossRef]
- Khalifa, M.; Eltahir, E.A.B. Assessment of global sorghum production, tolerance, and climate risk. Front. Sustain. Food Syst. 2023, 7, 1184373. [Google Scholar] [CrossRef]
- USDA FAS. Sorghum Explorer. Available online: https://ipad.fas.usda.gov/cropexplorer/cropview/commodityView.aspx?cropid=0459200 (accessed on 1 January 2024).
- Demarco, P.A.; Mayor, L.; Rotundo, J.L.; Prasad, P.V.V.; Morris, G.P.; Fernandez, J.A.; Tamagno, S.; Hammer, G.; Messina, C.D.; Ciampitti, I.A. Retrospective study in U.S. commercial sorghum breeding: II. Physiological changes associated to yield gain. Crop Sci. 2023, 63, 867–878. [Google Scholar] [CrossRef]
- Dille, J.A.; Stahlman, P.W.; Thompson, C.R.; Bean, B.W.; Soltani, N.; Sikkema, P.H. Potential Yield Loss in Grain Sorghum (Sorghum bicolor) with Weed Interference in the United States. Weed Technol. 2020, 34, 624–629. [Google Scholar] [CrossRef]
- Tanwar, R.; Panghal, A.; Chaudhary, G.; Kumari, A.; Chhikara, N. Nutritional, Phytochemical and Functional Potential of Sorghum: A Review. Food Chem. Adv. 2023, 3, 100501. [Google Scholar] [CrossRef]
- Frankowski, J.; Przybylska-Balcerek, A.; Stuper-Szablewska, K. Concentration of Pro-Health Compound of Sorghum Grain-Based Foods. Foods 2022, 11, 216. [Google Scholar] [CrossRef]
- Khalid, W.; Ali, A.; Arshad, M.S.; Afzal, F.; Akram, R.; Siddeeg, A.; Kousar, S.; Rahim, M.A.; Aziz, A.; Maqbool, Z. Nutrients and bioactive compounds of Sorghum bicolor L. used to prepare functional foods: A review on the efficacy against different chronic disorders. Int. J. Food Prop. 2022, 25, 1045–1062. [Google Scholar] [CrossRef]
- Taylor, J.R.N.; Duodu, K.G. Resistant-Type Starch in Sorghum Foods—Factors Involved and Health Implications. Starch 2022, 75, 2100296. [Google Scholar] [CrossRef]
- Teferra, T.F.; Awika, J.M. Sorghum as a Healthy Global Food Security Crop: Opportunities and Challenges. Available online: https://www.cerealsgrains.org/publications/cfw/2019/September-October/Pages/CFW-64-5-0054.aspx (accessed on 16 September 2023).
- Mathur, S.; Umakanth, A.V.; Tonapi, V.A.; Sharma, R.; Sharma, M.K. Sweet sorghum as biofuel feedstock: Recent advances and available resources. Biotechnol. Biofuels 2017, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.; Abd El-Wahab, A.; Ahmed, M.F.E.; Buschmann, M.; Visscher, C.; Hartung, C.B.; Lingens, J.B. Nutrient Composition and In Vitro Fermentation Characteristics of Sorghum Depending on Variety and Year of Cultivation in Northern Italy. Foods 2022, 11, 3255. [Google Scholar] [CrossRef] [PubMed]
- Zarei, M.; Amirkolaei, A.K.; Trushenski, J.T.; Sealey, W.M.; Schwarz, M.H.; Ovissipour, R. Sorghum as a Potential Valuable Aquafeed Ingredient: Nutritional Quality and Digestibility. Agriculture 2022, 12, 669. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Pundir, R.P.S.; Dwivedi, S.L.; Gowda, C.L.L.; Reddy, V.G.; Singh, S. Developing a mini core collection of sorghum for diversified utilization of germplasm. Crop Sci. 2009, 49, 1769–1780. [Google Scholar] [CrossRef]
- Ahn, E.; Hu, Z.; Perumal, R.; Prom, L.K.; Odvody, G.; Upadhyaya, H.D.; Magill, C.W. Genome wide association analysis of sorghum mini core lines regarding anthracnose, downy mildew, and head smut. PLoS ONE 2019, 14, e0216671. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.D.; Vetriventhan, M.; Azevedo, V.C.R. Variation for photoperiod and temperature sensitivity in the global mini core collection of sorghum. Front. Plant Sci. 2021, 12, 571243. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, H.E.; Prom, L.K.; Rosa-Valentin, G. Population Structure of the NPGS Senegalese Sorghum Collection and Its Evaluation to Identify New Disease Resistant Genes. PLoS ONE 2018, 13, e0191877. [Google Scholar] [CrossRef]
- Ahn, E.; Fall, C.; Prom, L.K.; Magill, C. Genome-Wide Association Study of Senegalese Sorghum Seedlings Responding to a Texas Isolate of Colletotrichum Sublineola. Sci. Rep. 2022, 12, 13025. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.; Prom, L.K.; Hu, Z.; Odvody, G.; Magill, C. Genome-wide Association Analysis for Response of Senegalese Sorghum Accessions to Texas Isolates of Anthracnose. Plant Genome 2021, 14, e20097. [Google Scholar] [CrossRef]
- Cervantes, E.; Martín, J.J.; Saadaoui, E. Updated Methods for Seed Shape Analysis. Scientifica 2016, 2016, 5691825. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Guo, Q.; Li, H.; Li, J.; Zuo, W.; Long, C. Estimation of morphological variation in seed traits of Sophora moorcroftiana using digital image analysis. Front. Plant Sci. 2023, 14, 1185393. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Upadhyaya, H.D.; Zheng, J.; Liu, Y.; Singh, S.K.; Gowda, C.L.L.; Kumar, R.; Zhu, Y.; Wang, Y.-H.; Li, J. Genome-wide association mapping identifies novel panicle morphology loci and candidate genes in sorghum. Front. Plant Sci. 2021, 12, 743838. [Google Scholar] [CrossRef]
- Sakamoto, L.; Kajiya-Kanegae, H.; Noshita, K.; Takanashi, H.; Kobayashi, M.; Kudo, T.; Yano, K.; Tokunaga, T.; Tsutsumi, N.; Iwata, H. Comparison of shape quantification methods for genomic prediction, and genome-wide association study of sorghum seed morphology. PLoS ONE 2019, 14, e0224695. [Google Scholar] [CrossRef] [PubMed]
- Chopra, R.; Burow, G.; Burke, J.J.; Gladman, N.; Xin, Z. Genome-wide association analysis of seedling traits in diverse Sorghum germplasm under thermal stress. BMC Plant Biol. 2017, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Ahn, E.; Botkin, J.; Ellur, V.; Lee, Y.; Poudel, K.; Prom, L.K.; Magill, C. Genome-Wide Association Study of Seed Morphology Traits in Senegalese Sorghum Cultivars. Plants 2023, 12, 2344. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Hu, Z.; Olatoye, M.O.; Marla, S.; Morris, G.P. An integrated genotyping-by-sequencing polymorphism map for over 10,000 sorghum Genotypes. Plant Genome 2019, 12, 180044. [Google Scholar] [CrossRef] [PubMed]
- Upadhyaya, H.D.; Wang, Y.H.; Gowda, C.L.L.; Sharma, S. Association mapping of maturity and plant height using SNP markers with the sorghum mini core collection. Theor. Appl. Genet. 2013, 126, 2003–2015. [Google Scholar] [CrossRef]
- Browning, B.L.; Browning, S.R. Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 2016, 98, 116–126. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.S.; Zhang, Z. Iterative usage of fixed and random effect models for powerful and efficient genomewide association studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- McCormick, R.F.; Truong, S.K.; Sreedasyam, A.; Jenkins, J.; Shu, S.; Sims, D.; Kennedy, M.; Amirebrahimi, M.; Weers, B.D.; McKinley, B.; et al. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018, 93, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Diantina, S.; McGill, C.; Millner, J.; Nadarajan, J.; Pritchard, H.W.; Clavijo McCormick, A. Comparative Seed Morphology of Tropical and Temperate Orchid Species with Different Growth Habits. Plants 2020, 9, 161. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, H. Genetic Control of Morphological Traits Useful for Improving Sorghum. Breed. Sci. 2023, 73, 57–69. [Google Scholar] [CrossRef]
- Prom, L.K.; Ahn, E.; Isakeit, T.; Magill, C. Correlations among grain mold severity, seed weight, and germination rate of sorghum association panel lines inoculated with Alternaria alternata, Fusarium thapsinum, and Curvularia lunata. J. Agric. Crops 2022, 8, 7–11. [Google Scholar] [CrossRef]
- Ahn, E.; Prom, L.K.; Fall, C.; Magill, C. Response of Senegalese Sorghum Seedlings to Pathotype 5 of Sporisorium reilianum. Crops 2022, 2, 142–153. [Google Scholar] [CrossRef]
- Wright, P.J.; Fullerton, R.A.; Koolaard, J.P. Fungicide control of head smut (Sporisorium reilianum) of sweetcorn (Zea mays). N. Z. J. Crop Hortic. Sci. 2006, 34, 23–26. [Google Scholar] [CrossRef]
- Dong, N.Q.; Sun, Y.; Guo, T.; Shi, C.L.; Zhang, Y.M.; Kan, Y.; Xiang, Y.H.; Zhang, H.; Yang, Y.B.; Li, Y.C. UDP-glucosyltransferase regulates grain size and abiotic stress tolerance associated with metabolic flux redirection in rice. Nat. Comm. 2020, 11, 2629. [Google Scholar] [CrossRef]
- Xu, Q.K.; Yu, H.P.; Xia, S.S.; Cui, Y.J.; Yu, X.Q.; Liu, H.; Zeng, D.L.; Hu, J.; Zhang, Q.; Gao, Z.Y. The C2H2 zinc-finger protein LACKING RUDIMENTARY GLUME 1 regulates spikelet development in rice. Sci. Bull. 2020, 65, 753–764. [Google Scholar] [CrossRef]
- Sun, X.; Xie, Y.; Xu, K.; Li, J. Regulatory networks of the F-box protein FBX206 and OVATE family proteins modulate brassinosteroid biosynthesis to regulate grain size and yield in rice. J. Exp. Bot. 2023, 75, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Maruyama, K.; Seki, M.; Satou, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Leucine-Rich Repeat Receptor-Like Kinase1 Is a Key Membrane-Bound Regulator of Abscisic Acid Early Signaling in Arabidopsis. Plant Cell 2005, 17, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Shahollari, B.; Vadassery, J.; Varma, A.; Oelmüller, R. A Leucine-Rich Repeat Protein Is Required for Growth Promotion and Enhanced Seed Production Mediated by the Endophytic Fungus Piriformospora indica in Arabidopsis thaliana. Plant J. 2007, 50, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, X.; Guo, W. The Cytochrome P450 Superfamily: Key Players in Plant Development and Defense. J. Integr. Agric. 2015, 14, 1673–1686. [Google Scholar] [CrossRef]
- Hansen, C.C.; Nelson, D.R.; Møller, B.L.; Werck-Reichhart, D. Plant Cytochrome P450 Plasticity and Evolution. Mol. Plant 2021, 14, 1244–1265. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Wei, X.; Xiao, Z.; Wang, X.; Ma, S.; Lin, S.; Li, F.; Bu, S.; Liu, Z.; Zhu, H. GW10, a member of P450 subfamily regulates grain size and grain number in rice. Theor. Appl. Genet. 2021, 134, 3941–3950. [Google Scholar] [CrossRef]
- Bao, Y.; Song, W.-M.; Pan, J.; Jiang, C.-M.; Srivastava, R.; Li, B.; Zhu, L.-Y.; Su, H.-Y.; Gao, X.-S.; Liu, H.; et al. Overexpression of the NDR1/HIN1-Like Gene NHL6 modifies seed germination in response to abscisic acid and abiotic stresses in Arabidopsis. PLoS ONE 2016, 11, e0148572. [Google Scholar] [CrossRef] [PubMed]
- Ihsan, M.Z.; Ahmad, S.J.N.; Shah, Z.H.; Rehman, H.M.; Aslam, Z.; Ahuja, I.; Bones, A.M.; Ahmad, J.N. Gene Mining for Proline Based Signaling Proteins in Cell Wall of Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 233. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, L.; Cao, Y.; Wang, H.; Li, H. Association mapping and transcriptome analysis reveal the genetic architecture of maize kernel size. Front. Plant Sci. 2021, 12, 273. [Google Scholar] [CrossRef]
- Malzahn, A.; Lowder, L.; Qi, Y. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Petolino, J.F. Genome editing in plants via designed zinc finger nucleases. In Vitro Cell. Dev. Biol. 2015, 51, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Xu, Z.; Wang, F.; Huang, Y.; Xin, Y.; Liang, S.; Li, B.; Si, H.; Sun, L.; Wang, Q.; et al. Development of an efficient and precise adenine base editor (ABE) with expanded target range in allotetraploid cotton (Gossypium hirsutum). BMC Biol. 2022, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022, 40, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Lin, Q.; Gao, Q.; Gao, C. Optimized Prime Editing in Monocot Plants Using PlantPegDesigner and Engineered Plant Prime Editors (EPPEs). Nat. Protoc. 2023, 18, 831–853. [Google Scholar] [CrossRef] [PubMed]
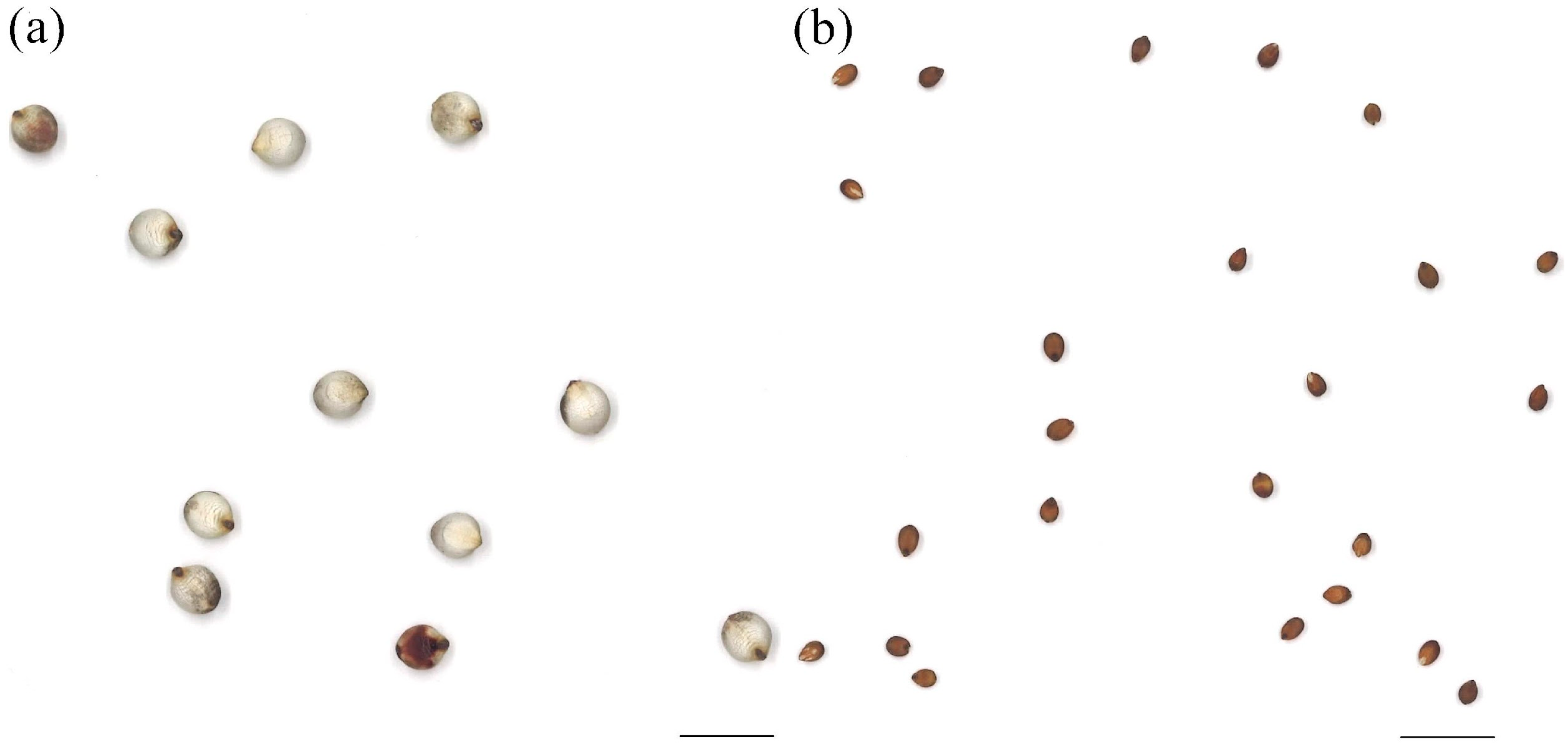
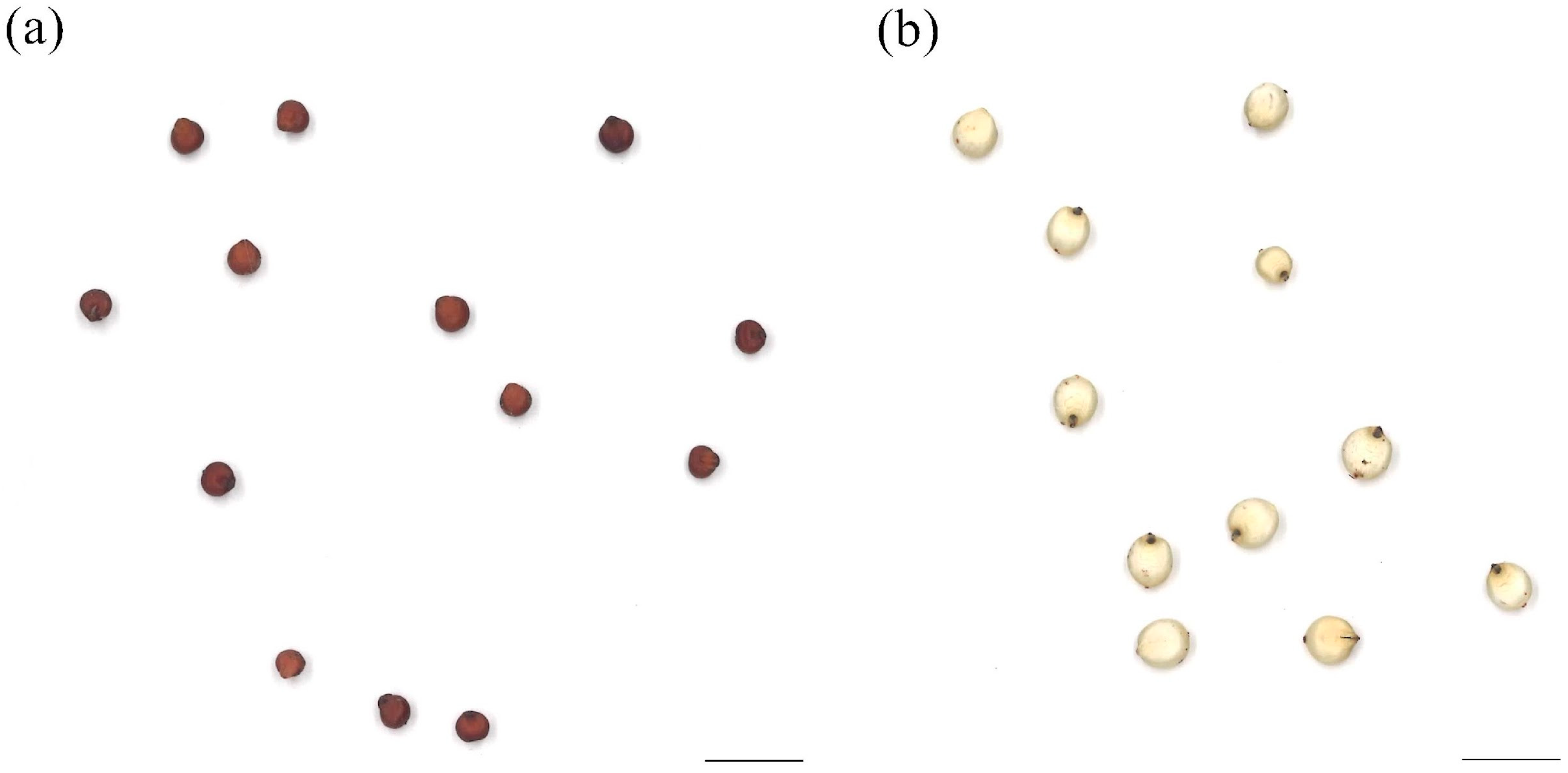
| Accession | Mean ± S.D. | Accession | Mean ± S.D. |
|---|---|---|---|
| Largest Area Size (mm2) | Smallest Area Size (mm2) | ||
| IS11473 | 25.60 ± 1.60 | IS12697 | 4.92 ± 0.83 |
| PI514404 | 21.61 ± 3.75 | IS13264 | 7.76 ± 1.26 |
| IS7987 | 21.24 ± 2.70 | PI514394 | 7.93 ± 1.00 |
| PI253986 | 20.04 ± 3.63 | PI514308 | 8.07 ± 0.89 |
| IS28141 | 19.32 ± 2.75 | PI514474 | 8.18 ± 0.78 |
| Longest perimeter (mm) | Shortest perimeter (mm) | ||
| IS11473 | 20.57 ± 0.65 | IS12697 | 8.54 ± 0.75 |
| PI514404 | 18.16 ± 1.70 | PI514394 | 11.04 ± 0.72 |
| IS7987 | 17.96 ± 1.21 | IS13264 | 11.08 ± 1.20 |
| PI253986 | 17.28 ± 1.54 | PI514308 | 11.13 ± 0.63 |
| IS28141 | 17.15 ± 1.21 | PI514474 | 11.25 ± 0.57 |
| Longest length (mm) | Shortest length (mm) | ||
| IS11473 | 6.32 ± 0.24 | IS12697 | 3.03 ± 0.29 |
| IS7987 | 5.73 ± 0.37 | PI514434 | 3.74 ± 0.23 |
| PI514404 | 5.59 ± 0.49 | PI514394 | 3.89 ± 0.24 |
| IS28141 | 5.55 ± 0.41 | PI514308 | 3.90 ± 0.20 |
| IS12804 | 5.46 ± 0.41 | IS9108 | 3.91 ± 0.28 |
| Longest width (mm) | Shortest width (mm) | ||
| IS11473 | 5.59 ± 0.23 | IS12697 | 2.17 ± 0.18 |
| PI514404 | 5.19 ± 0.48 | IS13264 | 2.61 ± 0.18 |
| IS7987 | 5.01 ± 0.35 | IS3121 | 2.76 ± 0.26 |
| IS28141 | 5.00 ± 0.39 | PI514394 | 2.79 ± 0.20 |
| IS11026 | 4.99 ± 0.29 | PI514474 | 2.82 ± 0.15 |
| Highest LWR | Lowest LWR | ||
| IS12804 | 1.73 ± 0.17 | IS10302 | 1.06 ± 0.03 |
| IS1233 | 1.59 ± 0.08 | IS11026 | 1.07 ± 0.05 |
| IS13264 | 1.58 ± 0.21 | PI514323 | 1.07 ± 0.04 |
| IS3121 | 1.47 ± 0.11 | PI514283 | 1.08 ± 0.05 |
| PI514471 | 1.46 ± 0.08 | PI514288 | 1.08 ± 0.04 |
| Highest circularity (0–1 scale) | Lowest circularity (0–1 scale) | ||
| IS13294 | 0.87 ± 0.01 | IS12804 | 0.73 ± 0.05 |
| IS2872 | 0.87 ± 0.01 | IS11473 | 0.76 ± 0.03 |
| IS13893 | 0.86 ± 0.01 | IS1233 | 0.78 ± 0.02 |
| IS9108 | 0.86 ± 0.01 | IS14090 | 0.79 ± 0.03 |
| IS12937 | 0.85 ± 0.01 | IS27034 | 0.79 ± 0.03 |
| The longest distance between IS and CG (mm) | The shortest distance between IS and CG (mm) | ||
| IS27034 | 0.40 ± 0.20 | PI514394 | 0.18 ± 0.12 |
| IS11026 | 0.39 ± 0.19 | IS12697 | 0.18 ± 0.10 |
| PI514288 | 0.37 ± 0.24 | PI514434 | 0.18 ± 0.10 |
| IS11473 | 0.37 ± 0.19 | IS2872 | 0.19 ± 0.11 |
| IS14090 | 0.36 ± 0.18 | PI514468 | 0.19 ± 0.12 |
| Brightest (0–255 scale) | Darkest (0–255 scale) | ||
| IS7987 | 237.14 ± 6.64 | IS9108 | 82.80 ± 10.00 |
| IS32439 | 234.96 ± 11.57 | IS11619 | 87.62 ± 13.74 |
| IS32349 | 234.14 ± 12.62 | IS9177 | 87.68 ± 9.00 |
| IS7305 | 232.78 ± 17.78 | IS13264 | 93.12 ± 22.34 |
| PI514446 | 230.68 ± 12.47 | PI11374 | 93.30 ± 12.03 |
| Area Size | Perimeter | Length | Width | LWR | Circularity | IS and CG | Brightness | |
|---|---|---|---|---|---|---|---|---|
| Area size (mm2) | 1.00 *** | 0.99 *** | 0.91 *** | 0.96 *** | −0.57 *** | −0.12 | 0.61 *** | 0.22 ** |
| Perimeter (mm) | 0.99 *** | 1.00 *** | 0.94 *** | 0.95 *** | −0.52 *** | −0.19 * | 0.62 *** | 0.24 ** |
| Length (mm) | 0.91 *** | 0.94 *** | 1.00 *** | 0.79 *** | −0.21 ** | −0.34 *** | 0.56 *** | 0.26 *** |
| Width (mm) | 0.96 *** | 0.95 *** | 0.79 *** | 1.00 *** | −0.75 *** | 0.01 | 0.60 *** | 0.17 ** |
| LWR | −0.57 *** | −0.52 *** | −0.21 ** | −0.75 *** | 1.00 *** | −0.44 *** | −0.31 *** | −0.04 |
| Circularity | −0.12 | −0.19 * | −0.34 *** | 0.01 | −0.44 *** | 1.00 *** | −0.42 *** | −0.08 |
| IS and CG | 0.61 *** | 0.62 *** | 0.56 *** | 0.60 *** | −0.31 *** | −0.42 *** | 1.00 *** | −0.07 |
| Brightness | 0.22 ** | 0.24 ** | 0.26 *** | 0.17 ** | −0.04 | −0.08 | −0.07 | 1.00 *** |
| Cluster | Members | R2 with Its Own Cluster | R2 with the Next Closest | 1 − R2 |
|---|---|---|---|---|
| 1 | Perimeter length | 0.98 | 0.06 | 0.02 |
| Area size | 0.97 | 0.07 | 0.03 | |
| Width | 0.9 | 0.2 | 0.12 | |
| Length | 0.86 | 0.07 | 0.15 | |
| Distance between IS and CG | 0.52 | 0.01 | 0.49 | |
| 2 | Circularity | 0.72 | 0.05 | 0.29 |
| Length-to-width ratio | 0.72 | 0.27 | 0.38 | |
| 3 | Brightness | 1 | 0.04 | 0 |
| Area Size (mm2) | Perimeter (mm) | Length (mm) | Width (mm) | LWR | Circularity (0–1 Scale) | IS and CG (mm) | Brightness (0–255 Scale) | |
|---|---|---|---|---|---|---|---|---|
| Resistant | 14.1 | 14.51 | 4.73 | 4 | 1.19 | 0.83 | 0.26 | 152.12 |
| Susceptible | 12.76 | 13.82 | 4.59 | 3.75 | 1.24 | 0.82 | 0.25 | 160 |
| Significance based on p-value | * | * | ND | * | * | * | ND | ND |
| Area Size (mm2) | Perimeter (mm) | Length (mm) | Width (mm) | LWR | Circularity (0–1 Scale) | IS and CG (mm) | Brightness (0–255 Scale) | |
|---|---|---|---|---|---|---|---|---|
| Anthracnose | NS | NS | NS | NS | NS | NS | * | NS |
| Head smut | * | * | NS | * | * | * | NS | NS |
| Downy mildew | NS | NS | NS | NS | NS | NS | * | NS |
| Traits | Genomic Region (Chr:Start:End) | Genbank Acc | Gene | Functional Description |
|---|---|---|---|---|
| Area size | 2:46,909,052:47,000,446 | XP_002462114.1 | LOC8063481 | Uncharacterized protein LOC8063481 |
| XP_021309157.1 | LOC8062049 | Obtusifoliol 14-alpha demethylase | ||
| XP_021307796.1 | LOC8062051 | Uncharacterized protein LOC8062051 | ||
| Brightness | 6:15,369,645:15,389,645 | XP_002447605.1 | LOC8086462 | Probable carbohydrate esterase At4g34215 |
| Circularity | 4:6,234,171:6,254,171 | XP_002451702.2 | LOC8079462 | Uncharacterized protein LOC8079462 |
| Distance between IS and CG | 1:9,022,825:9,058,954 | XP_002466572.1 | LOC8063379 | Exportin-2 |
| XP_002466573.1 | LOC8063380 | Mitogen-activated protein kinase kinase 9 | ||
| XP_002463995.1 | LOC8063381 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial | ||
| XP_021309701.1 | LOC8062905 | Uncharacterized protein LOC8062905 isoform X2 | ||
| XP_002463997.1 | LOC8062906 | Uncharacterized protein LOC8062906 | ||
| XP_021309690.1 | LOC110432893 | Dynamin-related protein 1C | ||
| XP_002466574.1 | LOC8062907 | Threonine dehydratase biosynthetic, chloroplastic | ||
| Length | 4:3,896,951:3,916,951 | XP_002451576.1 | LOC8074663 | Probable leucine-rich repeat receptor-like protein kinase At1g35710 |
| XP_002453340.2 | LOC8074664 | Opioid growth factor receptor | ||
| LWR | 7:59,797,465:59,817,465 | XP_002444553.1 | LOC8054827 | Zinc finger protein 593 homolog |
| XP_021320741.1 | LOC110437031 | Uncharacterized protein LOC110437031 | ||
| XP_021320742.1 | LOC110437032 | Protein MAINTENANCE OF MERISTEMS-like | ||
| Perimeter | 8:5,758,743:5,781,871 | XP_021321624.1 | LOC8076292 | Uncharacterized protein LOC8076292 |
| XP_002442907.1 | LOC8076293 | Transmembrane 9 superfamily member 1 | ||
| Width | 9:55,101,547:55,133,329 | XP_002440093.1 | LOC8067532 | Probable transcriptional regulatory protein At2g25830 |
| XP_002441399.1 | LOC8082991 | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3A | ||
| XP_021303951.1 | LOC8067925 | Probable peptidyl-tRNA hydrolase 2 isoform X3 | ||
| XP_002440094.1 | LOC8068943 | GATA transcription factor 12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, E.; Park, S.; Hu, Z.; Ellur, V.; Cha, M.; Lee, Y.; Prom, L.K.; Magill, C. A Genome-Wide Association Study of Seed Morphology-Related Traits in Sorghum Mini-Core and Senegalese Lines. Crops 2024, 4, 156-171. https://doi.org/10.3390/crops4020012
Ahn E, Park S, Hu Z, Ellur V, Cha M, Lee Y, Prom LK, Magill C. A Genome-Wide Association Study of Seed Morphology-Related Traits in Sorghum Mini-Core and Senegalese Lines. Crops. 2024; 4(2):156-171. https://doi.org/10.3390/crops4020012
Chicago/Turabian StyleAhn, Ezekiel, Sunchung Park, Zhenbin Hu, Vishnutej Ellur, Minhyeok Cha, Yoonjung Lee, Louis K. Prom, and Clint Magill. 2024. "A Genome-Wide Association Study of Seed Morphology-Related Traits in Sorghum Mini-Core and Senegalese Lines" Crops 4, no. 2: 156-171. https://doi.org/10.3390/crops4020012
APA StyleAhn, E., Park, S., Hu, Z., Ellur, V., Cha, M., Lee, Y., Prom, L. K., & Magill, C. (2024). A Genome-Wide Association Study of Seed Morphology-Related Traits in Sorghum Mini-Core and Senegalese Lines. Crops, 4(2), 156-171. https://doi.org/10.3390/crops4020012






