Abstract
Globalization propelled human migration and commercial exchanges at the global level, but woefully led to the introduction of non-indigenous organisms into several agroecological systems. These include pathogenic bacteria with devastating consequences for numerous crops of agronomical importance for food production worldwide. In the last decade, research efforts have focused on these noxious organisms, aiming to understand their evolutionary processes, degree of pathogenicity, and mitigation strategies, which have allowed stakeholders and policymakers to develop evidence-based regulatory norms to improve management practices and minimize production losses. One of these cases is the bacterium Pseudomonas syringae pv. actinidiae (Psa), the causal agent of the kiwifruit bacterial canker, which has been causing drastic production losses and added costs related to orchard management in the kiwifruit industry. Although Psa is presently considered a pandemic pathogen and far from being eradicated, the implementation of strict regulatory norms and the efforts employed by the scientific community allowed the mitigation, to some extent, of its negative impacts through an integrated pest management approach. This included implementing directive guidelines, modifying cultural practices, and searching for sources of plant resistance. However, bacterial pathogens often have high spatial and temporal variability, with new strains constantly arising through mutation, recombination, and gene flow, posing constant pressure to agroecosystems. This review aims to critically appraise the efforts developed to mitigate bacterial pathogens of agronomical impact, from orchard management to genome analysis, using Psa as a case study, which could allow a prompter response against emerging pathogens in agroecosystems worldwide.
1. Introduction
To meet the dietary needs of a constantly increasing global population, food security has become a major challenge. The need to access improved plant genotypes to increase production and market competitiveness frequently leads to the movement of plant material at regional and global scales, regrettably propelling plant–pathogen interactions outside their natural ecosystems. This subject is of particular concern in the predicted scenarios of climate change, which could alter the distribution and prevalence of plant pathogenic organisms [1].
The constant efforts in evidence-supported policymaking have played a paramount role in plant health management, particularly concerning the mitigation of the impact of bacterial pathogens that affect food crops. Nevertheless, the European and Mediterranean Plant Protection Organization (EPPO) presently recommends regulation as quarantine pests of a total of 26 bacterial organisms, apart from many fungal pathogens, viruses, and other organisms [2]. This list has been continuously changing in recent decades, mostly because emergent plant pathogens are constantly arising. This is the case of, e.g., the bacterium Curtobacterium flaccumfaciens pv. poinsettiae (which affects poinsettia plants), the fungus Raffaelea lauricola (the causing agent of laurel wilt), and the tomato mottle mosaic virus. In turn, the EPPO Alert List, which aims to draw attention to potentially emerging pests and to promote timely mitigation actions, currently includes 10 microorganisms, most of them with agronomical importance (affecting, e.g., maize, tomato, rice plants and grapevines). If effective action is not promptly pursued, these organisms may evolve from local to global dispersion, with severe impacts on the agronomic sector worldwide. So far, approximately 21 species of Pseudomonas have been described as plant pathogens. Among these, P. syringae is one of the most common, being one of the best-studied plant pathogens and frequently acting as a model for understanding plant–microorganism interactions [3]. P. syringae consists of a complex of gram-negative bacterial species formed by different pathogenic varieties that have been adapted to infect different plant species. Conversely, a specific P. syringae pathogenic strain can only induce disease in a limited number of host plants [3]. Based on the specific host range, degree of symptoms caused, metabolic requirements, and genetic analyses, the P. syringae complex has been divided into more than 60 intraspecific taxa called pathovars [4]. These include Pseudomonas syringae pv. actinidiae (Psa), whose spread sharply increased following its consideration as an emerging pathogen in the Mediterranean region in 2009, and which is now considered a quarantine pandemic organism and is mainly responsible for large production losses in the kiwifruit industry [5].
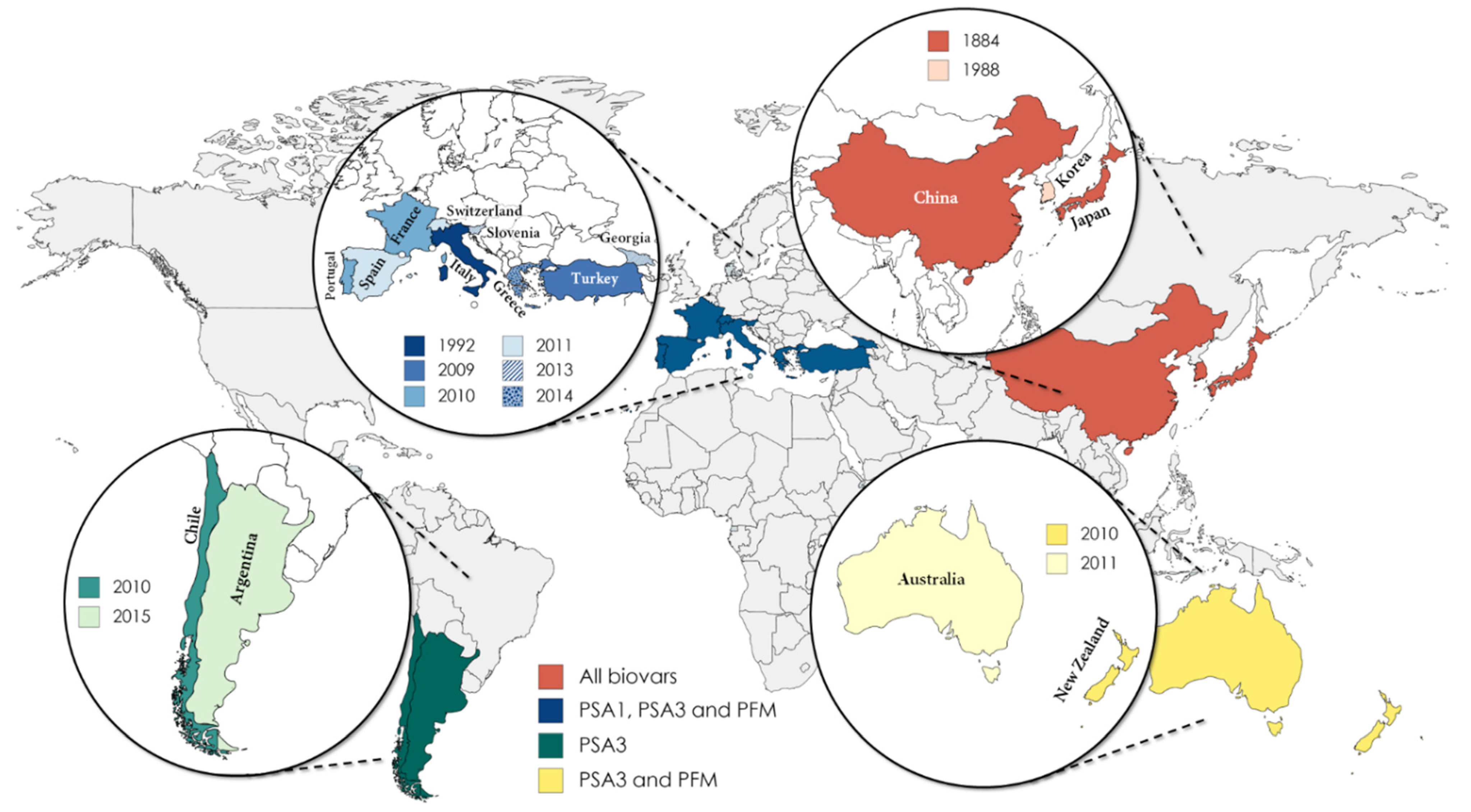
The kiwifruit bacterial canker (KBC), caused by Psa, was first recognized as a severe disease in 1984 following the first outbreaks in Japan in Actinidia chinensis var. deliciosa Hayward (green-fleshed kiwifruit) [6], but it was only during the late 2000s that it impacted European kiwifruit orchards of A. chinensis var. chinensis Hort16A (yellow-fleshed kiwifruit) in Italy [7,8]. As a strategy to prevent the further spreading of the pathogen, the European Commission ordered the member states to instate mandatory official annual surveys for the presence of Psa. As such, frequent prospections on orchards, nurseries, and gardens allowed to conclude that the pathogen was still absent in several European countries, including Austria, Estonia, Belgium, Lithuania, Finland, and the Netherlands [5], reporting negative results for the presence of Psa. In contrast, in countries such as China, Korea, Italy, New Zealand, Chile, Portugal, Spain, Switzerland, and France, the scenario was found to be dramatically different, with the wide spread of bacteria causing a strong economic impact [9]. Psa has also been detected in Turkey, Greece, and Slovenia, but its current distribution is restricted to a few orchards [5] (Figure 1).
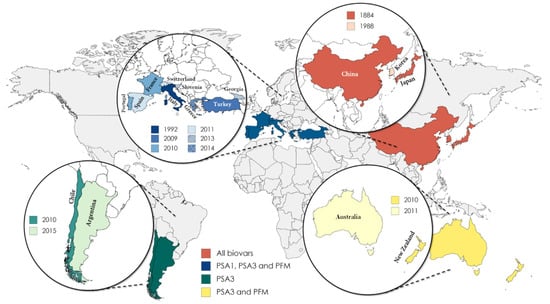
Figure 1.
Current dispersion of Pseudomonas syringae pv. actinidiae (Psa) biovars and Pseudomonas syringae pv. actinidifoliorum (Pfm) around the world and the year of the first occurrence in each country.
When dealing with agronomical diseases, constant surveys must be diligently pursued to allow a prompt response in cases of new disease outbreaks. However, one should be cautious when interpreting these data because the absence of a pathogen may simply result from low colonization levels or insufficient sampling points. Therefore, a higher frequency of more extensive environmental inspections should be greatly reinforced by national and international regulatory organizations so that timely containment measures can be employed against emergent pathogens in agroecosystems. In addition, several other aspects should be considered for an accurate stepwise decision-making scheme for disease management. These include, for example, the life cycle, virulence, and dissemination ability of the pathogen, including its adaptability to local orchard conditions, which should be taken into consideration for a holistic pest management strategy [10]. This review outlines and critically assesses the efforts employed to mitigate bacterial pathogens in crop plants, from orchard management to genome analysis and using Psa as a case study, aiming to support the control of emerging phytopathogens and foster integrated pest management strategies toward more sustainable and resilient agroecosystems.
2. Integrated Approaches to Orchard Management
Although the optimization of disease management strategies is generally lengthy in time and frequently done on a trial-and-error basis, the knowledge already available for other genetically close pathogens and plant species is undoubtedly valuable in shortening the pathway to achieving effective control measures. The approaches traditionally used for controlling pathogenic bacteria include their accurate identification, a thorough understanding of their life cycle, anticipation of dissemination potential and economic impacts, and adoption of basic good orchard management practices. In combination, they have proved to be the main drivers for more successful Psa management on a local and global scale.
2.1. Pathogen Detection and Identification
Diagnostic protocols represent the primary approach for disease monitoring, and in this regard, visual inspection is certainly the simplest method for pathogen detection. However, its efficacy is highly influenced by pathogen density within plant tissues and by other factors that affect the extent of disease symptoms (e.g., age of the plant). In the case of Psa, even in the absence of visual foliar symptoms, the bacterium can strive in plant saps [11], and it has been demonstrated that plant organs may remain asymptomatic until the epiphytic population reaches a quorum for plant infection, which, in turn, is dependent on plant susceptibility [12]. This time gap between infection and symptom appearance delays pathogen detection and decision making, thus potentiating pathogen dissemination within and between orchards. Moreover, some of the symptoms commonly associated with Psa (e.g., leaf spots) can be caused by other phytopathogenic bacteria, such as P. viridiflava. Therefore, an effective diagnosis can only be achieved through laboratory analysis, as visual inspections per se often show poor efficacy in detecting and identifying the pathogen.
Biochemical tests are among the most important methods for microbial identification and are relatively inexpensive and easy to perform. These allow the fast identification of relevant bacteria according to their enzymatic activity, usually related to the fermentation of carbohydrates or the catabolism of proteins or amino acids. For the identification and differentiation of Pseudomonas species, biochemical tests usually focus on their proteolytic and lipolytic activity, production of fluorescent pigments, nitrate, and glutamate utilization, and hemolytic reaction [13]. Other characteristics commonly associated with Pseudomonas spp. include the production of fluorescent siderophores, such as pyoverdine and thioquinolobactin [14]. The method most widely used for the discrimination of plant-pathogenic Pseudomonas spp. is biochemical profiling using the LOPAT tests (L: levan production; O: oxidase production; P: pectinolytic activity; A: arginine dihydrolase production; and T: tobacco hypersensitivity) [15]. Nevertheless, Pseudomonas spp. demonstrate a vast metabolic diversity, which often limits their discrimination through biochemical approaches. For example, while some species, such as P. syringae and P. viridiflava, show a negative reaction in the oxidase test, most species, including P. fluorescens, give a positive result [16]. In fact, LOPAT tests are not able to differentiate Psa from, e.g., P. syringae pv. syringae, and several emergent plant pathogens have shown an atypical LOPAT profile (e.g., Pseudomonas viridiflava) [17]. In addition, pathogen isolation from symptomatic tissues may be challenging if environmental conditions are not favorable for bacterial multiplication [18]. In doubtful cases or cases of first disease reports, pathogenicity tests may be useful in the diagnostic procedure involving the inoculation of the pathogen into its most common host plant (Actinidia spp., in the case of Psa), which can be performed in in vivo or in vitro conditions. In vivo tests involve the observation of symptom development in ten plantlets (ideally 15–20 cm tall) for up to 3 weeks after spray inoculation [18], whereas in vitro tests involve the use of leaf discs [19,20].
Ultimately, molecular tests are the most accurate for bacterial identification, although sufficient optimization must be achieved to avoid false-positive reactions with non-target bacteria. This was the case of the first molecular tests used for Psa detection, based on polymerase chain reaction (PCR) analysis of 16S rDNA, argK genes, and ITS, which yielded false positives due to the presence of DNA from other pathovars, such as P. syringae pv. syringae and P. syringae pv. theae [21,22,23]. Later, improved duplex PCR and real-time PCR (RT-PCR) and qualitative PCR (PCR-C) methodologies were developed. These techniques do not cross-react with genetically close pseudomonads, e.g., P. avellanae, P. viridiflava, and P. syringae pv. tomato [24,25] and are inclusively able to discriminate different Psa pathovars according to their main geographical area of origin based on the sequences of the effector gene HopZ3 and of a PPHGI-1-like genomic island [26]. The continuous development of novel genotyping technologies, such as multiple loci variable number of tandem repeats analysis (MLVA), which can discriminate bacterial populations even at an intra-pathovar level [27,28,29], and increased access to whole-genome sequencing data will certainly contribute to the efforts of accurate and timely bacterial identification [30,31]. More recently, quantitative PCR (qPCR) of Psa-specific genes, such as hopz5, was compared with classical plating assays, granting high-throughput Psa quantification, allowing sample storage, and higher sensitivity than conventional culturing methods in a shorter period [32,33,34]. Although these methods are specific for Psa biovar 3, they may be very useful for research studies aiming at, for example, host-pathogen interactions, and have the potential to be adapted for other pathogenic bacteria.
Rapid and accurate diagnosis is undoubtedly of utmost importance in applying effective measures of disease control. However, most methods for bacterial quantification and identification are either labor intensive, relying on colony counting methods, or are more expensive and require higher technical expertise, particularly those related to molecular profiling. The development of less complex and more accessible methods could propel their use among farmers and field technicians, particularly at the orchard level, thus avoiding the need to transport potentially infected samples to specific diagnostic sites. This could allow faster pathogen recognition, disease diagnosis, and deployment of mitigation strategies. In this regard, the loop-mediated isothermal amplification (LAMP) method based on colorimetric detection has been successfully tested in Psa identification [35,36] and could be used in the onset of novel in situ diagnostic tools. Nevertheless, their broader-scale application still needs to be achieved. Moreover, when distinct pathogens need to be surveyed in the same area, a similar epidemiological and inspection procedure should be used to optimize the survey program. This strategy has the potential to improve field inspections and disease management efforts by allowing for comprehensive pest surveys by cropping areas instead of individual pests.
2.2. Understanding Pathogen Epidemiology
Aside from the accurate identification of the bacterial pathogen, sufficient knowledge of its life cycle is of utmost importance to anticipate potential disease outbreaks and plan the most efficient management strategies to contain its local and global dissemination. In the case of Psa, its main hosts are the species from the genus Actinidia, with very few reports of its presence in non-kiwifruit plants [12,37,38]. As with other pathogenic pseudomonads, Psa infection begins with bacteria entering the host plant through natural openings (e.g., stomata, lenticels, broken trichomes and hydathodes), natural wounds (e.g., wind damage) and artificially made wounds (e.g., pruning, and girdling activities) [12,39,40]. After infection, Psa can colonize and multiply within the apoplastic tissues (asymptomatic biotrophic phase), or it can live epiphytically on leaf surfaces for up to three weeks [12,41,42]. Migration within the plant preferentially occurs through the xylem, often leading to vessel plugging, and through sieve elements and sclerenchyma fibers in the shoot cortex [12,43]. Thereafter, when it overcomes plant defenses (necrotrophic phase), a wide variety of disease symptoms appear. A. chinensis var. chinensis (yellow-fleshed kiwifruit) often shows flower wilt, stem, and cane death and/or bacterial cankers, with higher severity compared with A. chinensis var. deliciosa (green-fleshed kiwifruit), which generally demonstrates symptoms such as leaf spotting, flower wilt, stem, and cane death, and canker formation. In less susceptible Actinidia species, symptom appearance might be more sporadic, as in the case of A. eriantha (wild type) and A. arguta (kiwi berry), in which only a small number of genotypes have shown leaf spotting or leaf fall [7,44,45].
Disease symptoms begin primarily during spring with the appearance of dark-brown necrotic spots surrounded by a yellow halo in the leaves, which subsequently start to curl, wilt, and dry out. Flowers and buds can also wilt and decay, and canker formations can appear in trunks, branches, grafting zones, and wounds, frequently accompanied by the presence of reddish-brown discolorations under the bark and white or red-rusty exudates [12,40,46]. The release of these fluids, which are highly infectious to nearby plants and very characteristic of KBC, results from the elevated bacterial endophytic population that occurs during favorable periods. Branch wilting may compromise bud and flower development, sometimes leading to necrotic lesions in sepals, flower abortion, and deformed fruit [12]. During summer, an increase in average daily temperatures reduces the Psa population in plant tissues, and host infection seems to occur preferentially through stomata and hydathodes [8,12]. As a result of the plant scaring process, callous tissues may become visible [6]. During autumn, this process is further accelerated by an increase in air relative humidity (RH) and lower temperatures [47,48]. During this period, Psa takes advantage of both natural openings (leaf and fruit fall wounds, lenticels, and buds) and man-made wounds (related to harvest and pruning activities), aggravating the risk of infection and the release of infectious exudates [46]. In winter, bacteria can survive in the cortex of infected tissues, resuming their activity in late winter and early spring, and starting the disease cycle again [12]. During this season, periods of moderate rainfall and high RH can reduce the latency period that precedes the appearance of disease symptoms, which are correlated with the degree of infection present in the fall of the previous year [6,48,49]. Fruit quality parameters are also affected by Psa infection, both at harvest and during post-harvest storage. Fruit harvested from symptomatic plants had a lower weight and sugar content, particularly in A. chinensis var. chinensis, compared with A. chinensis var. deliciosa. In addition, fruit harvested from diseased plants showed increased ethylene (ET) production, which is known to reduce fruit storage capacity [50].
2.3. Improvement of Orchard Management
The spread of Psa within and between orchards mainly results from the presence of epiphytic bacterial populations, bacterial exudates, and the accumulation of infected plant material in the orchard. Therefore, the removal of infected material should be dully pursued and comprise not only plants showing disease symptoms but also their surrounding plants, as these may also harbor bacteria without showing disease symptoms [11]. Psa has been found to survive in fallen leaves and pruning wood litter for up to six months, especially under climatic conditions of high precipitation and mild temperatures [12,51]. As this represents a potential source of inoculum for the next growing season, plant litter should be promptly removed from the orchards [52]. Moreover, despite overhead irrigation being used to protect plants during frost episodes, it may facilitate pathogen dissemination between plants. Hence, its replacement with drip irrigation systems should be subject of consideration [53]. Pruning activities also proved to be of paramount importance in disease incidence because wounds can greatly favor the penetration of the pathogen within plant tissues. For example, a higher KBC incidence was observed in A. chinensis orchards of Hayward (female) compared with Matua (male), likely because the female cultivar is more frequently subjected to summer and winter pruning programs [54,55]. In addition, pruning activities performed at the beginning of the vegetative season induced the highest disease incidence and severity (largely due to sap bleeding), while plant pruning during the dormant period resulted in the lowest disease incidence and severity [50]. Furthermore, the hygiene of pruning tools is among the most critical factors in avoiding Psa spread from plant to plant, especially during periods of higher bacterial loads [56]. Therefore, during pruning or harvesting procedures, tools should be sanitized between plants using, e.g., 70% ethanol [57]. The use of wound protectants is also of great importance in controlling wound-infecting pathogens, such as Psa, by constituting a physical barrier, promoting healing, and preventing bacterial dispersal [58,59,60].
The absence of curative measures against bacterial diseases, such as Psa, has propelled the use of prophylactic treatments at strategic moments of plant growth to prevent infection. The use of antibiotics for plant disease control is still allowed in several countries, such as China, the USA, and New Zealand, but their use is generally limited and strongly regulated [61]. Although they are very effective in controlling plant pathogenic bacteria, including Psa, long-term exposure may lead to bacterial resistance [62,63]. The discovery of novel antibiotics for agricultural purposes is still a subject of research, but due to the great limitations involving their use, linked to environmental and human health concerns, their use in fruit production has decreased over the years. As such, during the last decades, the use of copper-based compounds to control microbial pathogens has been substituting antibiotics, mostly due to their high antibacterial activity, broad spectrum, cost-effectiveness, chemical stability, and prolonged effect after application [64]. Until the first Psa outbreaks in Italy and New Zealand, kiwifruit orchards did not require the regular application of phytosanitary products. However, since then, copper application has been adopted as a prophylactic measure against Psa worldwide. In Portugal, for example, governmental guidelines enforce at least six applications of copper-based formulations at strategic moments during the kiwifruit-growing season (e.g., after harvest, at flowering, and after winter pruning) [57]. Nevertheless, several factors affect the efficacy of copper treatments, mostly pertaining to the form of copper used in the formulation, particle size, degree of retention on the plant surface, and solubility [61,65]. More soluble forms (e.g., copper sulfates) pose increased problems because they are more easily washed out after raining, leading to less plant protection and a greater propensity to cause soil pollution [66]. Despite their undeniable success as a prophylactic measure, copper compounds do not have curative properties and show no systemic transport within plant tissues, thus failing in the broad protection of the infected plant [64]. In fact, the inexistence of curative solutions has been the most important handicap in the effective control of plant pathogens, including Psa. Combination of copper with other compounds, such as citric acid and zinc, humic acids, amino acids, or nitrates, has been shown to increase Psa control at lower copper concentrations, due to higher penetration capacity into plant tissues [67,68], having great potential in ensuring more durable plant protection at lower environmental costs.
2.4. Anticipating Pathogen Dispersal
To maintain the competitiveness of the kiwifruit sector, the challenges posed by KBC have been promptly tackled through the adoption of improved orchard management, with some degree of success. Nevertheless, assessing the potential of a pathogen to disseminate and establish in non-native areas is of utmost importance in the anticipation and mitigation of disease outbreaks and dissemination to new planting areas. In the case of Psa, the import of kiwifruit plants from, e.g., China and New Zealand, for the creation of new orchards for commercial exploitation, was likely the main pathway for Psa spread worldwide and may have been at the origin of the outbreaks in France, Spain, and Switzerland [5]. The large dissemination of Psa is also thought to be due to the application of contaminated pollen imported from China and New Zealand as a consequence of the high demand for artificial pollination procedures [69]. In Italy, micropropagated plants were also found to harbor Psa, raising concerns about the risk of pathogen introduction by exchanging asymptomatic plants on regional and global scales [53]. This problem is further worsened by discrepancies regarding national and international plant health directives and limitations in the plant passport system, which often have negligible biosecurity standards that challenge the effectiveness of regulatory guidelines. In this context, the implementation of a common international regulation and regional policy would be of utmost relevance for effective disease control on a global scale.
So far, Psa has severely affected countries such as China, Korea, Italy, New Zealand, Chile, Portugal, Spain, Switzerland, and France, but other favorable areas for its establishment have been identified using the CLIMEX model. These areas include central Africa, Madagascar, and East South America [53]. However, CLIMEX only considers the effects of climate on the distribution of bacterial pathogens, not including other important factors, such as human influence and competition with other microorganisms, limiting its accuracy in predicting potential areas of pathogen dispersal. To overcome this challenge, CLIMEX was recently combined with MaxEnt and a Multi-Model Framework that includes a Support Vector Machine in a study that highlights that climatically susceptible areas where Psa has not yet been established, such as the USA, Iran, Denmark, Belgium, and South Africa, might be affected in the near future [70]. Most importantly, the environmental changes predicted to occur in the next few decades also hinder efforts to anticipate the patterns of global dispersion of bacterial pathogens. Wang et al. [71] combined MaxEnt with ArcGIS to predict the potentially suitable areas and dissemination trends of Psa in China using future climate scenarios predicted by the Intergovernmental Panel on Climate Change (IPCC). The authors concluded that under certain scenarios, the habitat suitability of Psa will increase until the 2080s in China, but data for other regions throughout the globe are still lacking. These pieces of evidence strengthen the need to create improved multi-model approaches to accurately identify susceptible areas. Hence, policymaking could be promptly implemented to support agricultural management and avoid pathogen dispersal within and among geographic regions.
3. Biotic and Abiotic Factors Implicated in Disease Development
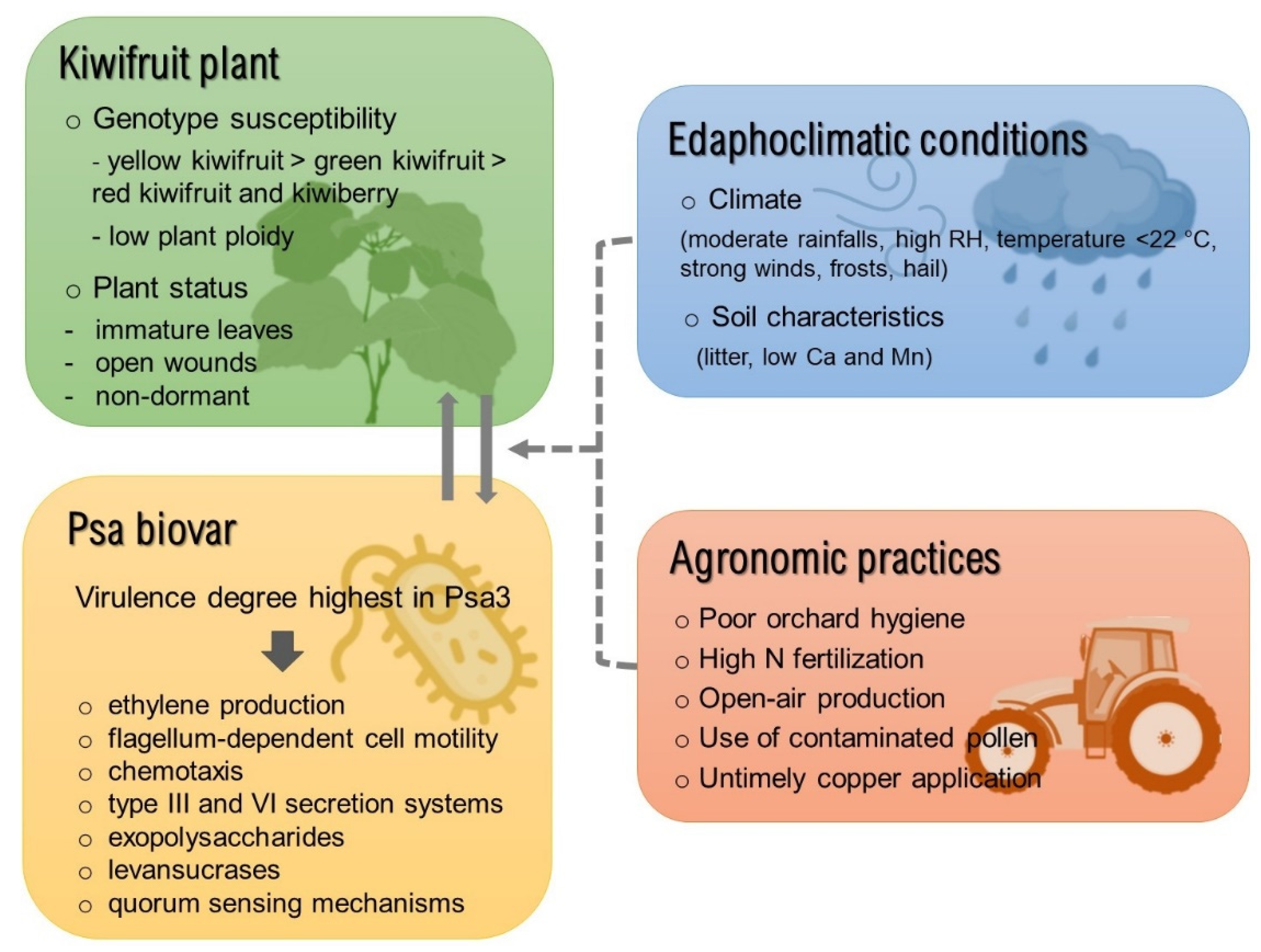
Although the combined development of timely diagnostic procedures, adoption of improved orchard hygiene practices, and thorough understanding of pathogen epidemiology are key aspects in disease management, the extent and outcomes of disease outbreaks are ultimately determined by the specific environment of each orchard. As such, for an effective risk assessment and disease mitigation strategy, factors related to plant genotype, specific strain of the bacterial pathogen, orchard edaphoclimatic conditions, and agronomical practices employed should be taken into consideration (Figure 2).
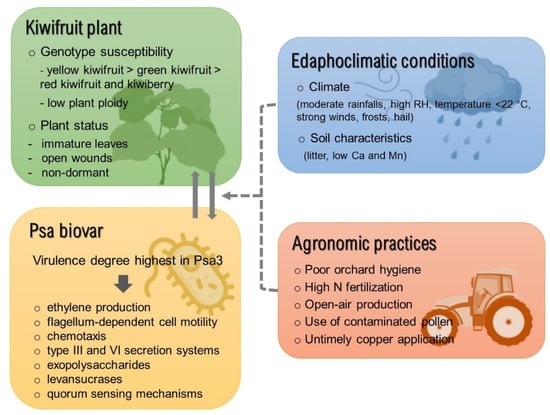
Figure 2.
Overview of the factors involved in the crosstalk between pathogen/host which promote the prevalence/severity of the kiwifruit bacterial canker. Full lines indicate aspects inherent to the pathosystem (Pseudomonas syringae pv. actinidiae/Actinidia spp.) and dashed lines refer to the external factors. Abbreviations: Ca—calcium; Mg—magnesium; RH—relative air humidity; N—nitrogen.
3.1. Plant Genotype
The identification of plant genotypes with a higher tolerance to pests and pathogens has been the cornerstone of more sustainable disease management. Intraspecific and interspecific variability in plant susceptibility to Psa is well known from field experience and observations, but literature references are scarce, and information is often reported in local publications. Moreover, this assessment is not always straightforward due to the complex genotypic variability of the host plant and the lack of standardized methods for tolerance screening. Nevertheless, it is generally acknowledged that all species of the genus Actinidia can be infected with Psa, although there seems to be substantial variation in their tolerance degree to this bacterium. Field and laboratory evidence has shown that cultivars of A. chinensis var. chinensis (such as Hort16A, Hongyang, Soreli, and Jin Tao) present the highest susceptibility to Psa, A. chinensis var. deliciosa cultivars (e.g., Hayward and Jinkui) have shown moderate susceptibility, and A. arguta (including Enza Red and Weiki) and A. rufa cultivars (e.g., Fuchu) were found to be the most tolerant ones [34,44,72,73,74,75,76,77]. Several studies, including a screening of a large number of orchards in New Zealand, also suggest a higher tolerance to Psa with an increase in plant ploidy level (tetraploidy and hexaploidy > diploidy): (i) tetraploid genotypes (A. arguta, A. chrysantha, A. kolomikta, A. macrosperma, A. polygama, A. tetramera, and A. valvata) had higher tolerance to Psa, and (ii) tetraploid and hexaploid genotypes of A. chinensis had a generally higher tolerance than diploid genotypes of the same species [72,74,76,78]. Additionally, male cultivars of A. chinensis have been pointed out as having a lower tolerance to Psa than female cultivars, showing higher levels of Psa endophytic density [12]. Nevertheless, despite these reports consistently attesting for the higher tolerance of some kiwifruit species and varieties over others, some genotypes of a given species considered tolerant can perform drastically differently in their response to Psa infection. For example, distinct genotypes of A. eriantha, considered a tolerant species, behaved from highly tolerant to highly susceptible in an in vitro assay [76]. This hinders the identification of tolerant genotypes, imposing additional efforts on crop improvement strategies.
The full characterization of the vast array of currently existing commercial, wild, and crossbred plant genotypes according to their susceptibility to a specific pathogen may seem a never-ending work, but it has already proven to be successful in overcoming the Psa limitations of the commercial exploration of kiwifruit. After the first devastating outbreaks occurred in New Zealand orchards of Hort16A in 2011, trials were conducted using the library of cultivars already available to find equally interesting commercial ones but with increased tolerance to Psa. Based on the observation of less prevalent and less severe Psa symptoms, decreased bud rot incidence, and rare production of bacterial exudate, affected orchards of Hort16A have been largely replaced by Gold3 [9]. In addition, Actinidia guilinensis, tolerant to Psa, was successfully used as a rootstock to increase the tolerance of highly susceptible A. chinensis Hongyang in an in vitro assay [79]. This evidence opens up the possibility of using grafting-induced resistance to Psa, which can become a long-term sustainable strategy to mitigate the effects of KBC.
Crop improvement through the identification of tolerant genotypes is undoubtedly one of the foundations for more successful management of agronomical pathogens, but potential limitations on, e.g., fruit organoleptic properties, should be thoroughly addressed prior to their large-scale cultivation. Moreover, one should consider that most of the screening programs are developed under greenhouse or microcosm conditions (which allow the screening of a larger number of cultivars at the same time), not accounting for outcomes regarding plant productivity and adaptability to more challenging environmental conditions.
3.2. Bacterial Strain
Aside from plant genotypic variability, the phylogeny of an emergent pathogen is also a key aspect determining the outcome of the disease. Understanding the history of the evolution of the potentially noxious microorganism is, therefore, essential to anticipate its devastating potential and backtracking the origin of emergent virulent isolates, which could help predict the appearance of new strains with higher virulence. As a first step in mitigating the effects of KBC, numerous studies have focused on assessing the genetic diversity and virulence of distinct Psa populations isolated worldwide. So far, six Psa populations (or biovars) have been identified and named according to the chronological order they were characterized, taking into consideration several factors, including their geographic distribution patterns, analysis of whole-genome sequencing, multi-locus sequence of housekeeping and virulence genes, and toxin production [28,30,80,81,82,83].
Until the present moment, Japan has the most diverse bacterial populations associated with KBC [30], whereas lower diversity has been reported in other countries (Figure 1). Psa population 1 (Psa1) is widely distributed in Japan (not only in commercially cultivated kiwifruit plants but also in wild Actinidia spp. from mountain regions) in Italy and Korea [83,84,85,86], and presents a genomic island containing an argK-tox cluster, which is involved in the production of a phaseolotoxin that induces yellow halos around leaf lesions and wilting of shoots and branches [87]. However, not all Psa1 populations can produce this phytotoxin, probably due to mutations in the argK-tox cluster or to the lack of its acquisition from their ancestors [86]. In contrast, Psa2 has only been found in Korea and possesses the gene cluster cor responsible for the synthesis of coronatine, which induces chlorosis, hypertrophy, and stunting of plant tissues [30,88]. Unlike Psa1 and Psa2, Psa population 3 (Psa3) is unable to produce phytotoxins but is the only one capable of producing ET when grown on fresh plant extracts, a feature previously reported for other P. syringae pathovars, such as pvs. glycinea and phaseolicola, but not pv. actinidiae [89]. Psa3 is generally acknowledged as the most virulent among all biovars and is responsible for the most recent and severe outbreaks associated with KBC; being present in all main kiwifruit-producing regions [7,56,90]. Transcriptomic analysis demonstrated that Psa3 pathogenicity results from a strong activation of hypersensitive reaction and pathogenicity (hrp) and hrp conserved (hrc) cluster genes, regulated by bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP), a ubiquitous bacterial second messenger involved in the molecular regulation of bacterial motility [91]. The authors suggested that the lower pathogenicity of Psa2 and Psa1 may result from potential repression of the hrp/hrc cluster or from the activation of flagellum-dependent cell motility and chemotaxis genes, which leads to a weaker induction of type III secretion system (T3SS) responses [91].
Like many other phytopathogenic P. syringae, T3SS plays a central role in Psa3 virulence [92]. Ishiga et al. [93] observed that kiwifruit seedlings inoculated with Psa3 showed severe necrosis within one week after infection, associated with the expression of genes involved in T3SS in planta, which did not occur in seedlings inoculated with a Psa3 T3SS-deficient hrcN mutant. A subsequent study reported that among several T3SS genes, avrE1 and hopR1 are additively required for Psa3 in planta growth and lesion production [94], whereas disruptions in the T3SS genes hopBB1-1 and hopBB1-2, involved in the induction of plant hypersensitive response (HR), resulted in Psa3 populations with lower pathogenicity [95]. Other avirulence genes associated with A. arguta resistance to Psa3 (hopAW1a, avrRpm1a, hopF1c, and hopZ5a) were also identified through effector knockout assays [96]. However, recent studies have pointed out that the T3SS-regulated pathogenicity of Psa3 may be affected by the type VI secretion system (T6SS) through a complex and still poorly understood crosstalk [97]. Two T6SS core genes, tssM and tssJ, seem to be necessary for hemolytic co-regulatory protein secretion, bacterial competition, biofilm formation, hydrogen peroxide tolerance, proteolytic activity, and pathogenicity. In the later stages of infection, T6SS genes seem to be required for the expression of T3SS structural genes (e.g., hrpZ, hrcC, and hopP1), transcriptional regulatory genes (such as hrpR), and effector genes (including hopH1 and hopM1), indicating a potential synergy between T3SS and T6SS during plant infection with Psa3 [97]. In addition, the regulatory system hrpR/hrpS, seems to have a central role in the pathogenicity of Psa3. Insertion events of the transposons ISPsy31 and ISPsy36 in the hrpS and hrpR genes, respectively, impaired Psa3 ability to induce HR and multiply in plant leaves, leading to low virulence [95,98]. Exogenous ET treatments increased the expression of virulence effectors (avrPto1, hopD1, hopR1) and genes related to pilum formation (pilC, pilO), also highlighting the role of plant phytohormones in conditioning bacterial virulence [89]. Moreover, T3SS-mediated HR has recently been demonstrated to play a central role in Psa3-induced cell death, plant immunity activation, and regulation of calcium-sensing receptors (CaS), leading to the upregulation of immune-related genes, accumulation of reactive oxygen species (ROS), and callose deposition [99]. Further knowledge of the highly specialized role of T3SS and T6SS genes during plant infection with phytopathogenic bacteria, including Psa3, could allow for the identification of resistance proteins, supporting effector-assisted breeding in crop improvement programs. For example, the silencing of CaS by RNAi in Nicotiana benthamiana greatly attenuated hopAU1-triggered cell death, impairing plant resistance against Sclerotinia sclerotiorum and Phytophthora capsica [99]. The authors proposed that hopAU1 may be an immune elicitor targeting CaS to trigger plant immunity, and thus CaS could be a promising resistance-related gene for plant breeding against Psa3 [99].
Psa3 can also produce exopolysaccharides, such as alginate and levan, which seem noxious to kiwifruit plants by inducing biofilm production and pathogenicity [100,101]. Recent works have demonstrated that Psa3 possesses at least three genes coding for levansucrases (lscα, lscβ and lscγ) [102]. While lscγ may facilitate Psa’s early infection stages (biotrophic phase) due to the quick polymerization of levan, which allows the pathogen to disguise and protect itself while providing a form of nutrient storage, lscβ promotes pathogen multiplication (necrotrophic phase) through the production of large amounts of glucose for its own metabolism [102]. The capacity of Psa for biofilm formation, motility, and endophytic colonization is also regulated by a putative LuxR solo (PsaR2) involved in quorum sensing [103]. Unlike PsaR2, which seems to be regulated by plant-derived signals, PsaR1 and PsaR3 respond to AHLs produced by neighboring bacteria, putatively regulating Psa interactions with the environment, including other bacteria present in the phyllosphere [104]. Psa3 can also be found in syndemic associations with two other kiwifruit pathogens, e.g., P. syringae pv. syringae and P. viridiflava, suggesting that the establishment of a pathogenic consortium enhances Psa3 pathogenesis [105]. It also induces profound changes in leaf bacterial communities, reducing the relative abundance of, e.g., Hymenobacter, Sphingomonas, and Massilia spp., particularly in male kiwifruit plants [106]. Unravelling the quorum sensing and chemotaxis mechanisms involved in Psa’s perception of the environmental cues that contribute to its virulence could allow the development of novel microbiological strategies to mitigate KBC.
In addition to the Psa biovars previously identified, two other distinct populations, Psa5 and Psa6, were described in Japan in subsequent years [82,83], although they have not been linked to severe disease outbreaks. Conversely, Psa4 was considered a Psa biovar until 2015, having been reclassified as Pseudomonas syringae pv. actinidifoliorum (Pfm) [81,107]. This pathovar encompasses the low virulent strains present in Japan, New Zealand, Australia, France, Spain, and Switzerland [8,11,80,81] and has not been linked with significant symptomatology or production losses [75,108,109]. For these reasons, Pfm has not been subjected to the strict surveying and control measures applied to Psa.
Given that the number of described Psa populations continues to increase, there may be vast bacterial diversity still to be identified. This increase is mostly a result of the extensive survey campaigns implemented in several countries as part of local disease mitigation programs and the development of novel genotyping technologies [110]. However, the missing pieces regarding the genomes of bacterial pathogens greatly limit the full understanding of their origin and temporal development, hindering the knowledge of the evolutionary processes that lead to the appearance of current bacterial populations and the prediction of future evolutionary events.
3.3. Edaphoclimatic Conditions
Regardless of the plant genotype and bacterial strain, orchard edaphoclimatic conditions, mainly concerning rainfall, RH, wind, temperature, and the occurrence of frost and hail, have been considered pivotal modulators of plant responses against Psa infection (Figure 2). The vast multiplicity of these factors is the main driver of distinct disease outcomes among affected regions. For example, it has been observed that rainfall plays a major role in long-term Psa prevalence in open-air orchards, whereas in protected orchards (i.e., installed under covering structures with no exposure to rainfall), successful pathogen eradication was accomplished with the removal of infected plants [111]. However, this climatic parameter seems to have a dual effect, depending on its intensity, as it can promote Psa dispersion and penetration into plant tissues when in small amounts, whereas heavy rainfall can lead to inoculum washing from the leaf canopy, thus decreasing Psa penetration [112]. Long-term high RH also seems to favor bacterial multiplication and movement inside plant tissues. Vanneste et al. [113] showed that maintaining Psa-infected kiwifruit seedlings at an RH above 95% for 21 days can lead to extensive symptoms, including necrosed stem tissues, dead leaves, wilting of plant tips, and colonization of the vascular system (with Psa being detected throughout the stem). However, if the high RH was maintained for only two days, followed by 19 days at 60% RH, bacterial dispersal and visual symptoms were mostly restricted to the point of inoculation [113]. The predicted optimal RH for Psa epiphytic and endophytic growth was estimated to be 60% and 80%, respectively, and it was demonstrated that RH significantly influenced Psa’s maximum population size during its endophytic phase [12].
Along with rainfall and RH, average daily temperatures may also affect the extent of disease symptoms. Reports from several countries have described decreased Psa symptoms during the summer months [113,114,115]. For instance, it was shown that Psa strains from different geographic origins have unique growth patterns at different temperatures, with Psa3 strains being completely inhibited above 30 °C [116]. Other environmental events, such as frost and strong winds, also favor the development of the disease, mostly due to the appearance of injuries in plant tissues that allow bacterial entry [6,117]. Additionally, disease incidence has also been associated with soil moisture, since it has been demonstrated that irrigation targeting soil moisture at field capacity can result in decreased plant infection when compared to lower irrigation levels [50].
To decrease the influence of environmental conditions that favor pathogen growth, several solutions have been suggested during the last decade, such as the use of fans, under-vine shelters, and overhead irrigation [61,118,119]. Protection against rain, wind, and hail can be provided by the installation of windbreaks (either artificial or natural) and coverings (made of plastic or cloth) to diminish bacterial spread and the incidence of plant wounding [118,119,120]. However, these structures pose added costs to orchard management, and very little is known about how they may affect plant productivity. The influence of orchard edaphoclimatic conditions on the outcome of bacterial infections is of undeniable importance, but further research (particularly involving the combination of multiple environmental factors) is still needed to better understand how pathogen multiplication and plant tolerance mechanisms are affected in different cultivation areas. This knowledge could lead to the identification of areas less prone to infection and to the identification of more suitable agronomical practices for each specific region.
3.4. Agronomical Practices
The agronomical practices employed in orchard management are constantly evolving, not only to promote plant productivity but also to cope with undesirable plant pathogenic agents. For example, in kiwifruit orchards, vine architectures favoring open canopies have been proposed to substitute for the most traditional pergola structures in areas with a higher pressure of bacterial infection [8,50]. The implementation of multiple leaders, either through training new plants or through grafting bud sticks into well-established trunks, is also a promising approach to obtaining less dense canopies with a lesser proportion of woody material, thus hampering the spread of bacterial pathogens, including Psa [61,121]. Binding sites of plant branches to poles, touching points of irrigation tubes and shoots, and plant production under high density also seem to facilitate bacterial dispersal and colonization [8]. Therefore, improved management of plant canopies and cultivation density could be adopted to disrupt the favorable conditions of bacterial growth by promoting aeration, light penetration, low overlapping of canes and foliage, and facilitating the penetration and uniformization of chemical treatments [122]. Special attention should also be paid to other plant species present in and around infected orchards, as they can also constitute a source of inoculum. For example, Psa was found living epiphytically on Cryptomeria japonica, often used as shelterbelts in kiwifruit orchards in New Zealand, surviving up to 14 days when artificially inoculated [123]. Psa was also isolated from Setaria viridis, Alternanthera philoxeroides, Paulownia tomentosa, and Broussonetia papyrifera, which inclusively presented the same characteristic disease symptoms [37,38]. Several non-host plants collected in proximity to infected kiwifruit plants, including Calystegia sylvatica and Capsella bursa-pastoris, have also shown potential for harboring Psa [12]. In addition, although artificial pollination is frequently used to overcome potential limitations in plant productivity, flower pollen and the pollinators Apis mellifera and Bombus terrestris are passible for Psa contamination, raising concerns about the possibility of pollinators representing a risk factor for pathogen dissemination [69,124].
Nutrition has also been implicated in plant tolerance against several pathogens, mainly due to alterations in growth, tissue composition, and the expression of disease tolerance-related genes [125]. On the other hand, pathogens may interfere with plants’ nutrient translocation or utilization efficiency, potentially causing nutrient deficiency, hyperaccumulation, or even toxicity [126]. So far, there is still a limited number of studies have addressed the importance of plant nutrition/fertilization in mitigating kiwifruit pathogens such as Psa. For example, the role of N in plant defense against Psa is not straightforward. It has been suggested that high nitrogen fertilization favors the microclimatic conditions needed for Psa epiphytic growth, also leading to reduced penetration of copper treatments due to denser canopies, and exogenous N-supply (as 0.5 mM urea) enhanced plant sensitivity to Psa, increasing bacterial endophytic density [127]. On the other hand, its deficiency also seems to promote Psa growth in planta, probably due to the induction of early leaf senescence, impairment of plant defenses, and depletion of nutrient storage [128]. In addition, the type of N source seems to impact the extent of disease progression. For instance, it was found that in A. chinensis var. deliciosa Hayward, high nitrate rates (24 ppm, two times/week) positively influenced plant responses against Psa, whereas ammonium nitrate showed higher disease incidence and severity, increasing endophytic Psa populations [50]. Recently, it was demonstrated that Hayward plants grown with nitrate as the sole N source showed no visual disease symptoms and had a lower Psa endophytic population in comparison with plants receiving ammonium, which led to the appearance of typical Psa-induced leaf spotting, lower total chlorophyll content, and impaired photosynthetic capacity [129]. With nitrate supply, plants accumulated higher phosphorus (P), potassium (K), magnesium (Mg), calcium (Ca), and iron (Fe) contents, which possibly contributed to a more balanced mineral nutrition and increased tolerance to Psa, whereas with ammonium, the higher N content observed in plant tissues may have favored bacterial multiplication [129]. In contrast, in A. chinensis var. chinensis Hort16A, ammonium application (24 ppm, two times/week) proved to be more effective in increasing plant survival rates than nitrate [130]. Depletion of Ca and manganese (Mn) in Psa-inoculated Hayward micropropagated plants also led to a slight increase in symptom severity, whereas a deficiency of Ca, Fe, and boron (B) significantly increased the Psa endophytic population. Moreover, a surplus of B and Ca slightly increased symptom severity, and an excess of B and Mn significantly reduced the plant’s photosynthetic quantum yield [50]. Iodine (I) application to Hort16A seedling media also resulted in a lower disease incidence, which was attributed by the authors to its antioxidant activity, but in Hayward seedlings, it led to phytotoxicity symptoms [130,131]. The optimization of I applications to kiwifruit plants may be successful in increasing plant tolerance to bacterial pathogens, and should, therefore, be the subject of further investigation. More recently, the importance of sulfur (S) nutrition on plant tolerance against Psa was demonstrated, with S rates of 1.5–2.0 kg m−3 resulting in the lowest infection rates [132,133]. The authors proposed that adequate S application delivers higher plant tolerance due to an increase in polyphenolic and enzymatic activity mediated by the salicylic acid (SA) pathway [132,133]. As such, kiwifruit fertilization strategies to improve plant resilience to Psa should move from regimens focusing on the primary macronutrients (N, P, and K) to nutritional recipes in which special attention is paid to chemical elements that are often overlooked in plant nutrition, such as S and I. Also, as regular copper foliar spraying is currently a staple in orchard management for the control of several fungal and bacterial diseases, including KBC [62], its application for nutritional purposes should ideally take into consideration the phytosanitary treatments already undertaken to avoid potential phytotoxic or leaching events. Nevertheless, the role of copper in plant nutrition and its response to bacterial infection has been poorly explored. For example, copper application (10 ppm, two times/week) to Hort16A and Hayward seedlings reduced the levels of disease incidence but resulted in copper accumulation in plant tissues, whereas treatment with copper phosphite produced a marked phytotoxic effect on Hort16A [130].
A deeper understanding of the environmental conditions associated with bacterial epidemics and how they interplay with each other is a key aspect of the development of effective strategies to control disease outbreaks and further dissemination. It is clear that the relationship between plant fertilization and tolerance is highly complex since, besides the studied mineral concentrations, the mode and frequency of application, as well as possible interactions with the edaphoclimatic conditions of the orchard, could result in different disease incidences. Further research could allow for reducing disease incidence while promoting nutrient use efficiency to ensure plant tolerance at lower environmental costs.
4. Exploring Plant Genetic Resources for Increased Tolerance
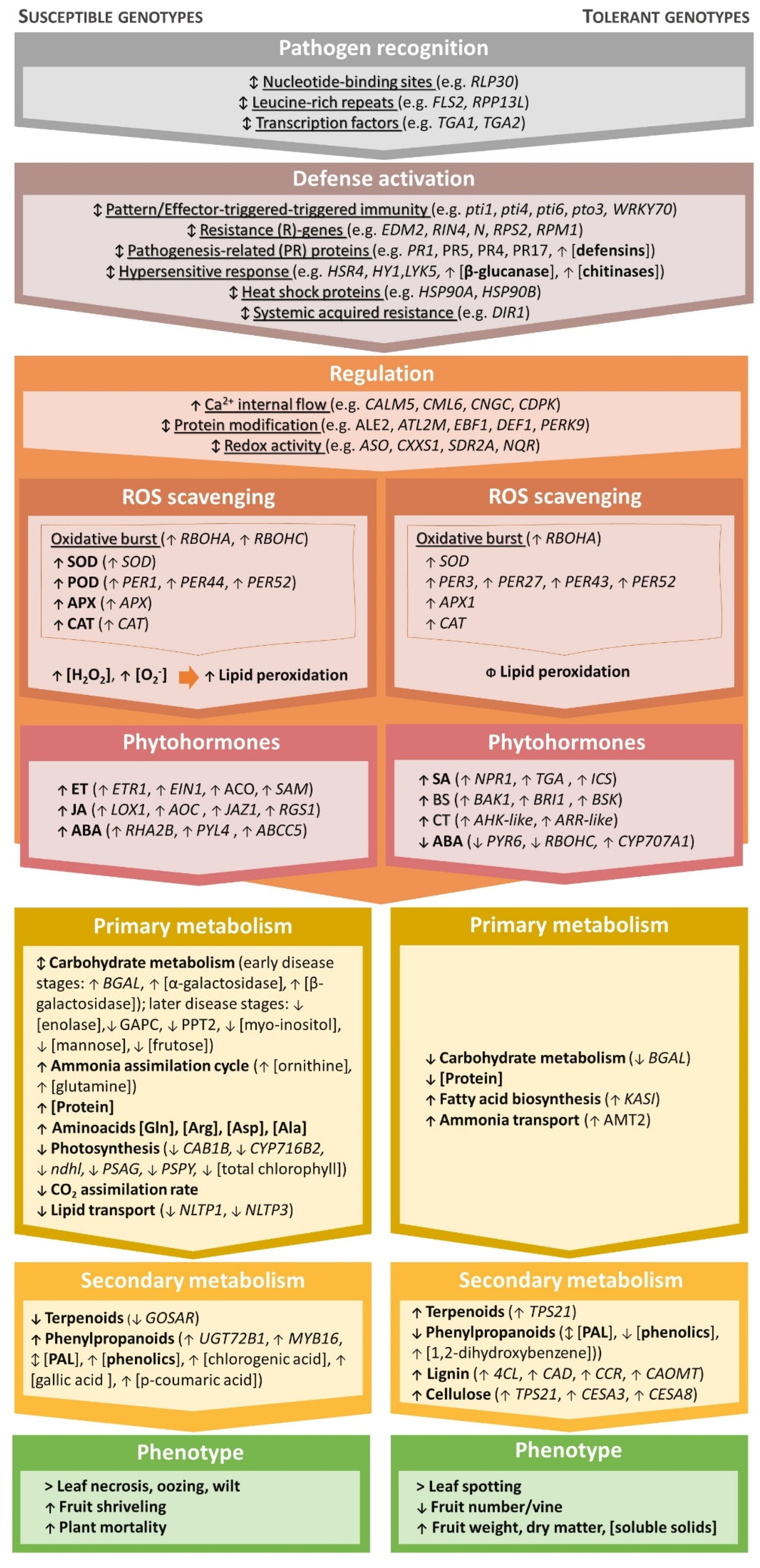
Apart from considering the specific edaphoclimatic conditions of each orchard and adopting more efficient fertilization strategies, breeding for resistance is an integral part of an integrated crop protection approach that is easy to implement with little environmental impact. However, a good comprehension of plant regulatory mechanisms upon infection is essential for identifying tolerance traits and supporting genotype selection. Thus far, several groups of transcripts, specific genes, and metabolites have been found to be involved in the response of kiwifruit plants during Psa infection, which could be used to propel the understanding of plant resistance mechanisms (Figure 3).

Figure 3.
Overview of the pathways reported to be affected in susceptible (left) and tolerant (right) Actinidia genotypes following infection by Pseudomonas syringae pv. actinidiae (Psa). Arrows indicate: ↑ increase, ↓ decrease, or ↨ distinct responses between genotypes and disease stages. Families of transcripts are underlined, differentially expressed genes appear in italics, classes of metabolites are marked in bold case, and individual metabolites appear without any formatting [37,70,84,86,88,103,122,129,130,131,132,133,134,135,136,137,138,139,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162,163,164,165,166,167].
4.1. Pathogen Recognition and Triggered Immunity
The first line of plant defense against pathogenic bacteria, called PAMP-Triggered Immunity (PTI), is activated through plant cell pattern recognition receptors (PRRs), which recognize the specific pathogen-associated molecular patterns (PAMP) characteristic of each pathogen. The second line of plant defense mechanisms, called effector-triggered immunity (ETI), is activated after recognition by plant resistance (R) proteins of bacterial effectors injected by the pathogen into the cytoplasm of plant cells. PTI and ETI can subsequently develop into a hypersensitive response (HR) at the infection site, which involves, e.g., production of reactive oxygen species (ROS), increased enzymatic activity, changes in structural defenses, and overexpression of R-genes [75,134,135,136,137,138,139,140]. It has been demonstrated that Psa is perceived by infected plants through flagellin (FLS2), lipopolysaccharides, or nucleotide-binding site leucine-rich repeats (NBS-LRR) proteins, including CC-NBS-LRR [136,137,138,140,141]. Pto3, also involved in pathogen recognition and activation of plant defenses, was found to increase 2 dpi with Psa in A. arguta var. arguta Weiki (tolerant to the pathogen), but only at 14 dpi in A. chinensis var. deliciosa Hayward (susceptible), putatively indicating that the expression of specific genes involved in pathogen recognition in the early stages of infection underpins the higher tolerance of some kiwifruit genotypes, resulting from an earlier activation of plant defenses [75]. After Psa recognition, kiwifruit plants show upregulation of several genes directly involved in PTI and ETI, such as Pti1, Pti4, RPP13, RPM1, EDM2, GFS12, PSL5, CERK1, WRKY70, and RPS2 [127,135,137,139,140,142]. Other downstream defensive mechanisms against Psa have also been identified through the upregulation of specific genes shortly after infection, including PLDDELTA and PLDBETA1, which encode phospholipases involved in basal defense and nonhost resistance against bacterial and fungal pathogens, AGO1, AGO5, DCL4, and RDR1, related to antiviral RNA silencing, and R-proteins (GLOX, RFL1, TAO1), which contribute to disease resistance induced by, for example, Type III effectors from P. syringae spp. [140]. PRR signaling eventually leads to the activation of defense-related mitogen-activated protein kinases (MAPKs), accumulation of reactive oxygen species (ROS), and transcriptional reprogramming by specific transcription factors (TFs) (such as AP2/ERF, bZIP, MYB, and WRKY). Recent whole-transcriptome analysis of resistant cultivars of A. chinensis var. deliciosa, such as Jinkui, resistant hybrids from A. chinensis var. chinensis Hort16A and of A. arguta, also pinpoint the key role of several of these TFs, including the R2R3-MYB and MYB48 families [139,140,143], in the regulation of ROS- and phytohormone-mediated signaling to finally determine plant tolerance [34,141,144,145].
4.2. Plant Antioxidant Response
Plant proteins, particularly those related to antioxidant response, have been found to play a major role in the regulation of plant defenses against Psa shortly following pathogen recognition. The content of soluble proteins in annual branches of tolerant cultivars was also found to be significantly higher than in susceptible ones before natural infection, and the protein content increased in susceptible cultivars and decreased in tolerant cultivars after infection [146]. Further proteomic and genomic analyses of Psa-infected kiwifruit plants have revealed distinct groups of proteins involved in plant defense, such as heat-shock, pathogenesis, transport, signaling processes and, importantly, tolerance to oxidative stress [42,108,135,137,141,147]. In fact, following pathogen recognition and the activation of plant defenses, several cellular changes occur, particularly in susceptible genotypes, including the formation of ROS and lipid peroxidation, which threaten cellular homeostasis [75,134]. To counteract the potentially harmful effects of these processes, plants produce several antioxidant enzymes, such as peroxidases, to a higher extent in more tolerant varieties than in susceptible ones [146]. Accordingly, transcriptomic analysis revealed that following Psa infection, APX, SOD, and CAT transcriptional levels were upregulated earlier in the tolerant A. arguta than in A. chinensis [75,108,148]. Tahir et al. [139] also observed upregulation of a gene encoding a putative thioredoxin-like protein (CXXS1) in hybrids of A. chinensis var. chinensis Hort16A tolerant to Psa infection but not in less tolerant genotypes. Tolerant species, such as A. arguta, also showed a higher number of upregulated genes involved in ROS scavenging in response to Psa infection, including RBOHA, related to a rapid and transient Phase I oxidative burst induced by pathogen infection, FOX1, which encodes an important oxidoreductase involved in Arabidopsis thaliana defenses against P. syringae, and several peroxidases (e.g., PER1, PER27, PER43, PER52) [140]. In addition, in sulfur-treated kiwifruit plants, tolerance increased along with the activities of not only peroxidase (POD) but also polyphenol oxidase (PPO) [132]. Taken together, these pieces of evidence suggest that in genotypes with a higher tolerance to Psa, the antioxidant system is more complex, potentially preventing imbalances in cellular homeostasis, promoting plant fitness, and sustaining a more effective defensive response.
4.3. Phytohormone Regulation
Plant responses against pathogens are regulated by complex phytohormone-mediated networks in which ET, SA, jasmonic acid (JA), and abscisic acid (ABA) counteract and interact with each other, modulating defense responses according to the type of pathogen and plant species [149]. Ethylene regulates multiple physiological and developmental processes in plants, such as senescence, and it is also involved in plant responses to abiotic and biotic stresses. Enhanced ET production is regarded as an early response to pathogen recognition and defense induction in infected plants, but several plant pathogenic bacteria produce ET as a virulence factor to improve their ability to colonize plant tissues (e.g., by means of increased stomatal aperture) [150]. This seems to be the case with Psa, as plant infection with an ET-producing strain induced the expression of genes related to ET signaling in the host plant (including ETR1, EIN2, EIN3, ERF1, and ERF2), but genes responsible for ET production (such as ACS and ACO) were little affected during infection [89]. Genes involved in ET biosynthesis and ET-mediated signaling, such as AP2/ERF, ERF2, SAM, and EIN2, were found to be upregulated shortly after Psa infection of susceptible A. chinensis plants [75,135,141]. Moreover, treatment with ET increased disease development, while application of the ET receptor blocker 1-methylcyclopropene (1-MCP) decreased disease incidence [134]. As such, the ET pathway is regarded as an antagonist of plant defenses against Psa and seems to be co-regulated by the JA pathway. In fact, genes related to ET biosynthesis, such as ETR1, SAM, and ACAS1, were upregulated in plants treated with JA-inducer methyl jasmonate (MJ), demonstrating that a synergistic effect occurs between ET and JA in the Actinidia-Psa pathosystem [151]. In tolerant genotypes, including some cultivars belonging to the species A. arguta, the expression of LOX1, involved in the first step of the JA pathway, increases just 1 day after inoculation with Psa, decreasing thereafter, while in susceptible genotypes (A. chinensis), LOX1 is upregulated in later moments of disease development [75]. Transcriptome analysis of A. chinensis also revealed upregulation of several genes related to JA-mediated defenses (e.g., SYD, COI1) and JA-mediated signaling (such as UBP12, UBP13, and RGS1) shortly after infection [140]. This most likely occurs as a strategy used by the pathogen to antagonize SA responses through the enhancement of the JA pathway or as a consequence of the activation of other metabolic pathways, such as ET [134]. In fact, JA accumulated in the susceptible cultivar Actinidia chinensis Hongyang, leading to the upregulation of JAR1, COI1, JAZ, and MYC2, but it decreased in the resistant A. chinensis var. deliciosa Jinkui in response to Psa. [143]. JIH, a negative marker of the JA pathway that allows plants to modulate SA-mediated responses according to their needs, was upregulated in Psa-infected inoculated plants, probably as a strategy to antagonize JA-mediated defenses upon Psa infection [151]. Moreover, the transcription-factor-encoding gene MYB16, which possesses a methyl jasmonate responsiveness cis-acting regulatory element (CRE), was downregulated in the resistant cultivar A. chinensis var. deliciosa Jinkui but upregulated in the susceptible cultivar A. chinensis var. chinensis Hongyang in response to Psa, putatively indicating that MYB16 acts as a negative regulatory gene in response to JA in kiwifruit infected with Psa [143]. Concordantly, a significant increase in disease development was observed in plants treated with methyl jasmonate [134,151,152], and thus, it is currently acknowledged that Actinidia spp. defense responses against Psa are mainly regulated by SA, whereas elicitation of the JA and ET pathways decreases plant tolerance to the pathogen. [135,143]. Nevertheless, the commonly antagonistic SA and JA pathways seem to be concertedly activated within a short period after infection as the concentration of both hormones increases in both tolerant and susceptible genotypes in the early stages of Psa infection [135,143]. It has been suggested that during plant infection with biotrophic pathogens, SA activates JA signaling as a coping mechanism against potential attacks by necrotrophic agents. During this process, instead of being activated by the receptor COI1, de novo JA synthesis and JA-responsive genes are induced through the SA receptors NPR3 and NPR4, also resulting in the accumulation of SA in plant tissues [153]. Genes involved in SA signaling pathways, such as PAD4, TGA, and PR1, were overexpressed in A. chinensis plants after inoculation with Psa [141,143], and elicitation of the SA pathway through exogenous application of acibenzolar-S-methyl (ASM) also led to transient upregulation of important genes involved in the regulation of basal resistance against pathogens, including P. syringae pv. tomato (e.g., genes BAD and NPR3), ETI (e.g., EDS1A), and systemic acquired resistance (e.g., NIMIN2) [154]. Treatment of Psa-infected plants with ASM also resulted in the upregulation of PR1, PR2, PR5, and PR8 [134,152,155], supporting the important role of the SA pathway in the regulation of resistance gene-mediated signaling [154]. In addition, NPR1, a key component of the SA-dependent signaling pathway, was overexpressed in tolerant and susceptible genotypes shortly following infection with Psa [89,143,156]. Constitutive expression of this gene in transgenic tobacco plants induced the expression of PR genes and promoted tolerance against bacterial infection, restoring basal tolerance against P. syringae pv. tomato DC3000 in an A. thaliana npr1-1 mutant. As such, these authors suggested that NPR1 may have a central role in SA-mediated kiwifruit defense responses against Psa [156]. Aside from SA, ABA is also a key hormone involved in plant defenses against pathogens, as stomatal closure, regulated through the ABA pathway, frequently acts as a mechanical barrier against pathogenic agents [157,158,159]. A gene involved in the positive regulation of ABA, RHA2B, was found to be upregulated during Psa infection, indicating that ABA regulation may be important in the first stages of plant infection [135]. Further research revealed that in susceptible genotypes, ABA concentration increases following Psa infection, accompanied by the accumulation of related proteins (PP2C, SnRK2, ABF) [143]. In contrast, in tolerant genotypes, such as A. arguta and A. chinensis var. deliciosa Jinkui, it only slightly increased or inclusively decreased, along with the downregulation of genes related to ABA-biosynthesis (e.g., NCED1 and ZEP) and the upregulation of genes related to ABA catabolism (such as CYP707A1 and CYP707A3) [140,143].
Although ABA is most commonly associated with stomatal regulation, its regulation has important implications for the JA (synergism) and SA (antagonism) pathways during plant infection [134,157]. Exogenous application of ABA slightly increased disease development in several A. chinensis varieties [134], and plant infection with Psa after SA-elicitation significantly decreased the ABA concentration in plant tissues, whereas plants treated with MJ showed increased ABA levels [151]. As such, the downregulation of the ABA pathway shortly following infection could facilitate stomatal closure and promote SA-mediated defenses and, by extent, antagonize the JA and ET-signaling pathways, impairing Psa colonization and limiting disease development, resulting in higher tolerance [89]. Thus, the tolerant character of some kiwifruit genotypes to Psa probably results from the upregulation of the SA pathway, which, in turn, negatively regulates the JA, ET, and ABA pathways. These pieces of evidence have already propelled the use of plant elicitors composed of the SA-inducer ASM, commercialized as Bion® or Actigard®, although their efficacy against Psa seems to be highly dependent on the kiwifruit genotype [134,154,155,160,161]. Further understanding of plant hormonal regulation in plant responses to bacterial pathogens would greatly benefit the development of tailored elicitor formulations for each plant genotype and/or pathogen, a reason why it should be the subject of additional research.
4.4. Primary and Secondary Metabolism
Plant infection by noxious pathogens frequently leads to impairments in both primary and secondary metabolism. For example, loss of chloroplast functions often occurs following pathogen invasion, consequently leading to tissue chlorosis or necrosis and changes in the photosynthetic metabolism of the host plant, further contributing to disease development [162]. In kiwifruit plants, transcripts related to photosynthesis were downregulated in susceptible A. chinensis cultivars but highly expressed in the more tolerant A. eriantha ones [163]. Decreased total chlorophyll was also observed in Psa-infected A. chinensis, including in plants with lower bacterial colonization resulting from plant elicitation with SA [151]. Moreover, lower levels of chlorophylls a and b were also associated with higher disease severity [133]. This pinpoints that Psa infection leads to the loss of chlorophylls, likely due to tissue destruction and nutrient depletion within leaf tissues resulting from bacterial infection and multiplication in plant tissues, but that the extent of impairments to these metabolites is not directly correlated with bacterial density inside plant tissues.
Impairments in several amino acids, including L-serine and aspartate, were also observed in Psa-infected plants, leading to extensive reprogramming of carbon (C) and N cycling in various metabolic pathways, including photosynthesis, C fixation, and the tricarboxylic acid cycle [127,140,164]. Several sugars, e.g., sucrose and fructose, were differently impaired in plant leaves, stems, or bleeding saps in later stages of Psa infection (20 days after infection) [127]. Moreover, genes related to sucrose metabolism and transport (SUS2, CINV1, INVB, SUT, and SUT2) were upregulated in susceptible A. chinensis var. deliciosa, but not in tolerant A. arguta [140]. Within the first 2 days of infection, Psa infection led to a downregulation of glucose-related genes in both plant species, while genes involved in the downstream steps of glycolysis (particularly pertaining to glucose 6-phosphate, glyceraldehyde 3-phosphate, 3-phosphoglyceric acid and phosphoenolpyruvic acid, PEP) were mainly upregulated [140]. In contrast, in later disease stages (10 days), enolase, involved in the conversion of 2-phosphoglycerate to PEP during the glycolytic process, decreased during shoot systemic infection caused by Psa [147], putatively suggesting a higher energetic demand during the first stages of the plant defensive process against Psa and/or the exploitation of plant glycolic resources by Psa. In fact, a gene encoding a putative beta-galactosidase (BGAL), linked to carbohydrate metabolism, was also upregulated in moderately tolerant genotypes but downregulated in highly tolerant ones [139], also indicating that carbohydrate metabolism is impaired to different extents, depending on the tolerance degree to Psa. Although carbohydrates serve as signaling molecules and provide energy to support demanding defense-related pathways, a high concentration of sugars may propel bacterial proliferation and progression inside plant tissues [127]. Thus, tolerant genotypes seem to be able to cope more efficiently with Psa infection through a more efficient regulation of sugar metabolism in the early stages of infection, and by restricting bacterial access to key carbohydrates, such as glucose, throughout the infection process. Conversely, ornithine and glutamine, involved in N cycling, increased in plant tissues following Psa infection in A. chinensis var. deliciosa (susceptible), but not in A. arguta (tolerant), and were accompanied by the upregulation of several genes related to the ammonia-assimilation cycle, pertaining not only to ornithine and glutamine but also to arginine and glutamic acid (e.g., DIT1, ogdh, CARB, GAD1, GAD4, and GLT1) [140]. Higher concentrations of gamma-aminobutyric acid (GABA), caffeic acid, and chlorogenic acid have also been proposed to provide a defensive advantage to A. arguta against Psa [65]. In fact, GABA is involved in ROS scavenging and antioxidant activity, membrane stability, and crosstalk among phytohormones [165], and it could be a key player in plant tolerance against Psa. Overall, tolerant genotypes seem to regulate primary metabolism in a more efficient manner, which may constitute an important coping mechanism against the pathogen by (i) restricting its access to crucial plant metabolic resources, such as amino acids, sugars, and nitrogen, and (ii) producing key metabolites that impair bacterial colonization by promoting plant cellular homeostasis and defense mechanisms (as is the case with GABA).
The phenylpropanoid pathway, responsible for synthesizing a wide variety of phenolic compounds, such as lignin, phenolic acids, stilbenes, and flavonoids, also seems to be of particular importance for plant tolerance against bacterial pathogens, including Psa [137]. Miao et al. [166] compared the content of phenolic compounds in shoots and leaves of kiwifruit cultivars with different tolerances to Psa and observed that, before inoculation, phenolic compounds in tolerant cultivars were significantly higher than in susceptible cultivars, and that after inoculation their concentration increased in tolerant and susceptible cultivars. The expression of PAL, encoding the enzyme phenylalanine ammonia-lyase involved in the first step of the phenylpropanoid pathway, was significantly increased in Psa-inoculated plants elicited with ASM, highlighting the role of phenolic biosynthesis in plant tolerance to Psa [134,141]. Nevertheless, phenolic compounds have a dual role in plant defenses against Psa, with tolerant and susceptible genotypes showing important qualitative differences during infection. For example, in cultivars with higher tolerance to this pathogen, the main phenolic compounds identified included 1,2-dihydroxybenzene, epicatechin, and trans-cinnamic acid, whereas in cultivars with lower tolerance, epicatechin, trans-cinnamic acid, p-coumaric acid, and gallic acid represented the major phenolic compounds [127]. Most importantly, recent evidence suggests that susceptible genotypes accumulate soluble polyphenols during infection, whereas in tolerant genotypes, such as A. chinensis var. deliciosa Jinkui, phenolic substances are used to synthesize lignin as a strategy to restrain Psa infection. Accumulation of soluble phenolic compounds was reported after inoculation with Psa in the susceptible A. chinensis cultivars Hayward and Hongyang, accompanied by a higher concentration of PAL, phenolics, gallic acid, and chlorogenic acid and the upregulation of genes and proteins related to phenylpropanoid biosynthesis, including UGT72B1 and PAL, and of the transcription factor gene MYB16 [132,139,141,143,151]. In contrast, in the tolerant A. chinensis var. deliciosa Jinkui, PAL activity decreased, but lignin was constitutively higher or increased in several tolerant genotypes, including Jinkui, A. rufa, and A. arguta [75,127,143,164]. Considering these pieces of evidence, it has been hypothesized that plant responses against Psa are regulated by the allocation of phenolic substances into lignin biosynthesis through a mechanism involving the downregulation of MYB16 [143]. Considering the intricacy of the Actinidia spp./Psa pathosystem, the use of big data analysis as a tool to develop models of plant resistance has not yet been undertaken, limiting the options to translate complex metabolomic and genomic data toward supporting farmers and breeders in their decision-making.
5. Future Prospects
Host resistance seems to be the best long-term approach to controlling plant diseases, but it will only be attainable when sufficient knowledge of plant regulatory mechanisms against pathogenic bacteria is available. However, the still scarce knowledge regarding the genetic and metabolic traits that underpin the higher tolerance of certain plant genotypes against bacterial pathogens, such as Psa, greatly hinders the possibility of identifying resistance markers, developing novel genotypes, and promoting nonhost resistance. Overall, the current findings suggest that metabolic pathways associated with sugars, N, and phenolic compounds may play a central role in plant tolerance against Psa. Genome editing and biotechnology approaches could be a powerful ally for overcoming these lock-ins through the modification of key genes. For instance, the CRISPR/Cas9 technology was already used to induce disease resistance in A. chinensis var. chinensis through a site-specific modification of a gene belonging to AP2/ERF transcription factors, involved in plant–pathogen interaction and defense induction [145]. However, these molecular approaches are still at an embryonic stage, and effective proof of concept is still lacking. Moreover, this path should be pursued with caution, as crop improvement for resistance may propel bacterial evolutionary events, leading to the appearance of more virulent strains, or resulting in loss of plant productivity and/or fruit quality. Moreover, the growing environment may modify the degree of resistance of the plant genotype, posing additional challenges for farmers and researchers. Most importantly, plant disease management must be temporally and spatially dynamic to accompany the constantly shifting genetics of bacterial populations and their respective plant hosts. Despite the multiplicity of factors involving bacterial pathogens and their plant hosts that are currently challenging agricultural production worldwide, scientific, governmental, and agricultural approaches to a given pathosystem may have the potential to provide guidelines in addressing other phytosanitary problems of microbial origin. Thus, with timely interdisciplinary efforts based on knowledge transfer, it may be possible to foster the effective control of noxious organisms in agroecosystems, while supporting plant productivity and fruit quality [167].
6. Conclusions
Although knowledge concerning plant protection has greatly increased over the past decades, the constant introduction of plant and microbial organisms into non-native areas poses a constant challenge for producers and researchers worldwide due to the appearance of new pathosystems and the occurrence of unpredicted disease outbreaks. Research efforts employed in the last decade aimed at classifying Psa populations and understanding Psa pathogenesis (mainly targeting Psa3 as a causal agent of the current KBC pandemic) and plant defense mechanisms allowed to: (i) identify distinct virulence degrees among the several bacterial populations identified so far; (ii) improve orchard management routines; and (iii) unravel phenotypic and genotypic traits that may be responsible for increased tolerance to KBC. Ultimately, the key to sustainable disease management is to prevent the introduction of the pathogen into new or recovered areas, and subsequently minimize its impact and spread through optimized agricultural practices combining early detection, adequate crop protection, and enhanced plant tolerance. This should take into consideration not only the genotype of the cultivated plant, but also the particular strain of the pathogen, and local orchard edaphoclimatic conditions. Although the eradication of highly virulent bacterial phytopathogens, such as Psa, is extremely challenging, a steady co-existence aimed at low impact on food production and using natural resources in a sustainable fashion can be achieved through timely cooperation between producers, researchers, and policymakers.
Author Contributions
M.N.d.S. and M.G.S. performed the bibliographic search and prepared the main manuscript. S.M.P.C. and M.W.V. coordinated the outline of the paper and performed critical revision throughout the writing process. S.M.P.C. acquired the funding that allowed the development of the work. All authors have read and agreed to the published version of the manuscript.
Funding
The authors gratefully acknowledge funding by Fundação para a Ciência e a Tecnologia (FCT) through project PSAlert (PTDC/AGRPRO/6156/2014) and the PhD scholarships of MNS (SFRH/BD/99853/2014) and MGS (2020.08874.BD).
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors would like to thank FCT for the scientific collaboration under R&D support projects (UIDB/05748/2020, UIDP/05748/2020, UIDB/50016/2020) and Norte2020—Sistema de Apoio à Investigação Científica e Tecnológica—‘Projetos Estruturados de I&D&I’ for the scientific collaboration under the project AgriFoodXXI (NORTE-01-0145-FEDER-000041).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Hunjan, M.S.; Lore, J.S. Climate Change: Impact on Plant Pathogens, Diseases, and Their Management. In Crop Protection under Changing Climate; Jabran, K., Florentine, S., Chauhan, B.S., Eds.; Springer International Publishing: Cham, The Netherlands, 2020; pp. 85–100. [Google Scholar]
- European and Mediterranean Plant Protection Organization. EPPO A1 and A2 Lists of Pests Recommended for Regulation as Quarantine Pests, Version 2021-09; Available online: https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list (accessed on 1 August 2022).
- Xin, X.-F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Bull, C.T.; De Boer, S.; Denny, T.; Firrao, G.; Saux, M.F.-L.; Saddler, G.; Scortichini, M.; Stead, D.; Takikawa, Y. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J. Plant Pathol. 2010, 551–592. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Pseudomonas syringae pv. actinidiae—Distribution; Available online: https://gd.eppo.int/taxon/PSDMAK/distribution (accessed on 1 August 2022).
- Serizawa, S.; Ichikawa, T.; Takikawa, Y.; Tsuyumu, S.; Goto, M. Occurrence of bacterial canker of kiwifruit in Japan description of symptoms, isolation of the pathogen and screening of bactericides. Jpn. J. Phytopathol. 1989, 55, 427–436. [Google Scholar] [CrossRef]
- Ferrante, P.; Scortichini, M. Identification of Pseudomonas syringae pv. actinidiae as causal agent of bacterial canker of yellow kiwifruit (Actinidia chinensis Planchon) in Central Italy. J. Phytopathol. 2009, 157, 768–770. [Google Scholar] [CrossRef]
- Scortichini, M.; Marcelletti, S.; Ferrante, P.; Petriccione, M.; Firrao, G. Pseudomonas syringae pv. actinidiae: A re-emerging, multi-faceted, pandemic pathogen. Mol. Plant Pathol. 2012, 13, 631–640. [Google Scholar] [CrossRef]
- Vanneste, J.L. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Guidelines on Pest Risk Analysis; EPPO: Paris, France, 1993; pp. 191–198. [Google Scholar]
- Abelleira, A.; Ares, A.; Aguín, O.; Mansilla, P.; López, M.C. Distribution, detection protocols and characterization of Pseudomonas syringae pv. actinidiae from kiwifruit in Galicia (northwest Spain). Acta Hortic. 2015, 1095, 49–56. [Google Scholar] [CrossRef]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balestra, G.M.; Spinelli, F. Pseudomonas syringae pv. actinidiae: Ecology, infection dynamics and disease epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Al-Saffar, M.F.; Jarallah, E.M. Isolation and characterization of Pseudomonas aeruginosa from Babylon Province. Biochem. Cell. Arch. 2019, 19, 203–209. [Google Scholar]
- Cornelis, P.; Matthijs, S. Pseudomonas Siderophores and Their Biological Significance. In Microbial Siderophores; Varma, A., Chincholkar, S.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 193–203. [Google Scholar]
- Kałużna, M. Characterization and phylogeny of the novel taxon of Pseudomonas spp., closely related to Pseudomonas avellanae as causal agent of a bacterial leaf blight of cornelian cherry (Cornus mas L.) and Pseudomonas syringae pv. syringae as a new bacterial pathogen of red dogwood (Cornus sanguinea L.). J. Plant Pathol. 2019, 101, 251–261. [Google Scholar] [CrossRef]
- Nepali, B.; Bhattarai, S.; Shrestha, J. Identification of Pseudomonas fluorescens using different biochemical tests. Int. J. Appl. Biol. 2018, 2, 27–32. [Google Scholar] [CrossRef]
- González, A.J.; Rodicio, M.R.; Mendoza, M.C. Identification of an emergent and atypical Pseudomonas viridiflava lineage causing bacteriosis in plants of agronomic importance in a Spanish region. Appl. Environ. Microbiol. 2003, 69, 2936–2941. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. PM 7/120 (1) Pseudomonas syringae pv. actinidiae; pp. 360–375. Available online: https://onlinelibrary.wiley.com/doi/pdf/10.1111/epp.12171 (accessed on 1 August 2022).
- Prencipe, S.; Gullino, M.L.; Spadaro, D. Pseudomonas syringae pv. actinidiae isolated from Actinidia chinensis var. deliciosa in Northern Italy: Genetic diversity and virulence. Eur. J. Plant Pathol. 2018, 150, 191–204. [Google Scholar] [CrossRef]
- Brunetti, A.; Pucci, N.; Modesti, V.; Lumia, V.; Latini, A.; Loreti, S.; Pilotti, M. In vitro and in planta screening of compounds for the control of Pseudomonas syringae pv. actinidiae in Actinidia chinensis var. chinensis. Eur. J. Plant Pathol. 2020, 158, 829–848. [Google Scholar] [CrossRef]
- Sawada, H.; Takeuchi, T.; Matsuda, I. Comparative analysis of Pseudomonas syringae pv. actinidiae and pv. phaseolicola based on phaseolotoxin-resistant ornithine carbamoyltransferase gene (argK) and 16S-23S rRNA intergenic spacer sequences. Appl. Environ. Microbiol. 1997, 63, 282–288. [Google Scholar] [CrossRef]
- Koh, Y.J.; Nou, I.S. DNA markers for identification of Pseudomonas syringae pv. actinidiae. Mol. Cells 2002, 13, 309–314. [Google Scholar]
- Rees-George, J.; Vanneste, J.; Cornish, D.; Pushparajah, I.; Yu, J.; Templeton, M.; Everett, K. Detection of Pseudomonas syringae pv. actinidiae using polymerase chain reaction (PCR) primers based on the 16S–23S rDNA intertranscribed spacer region and comparison with PCR primers based on other gene regions. Plant Pathol. 2010, 59, 453–464. [Google Scholar] [CrossRef]
- Gallelli, A.; L’Aurora, A.; Loreti, S. Gene sequence analysis for the molecular detection of Pseudomonas syringae pv. actinidiae: Developing diagnostic protocols. J. Plant Pathol. 2011, 93, 425–435. [Google Scholar] [CrossRef]
- Gallelli, A.; Talocci, S.; Pilotti, M.; Loreti, S. Real-time and qualitative PCR for detecting Pseudomonas syringae pv. actinidiae isolates causing recent outbreaks of kiwifruit bacterial canker. Plant Pathol. 2014, 63, 264–276. [Google Scholar] [CrossRef]
- Balestra, G.M.; Taratufolo, M.C.; Vinatzer, B.A.; Mazzaglia, A. A multiplex PCR assay for detection of Pseudomonas syringae pv. actinidiae and differentiation of populations with different geographic origin. Plant Dis. 2013, 97, 472–478. [Google Scholar] [CrossRef]
- Ciarroni, S.; Gallipoli, L.; Taratufolo, M.C.; Butler, M.I.; Poulter, R.T.; Pourcel, C.; Vergnaud, G.; Balestra, G.M.; Mazzaglia, A. Development of a multiple loci variable number of tandem repeats analysis (MLVA) to unravel the intra-pathovar structure of Pseudomonas syringae pv. actinidiae populations worldwide. PLoS ONE 2015, 10, e0135310. [Google Scholar] [CrossRef] [PubMed]
- Cunty, A.; Cesbron, S.; Poliakoff, F.; Jacques, M.A.; Manceau, C. Origin of the outbreak in France of Pseudomonas syringae pv. actinidiae biovar 3, the causal agent of bacterial canker of kiwifruit, revealed by a multilocus variable-number tandem-repeat analysis. Appl. Environ. Microbiol. 2015, 81, 6773–6789. [Google Scholar] [CrossRef] [PubMed]
- Mazzaglia, A.; Turco, S.; Taratufolo, M.C.; Tatì, M.; Rahi, Y.J.; Gallipoli, L.; Balestra, G.M. Improved MLVA typing reveals a highly articulated structure in Pseudomonas syringae pv. actinidiae populations. Physiol. Mol. Plant Pathol. 2021, 114, 101636. [Google Scholar] [CrossRef]
- Sawada, H.; Fujikawa, T. Genetic diversity of Pseudomonas syringae pv. actinidiae, pathogen of kiwifruit bacterial canker. Plant Pathol. 2019, 68, 1235–1248. [Google Scholar] [CrossRef]
- Pan, X.; Zhao, S.; Wang, Y.; Li, M.; He, L.; Zhuang, Q. Complete genome sequencing of Pseudomonas syringae pv. actinidiae Biovar 3, P155, kiwifruit pathogen originating from China. Biosci. J. 2020, 36, 2220–2228. [Google Scholar] [CrossRef]
- Andersen, M.T.; Templeton, M.D.; Rees-George, J.; Vanneste, J.L.; Cornish, D.A.; Yu, J.; Cui, W.; Braggins, T.J.; Babu, K.; Mackay, J.F.; et al. Highly specific assays to detect isolates of Pseudomonas syringae pv. actinidiae biovar 3 and Pseudomonas syringae pv. actinidifoliorum directly from plant material. Plant Pathol. 2018, 67, 1220–1230. [Google Scholar] [CrossRef]
- Barrett-Manako, K.; Andersen, M.; Martínez-Sánchez, M.; Jenkins, H.; Hunter, S.; Reese-George, J.; Montefiori, M.; Wohlers, M.; Rikkerink, E.; Templeton, M.; et al. Real-time PCR and droplet digital PCR are accurate and reliable methods to quantify Pseudomonas syringae pv. actinidiae biovar 3 in kiwifruit infected plantlets. Plant Dis. 2021, 105, 1748–1757. [Google Scholar] [CrossRef]
- Jayaraman, J.; Chatterjee, A.; Hunter, S.; Chen, R.; Stroud, E.A.; Saei, H.; Hoyte, S.; Deroles, S.; Tahir, J.; Templeton, M.D.; et al. Rapid methodologies for assessing Pseudomonas syringae pv. actinidiae colonization and effector-mediated hypersensitive response in kiwifruit. Mol. Plant-Microbe Interact. 2021, 34, 880–890. [Google Scholar] [CrossRef]
- Ruinelli, M.; Schneeberger, P.H.H.; Ferrante, P.; Bühlmann, A.; Scortichini, M.; Vanneste, J.L.; Duffy, B.; Pothier, J.F. Comparative genomics-informed design of two LAMP assays for detection of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae and discrimination of isolates belonging to the pandemic biovar 3. Plant Pathol. 2017, 66, 140–149. [Google Scholar] [CrossRef]
- Suzaki, K.; Sawada, H.; Kisaki, G. Loop-mediated isothermal amplification of bacterial effector genes to detect Pseudomonas syringae pv. actinidiae biovars 1 and 3. J. Gen. Plant Pathol. 2022, 88, 2–9. [Google Scholar] [CrossRef]
- Liu, P.; Xue, S.; He, R.; Hu, J.; Wang, X.; Jia, B.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M.; Zhu, L. Pseudomonas syringae pv. actinidiae isolated from non-kiwifruit plant species in China. Eur. J. Plant Pathol. 2016, 145, 743–754. [Google Scholar] [CrossRef]
- Li, L.; Pan, H.; Deng, L.; Feng, D.D.; Zhong, C.H. First report of bacterial leaf spot disease of Broussonetia papyrifera caused by Pseudomonas syringae pv. actinidiae in China. Plant Dis. 2021, 105, 696. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, F.; Donati, I.; Vanneste, J.; Costa, M.; Costa, G. Real time monitoring of the interactions between Pseudomonas syringae pv. actinidiae and Actinidia species. Acta Hortic. 2010, 913, 461–465. [Google Scholar] [CrossRef]
- Ferrante, P.; Fiorillo, E.; Marcelletti, S.; Marocchi, F.; Mastroleo, M.; Simeoni, S.; Scortichini, M. The importance of the main colonization and penetration sites of Pseudomonas syringae pv. actinidiae and prevailing weather conditions in the development of epidemics in yellow kiwifruit, recently observed in Central Italy. J. Plant Pathol. 2012, 94, 455–461. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit 1. Infection and bacterial movement in tissue of new canes. Jpn. J. Phytopathol. 1993, 59, 452–459. [Google Scholar] [CrossRef]
- Petriccione, M.; Salzano, A.M.; Di Cecco, I.; Scaloni, A.; Scortichini, M. Proteomic analysis of the Actinidia deliciosa leaf apoplast during biotrophic colonization by Pseudomonas syringae pv. actinidiae. J. Proteom. 2014, 101, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Renzi, M.; Copini, P.; Taddei, A.R.; Rossetti, A.; Gallipoli, L.; Mazzaglia, A.; Balestra, G.M. Bacterial canker on kiwifruit in Italy: Anatomical changes in the wood and in the primary infection sites. Phytopathology 2012, 102, 827–840. [Google Scholar] [CrossRef]
- Vanneste, J.L.; Cornish, D.A.; Yu, J.; Stokes, C.A. First report of Pseudomonas syringae pv. actinidiae the causal agent of bacterial canker of kiwifruit on Actinidia arguta vines in New Zealand. Plant Dis. 2014, 98, 418. [Google Scholar] [CrossRef]
- Beatson, R.A.; Datson, P.M.; Ferguson, A.R.; Montefiori, M. Use of kiwifruit germplasm resources for genetic improvement. Acta Hortic. 2014, 1048, 25–34. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit, 3: The seasonal changes of bacterial population in lesions and of its exudation from lesion. Jpn. J. Phytopathol. 1993, 59, 469–476. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit. 2. The most suitable times and environments for infection on new canes. Jpn. J. Phytopathol. 1993, 59, 460–468. [Google Scholar] [CrossRef]
- Serizawa, S.; Ichikawa, T.; Suzuki, H. Epidemiology of bacterial canker of kiwifruit. 5. Effect of infection in fall to early winter on the disease development in branches and trunk after winter. Jpn. J. Phytopathol. 1994, 60, 237–244. [Google Scholar] [CrossRef][Green Version]
- Serizawa, S.; Ichikawa, T. Epidemiology of bacterial canker of kiwifruit. 4. Optimum temperature for disease development on new canes. Jpn. J. Phytopathol. 1993, 59, 694–701. [Google Scholar] [CrossRef]
- Mauri, S.; Cellini, A.; Buriani, G.; Donati, I.; Costa, G.; Spinelli, F. Optimization of cultural practices to reduce the development of Pseudomonas syringae pv. actinidiae, causal agent of the bacterial canker of kiwifruit. J. Berry Res. 2016, 6, 355–371. [Google Scholar] [CrossRef]
- Aguín, O.; Ares, A.; Abelleira, A.; Mansilla, P. Survival of Pseudomonas syringae pv. actinidiae in leaf-litter of Actinidia deliciosa in Galicia (northwest Spain). Acta Hortic. 2015, 1095, 111–115. [Google Scholar] [CrossRef]
- Tyson, J.; Curtis, C.; Manning, M.; Dobson, S.; McKenna, C. Preliminary investigations of the risk of plant debris as a Pseudomonas syringae pv actinidiae inoculum source. N. Z. Plant Prot. 2016, 69, 11–16. [Google Scholar] [CrossRef]
- European and Mediterranean Plant Protection Organization. Pest Risk Analysis for Pseudomonas syringae pv. actinidiae; Available online: https://gd.eppo.int/download/doc/1255_pra_exp_PSDMAK.pdf (accessed on 1 August 2022).
- Koh, Y.J.; Park, S.Y.; Lee, D.H. Characteristics of bacterial canker of kiwifruit occurring in Korea and its control by trunk injection. Plant Pathol. J. 1996, 12, 324–330. [Google Scholar]
- Kennelly, M.M.; Cazorla, F.M.; de Vicente, A.; Ramos, C.; Sundin, G.W. Pseudomonas syringae diseases of fruit trees: Progress toward understanding and control. Plant Dis. 2007, 91, 4–17. [Google Scholar] [CrossRef]
- Koh, Y.J.; Kim, G.H.; Jung, J.S.; Lee, Y.S.; Hur, J.S. Outbreak of bacterial canker on Hort16A (Actinidia chinensis Planchon) caused by Pseudomonas syringae pv. actinidiae in Korea. N. Z. J. Crop Hortic. Sci. 2010, 38, 275–282. [Google Scholar] [CrossRef]
- Direção-Geral de Alimentação e Veterinária, D. Plano de ação Nacional para o Controlo da Pseudomonas syringae pv. actinidiae do Kiwi (Psa); Available online: https://www.dgav.pt/wp-content/uploads/2021/01/Plano-de-Acao-Nacional-de-Controlo-da-Pseudomonas-syringae-pv-actinidiae-do-kiwi-2014.pdf (accessed on 1 August 2022).
- Cornish, D.A.; Yu, J.; Oldham, J.M.; Benge, J.R.; Max, W.; Vanneste, J.L. In vitro inhibition of Pseudomonas syringae pv. actinidiae by wound protectants. N. Z. Plant Prot. 2015, 68, 332–339. [Google Scholar] [CrossRef]
- Everett, K.R.; Pushparajah, I.P.S.; Vergara, M.; Shahjahan, K.; Parry, B. S C. Monitoring Effectiveness of Wound Protectants against Psa. Milestone N° 66873. A Report Prepared for ZESPRI Group Limited; Plant & Food Research: Auckland, New Zealand, 2016. [Google Scholar]
- Everett, K.R.; Shahjahan, K.; Pushparajah, I.P.S.; Ramos, L.; Parry, B.; Hasna, L.; Middleditch, C.; Vergara, M.J. Monitoring Effectiveness of Wound Protectants against Psa. Milestone N° 68497: VI1712-30-G. A Report Prepared for ZESPRI Group Limited; Plant & Food Research: Auckland, New Zealand, 2017. [Google Scholar]
- Woodcock, S.D. A Review of Research and Development Undertaken on Psa. A Report Prepared for ZESPRI Group Limited; Kiwifruit Vine Health (KVH), Mt.: Maunganui, New Zealand, 2016. [Google Scholar]
- Cameron, A.; Sarojini, V. Pseudomonas syringae pv. actinidiae: Chemical control, resistance mechanisms and possible alternatives. Plant Pathol. 2014, 63, 1–11. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, G.H.; Song, Y.-R.; Oh, C.-S.; Koh, Y.J.; Jung, J.S. Streptomycin resistant isolates of Pseudomonas syringae pv. actinidiae in Korea. Res. Plant Dis. 2020, 26, 44–47. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Osdaghi, E.; Behlau, F.; Köhl, J.; Jones, J.B.; Aubertot, J.-N. Thirteen decades of antimicrobial copper compounds applied in agriculture. A review. Agron. Sustain. Dev. 2018, 38, 28. [Google Scholar] [CrossRef]
- Sciubba, F.; Di Cocco, M.E.; Angori, G.; Spagnoli, M.; De Salvador, F.R.; Engel, P.; Delfini, M. NMR-based metabolic study of leaves of three species of Actinidia with different degrees of susceptibility to Pseudomonas syringae pv. actinidiae. Nat. Prod. Res. 2019, 34, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Ashrafzadeh, S.; Leung, D.W.M. In vitro breeding-shortcut to Pseudomonas syringae pv. actinidiae (Psa) tolerant kiwifruit. Arch. Phytopathol. Plant Prot. 2019, 52, 501–506. [Google Scholar] [CrossRef]
- Scortichini, M. Field efficacy of a zinc-copper-hydracid of citric acid biocomplex compound to reduce oozing from winter cankers caused by Pseudomonas syringae pv. actinidiae to Actinidia spp. J. Plant Pathol. 2016, 98, 651–655. [Google Scholar]
- Monchiero, M.; Gullino, M.L.; Pugliese, M.; Spadaro, D.; Garibaldi, A. Efficacy of different chemical and biological products in the control of Pseudomonas syringae pv. actinidiae on kiwifruit. Australas. Plant Pathol. 2015, 44, 13–23. [Google Scholar] [CrossRef]
- Kim, G.H.; Lee, Y.S.; Jung, J.S.; Koh, Y.J.; Poulter, R.T.M.; Butler, M. Genomic analyses of Pseudomonas syringae pv. actinidiae isolated in Korea suggest the transfer of the bacterial pathogen via kiwifruit pollen. J. Med. Microbiol. 2020, 69, 132–138. [Google Scholar] [CrossRef]
- Narouei-Khandan, H.A.; Worner, S.P.; Viljanen, S.L.; van Bruggen, A.H.; Balestra, G.M.; Jones, E. The potential global climate suitability of kiwifruit bacterial canker disease (Pseudomonas syringae pv. actinidiae (Psa)) using three modelling approaches: CLIMEX, Maxent and Multimodel Framework. Climate 2022, 10, 14. [Google Scholar] [CrossRef]
- Wang, R.; Li, Q.; He, S.; Liu, Y.; Wang, M.; Jiang, G. Modeling and mapping the current and future distribution of Pseudomonas syringae pv. actinidiae under climate change in China. PLoS ONE 2018, 13, e0192153. [Google Scholar] [CrossRef]
- Nardozza, S.; Boldingh, H.; Richardson, A.; Walter, M.; Kashuba, P.; Seelye, R.; Clearwater, M.; Gould, N. Kiwifruit xylem sap: Composition and in vitro growth of a virulent strain of Pseudomonas syringae pv. actinidiae. Acta Hortic. 2015, 1095, 123–128. [Google Scholar] [CrossRef]
- Kisaki, G.; Tanaka, S.; Ishihara, A.; Igarashi, C.; Morimoto, T.; Hamano, K.; Endo, A.; Sugita-Konishi, S.; Tabuchi, M.; Gomi, K.; et al. Evaluation of various cultivars of Actinidia species and breeding source Actinidia rufa for resistance to Pseudomonas syringae pv. actinidiae biovar 3. J. Gen. Plant Pathol. 2018, 84, 399–406. [Google Scholar] [CrossRef]
- Saei, A.; Hoeata, K.; Krebs, A.; Sutton, P.; Herrick, J.; Wood, M.; Gea, L. The status of Pseudomonas syringae pv. actinidiae (Psa) in the New Zealand kiwifruit breeding programme in relation to ploidy level. Acta Hortic. 2018, 1218, 293–298. [Google Scholar] [CrossRef]
- Nunes da Silva, M.; Vasconcelos, M.W.; Gaspar, M.; Balestra, G.M.; Mazzaglia, A.; Carvalho, S.M.P. Early pathogen recognition and antioxidant system activation contributes to Actinidia arguta tolerance against Pseudomonas syringae pathovars actinidiae and actinidifoliorum. Front. Plant Sci. 2020, 11, 1022. [Google Scholar] [CrossRef]
- Wang, F.M.; Mo, Q.H.; Ye, K.Y.; Gong, H.J.; Qi, B.B.; Liu, P.P.; Jiang, Q.S.; Li, J.W. Evaluation of the wild Actinidia germplasm for resistance to Pseudomonas syringae pv. actinidiae. Plant Pathol. 2020, 69, 979–989. [Google Scholar] [CrossRef]
- Thomidis, T.; Goumas, D.E.; Zotos, A.; Triantafyllidis, V.; Kokotos, E. Susceptibility of twenty-three kiwifruit cultivars to Pseudomonas syringae pv. actinidiae. Eng. Proc. 2021, 9, 33. [Google Scholar] [CrossRef]
- Cotruţ, R.; Renzi, M.; Taratufolo, M.C.; Mazzaglia, A.; Balestra, G.M.; Stănică, F. Actinidia arguta ploidy level variation in relation to Pseudomonas syringae pv. actinidiae susceptibility. Lucr. Ştiinţifice 2013, 56, 1–12. [Google Scholar]
- Wang, F.-M.; Li, J.-W.; Ye, K.-Y.; Gong, H.-J.; Liu, P.-P.; Jiang, Q.-S.; Qi, B.-B.; Mo, Q.-H. Preliminary report on the improved resistance towards Pseudomonas syringae pv. actinidiae of cultivated kiwifruit (Actinidia chinensis) when grafted onto wild Actinidia guilinensis rootstock in vitro. J. Plant Pathol. 2021, 103, 51–54. [Google Scholar] [CrossRef]
- Chapman, J.R.; Taylor, R.K.; Weir, B.S.; Romberg, M.K.; Vanneste, J.L.; Luck, J.; Alexander, B.J. Phylogenetic relationships among global populations of Pseudomonas syringae pv. actinidiae. Phytopathology 2012, 102, 1034–1044. [Google Scholar] [CrossRef]
- Cunty, A.; Poliakoff, F.; Rivoal, C.; Cesbron, S.; Fischer-Le Saux, M.; Lemaire, C.; Jacques, M.A.; Manceau, C.; Vanneste, J.L. Characterization of Pseudomonas syringae pv. actinidiae (Psa) isolated from France and assignment of Psa biovar 4 to a de novo pathovar: Pseudomonas syringae pv. actinidifoliorum pv. nov. Plant Pathol. 2015, 64, 582–596. [Google Scholar] [CrossRef]
- Fujikawa, T.; Sawada, H. Genome analysis of the kiwifruit canker pathogen Pseudomonas syringae pv. actinidiae biovar 5. Sci. Rep. 2016, 6, 21399. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Kondo, K.; Nakaune, R. Novel biovar (biovar 6) of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit (Actinidia deliciosa) in Japan. Jpn. J. Phytopathol. 2016, 82, 101–115. [Google Scholar] [CrossRef][Green Version]
- Sawada, H.; Miyoshi, T.; Ide, Y. Novel MLSA group (Psa5) of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit (Actinidia chinensis) in Japan. Jpn. J. Phytopathol. 2014, 80, 171–184. [Google Scholar] [CrossRef][Green Version]
- McCann, H.C.; Li, L.; Liu, Y.; Li, D.; Pan, H.; Zhong, C.; Rikkerink, E.H.A.; Templeton, M.D.; Straub, C.; Colombi, E.; et al. Origin and evolution of the kiwifruit canker pandemic. Genome Biol. Evol. 2017, 9, 932–944. [Google Scholar] [CrossRef]
- Fujikawa, T.; Hatomi, H.; Sawada, H. Draft genome sequences of 10 strains of Pseudomonas syringae pv. actinidiae biovar 1, a major kiwifruit bacterial canker pathogen in Japan. Microbiol. Resour. Announc. 2020, 9, e00759-20. [Google Scholar] [CrossRef]
- Genka, H.; Baba, T.; Tsuda, M.; Kanaya, S.; Mori, H.; Yoshida, T.; Noguchi, M.T.; Tsuchiya, K.; Sawada, H. Comparative analysis of argK-tox clusters and their flanking regions in phaseolotoxin-producing Pseudomonas syringae pathovars. J. Mol. Evol. 2006, 63, 401–414. [Google Scholar] [CrossRef]
- Gnanamanickam, S.S. Plant-Associated Bacteria, 1st ed.; Springer: Dordrecht, The Netherlands, 2006. [Google Scholar]
- Cellini, A.; Donati, I.; Farneti, B.; Khomenko, I.; Buriani, G.; Biasioli, F.; Cristescu, S.M.; Spinelli, F. A breach in plant defences: Pseudomonas syringae pv. actinidiae targets ethylene signalling to overcome Actinidia chinensis pathogen responses. Int. J. Mol. Sci. 2021, 22, 4375. [Google Scholar] [CrossRef]
- Donati, I.; Buriani, G.; Cellini, A.; Mauri, S.; Costa, G.; Spinelli, F. New insights on the bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). J. Berry Res. 2014, 4, 53–67. [Google Scholar] [CrossRef]
- Vandelle, E.; Colombo, T.; Regaiolo, A.; Maurizio, V.; Libardi, T.; Puttilli, M.-R.; Danzi, D.; Polverari, A. Transcriptional profiling of three Pseudomonas syringae pv. actinidiae biovars reveals different responses to apoplast-like conditions related to strain virulence on the host. Mol. Plant-Microbe Interact. 2021, 34, 376–396. [Google Scholar] [CrossRef]
- Bundalovic-Torma, C.; Lonjon, F.; Desveaux, D.; Guttman, D.S. Diversity, evolution, and function of Pseudomonas syringae effectoromes. Annu. Rev. Phytopathol. 2022, 60, 211–236. [Google Scholar] [CrossRef]
- Ishiga, T.; Sakata, N.; Nguyen, V.T.; Ishiga, Y. Flood inoculation of seedlings on culture medium to study interactions between Pseudomonas syringae pv. actinidiae and kiwifruit. J. Gen. Plant Pathol. 2020, 86, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, J.; Yoon, M.; Applegate, E.R.; Stroud, E.A.; Templeton, M.D. AvrE1 and HopR1 from Pseudomonas syringae pv. actinidiae are additively required for full virulence on kiwifruit. Mol. Plant Pathol. 2020, 21, 1467–1480. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Q.; Zhi, T.; Fan, R.; Xie, T.; Zhao, Z.; Long, Y.; Li, Z. Genetic causes of non-pathogenic Pseudomonas syringae pv. actinidiae isolates in kiwifruit orchards. Front. Microbiol. 2021, 12, 650099. [Google Scholar] [CrossRef] [PubMed]
- Hemara, L.M.; Jayaraman, J.; Sutherland, P.W.; Montefiori, M.; Arshed, S.; Chatterjee, A.; Chen, R.; Andersen, M.; Mesarich, C.H.; van der Linden, O.; et al. Effector loss drives adaptation of Pseudomonas syringae pv. actinidiae to Actinidia arguta. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, N.; Han, N.; Tian, R.; Chen, J.; Gao, X.; Wu, Z.; Liu, Y.; Huang, L. Role of the type VI secretion system in the pathogenicity of Pseudomonas syringae pv. actinidiae, the causative agent of kiwifruit bacterial canker. Front. Microbiol. 2021, 12, 627785. [Google Scholar] [CrossRef] [PubMed]
- Firrao, G.; Torelli, E.; Polano, C.; Ferrante, P.; Ferrini, F.; Martini, M.; Marcelletti, S.; Scortichini, M.; Ermacora, P. Genomic structural variations affecting virulence during clonal expansion of Pseudomonas syringae pv. actinidiae biovar 3 in Europe. Front. Microbiol. 2018, 9, 656. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, M.; Liu, W.; Nie, J.; Huang, L. Pseudomonas syringae pv. actinidiae effector HopAU1 interacts with calcium-sensing receptor to activate plant immunity. Int. J. Mol. Sci. 2022, 23, 508. [Google Scholar] [CrossRef]
- Andolfi, A.; Ferrante, P.; Petriccione, M.; Cimmino, A.; Evidente, A.; Scortichini, M. Production of phytotoxic metabolites by Pseudomonas syringae pv. actinidiae, the causal agent of bacterial canker of kiwifruit. J. Plant Pathol. 2014, 96, 169–175. [Google Scholar] [CrossRef]
- McAtee, P.A.; Brian, L.; Curran, B.; van der Linden, O.; Nieuwenhuizen, N.J.; Chen, X.; Henry-Kirk, R.A.; Stroud, E.A.; Nardozza, S.; Jayaraman, J.; et al. Re-programming of Pseudomonas syringae pv. actinidiae gene expression during early stages of infection of kiwifruit. BMC Genom. 2018, 19, 822. [Google Scholar] [CrossRef]
- Luti, S.; Campigli, S.; Ranaldi, F.; Paoli, P.; Pazzagli, L.; Marchi, G. Lscβ and lscγ, two novel levansucrases of Pseudomonas syringae pv. actinidiae biovar 3, the causal agent of bacterial canker of kiwifruit, show different enzymatic properties. Int. J. Biol. Macromol. 2021, 179, 279–291. [Google Scholar] [CrossRef]
- Cellini, A.; Buriani, G.; Correia, C.; Fiorentini, L.; Vandelle, E.; Polverari, A.; Santos, C.; Vanneste, J.L.; Spinelli, F. Host-specific signal perception by PsaR2 LuxR solo induces Pseudomonas syringae pv. actinidiae virulence traits. Microbiol. Res. 2022, 260, 127048. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Donati, I.; Fiorentini, L.; Vandelle, E.; Polverari, A.; Venturi, V.; Buriani, G.; Vanneste, J.L.; Spinelli, F. N-Acyl homoserine lactones and Lux solos regulate social behaviour and virulence of Pseudomonas syringae pv. actinidiae. Microb. Ecol. 2020, 79, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Purahong, W.; Orrù, L.; Donati, I.; Perpetuini, G.; Cellini, A.; Lamontanara, A.; Michelotti, V.; Tacconi, G.; Spinelli, F. Plant microbiome and its link to plant health: Host species, organs and Pseudomonas syringae pv. actinidiae infection shaping bacterial phyllosphere communities of kiwifruit plants. Front. Plant Sci. 2018, 9, 1563. [Google Scholar] [CrossRef] [PubMed]
- Ares, A.; Pereira, J.; Garcia, E.; Costa, J.; Tiago, I. The leaf bacterial microbiota of female and male kiwifruit plants in distinct seasons: Assessing the impact of Pseudomonas syringae pv. actinidiae. Phytobiomes J. 2021, 5, 275–287. [Google Scholar] [CrossRef]
- Ferrante, P.; Scortichini, M. Redefining the global populations of Pseudomonas syringae pv. actinidiae based on pathogenic, molecular and phenotypic characteristics. Plant Pathol. 2015, 64, 51–62. [Google Scholar] [CrossRef]
- Nunes da Silva, M.; Machado, J.; Balestra, G.M.; Mazzaglia, A.; Vasconcelos, M.W.; Carvalho, S.M.P. Exploring the expression of defence-related genes in Actinidia spp. after infection with Pseudomonas syringae pv. actinidiae and pv. actinidifoliorum: First steps. Eur. J. Hortic. Sci. 2019, 84, 206–212. [Google Scholar] [CrossRef]
- Vanneste, J.L.; Yu, J.; Cornish, D.A.; Tanner, D.J.; Windner, R.; Chapman, J.R.; Taylor, R.K.; Mackay, J.F.; Dowlut, S. Identification, virulence, and distribution of two biovars of Pseudomonas syringae pv. actinidiae in New Zealand. Plant Dis. 2013, 97, 708–719. [Google Scholar] [CrossRef]
- Ochoa-Díaz, M.M.; Daza-Giovannetty, S.; Gómez-Camargo, D. Bacterial genotyping methods: From the basics to modern. Methods Mol. Biol. 2018, 1734, 13–20. [Google Scholar] [CrossRef]
- Ko, S.-J.; Lee, Y.-H.; Cha, K.-H.; Park, K.-B.; Park, I.-J.; Kim, Y.-C. An improved method for testing pathogenicity of Pseudomonas syringae pv. actinidiae causing bacterial canker of kiwifruit. Res. Plant Dis. 2002, 8, 250–253. [Google Scholar] [CrossRef]
- Hirano, S.S.; Upper, C.D. Population biology and epidemiology of Pseudomonas syringae. Annu. Rev. Phytopathol. 1990, 28, 155–177. [Google Scholar] [CrossRef]
- Vanneste, J.; Reglinski, T.; Yu, J.; Cornish, D. Multiplication and movement of Pseudomonas syringae pv. actinidiae in kiwifruit plants. Acta Hortic. 2015, 1095, 117–122. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, H.; Fang, S.; Qian, Z. Prevalent forecast of kiwifruit bacterial canker caused by Pseudomonas syringae pv. actinidiae. Ying Yong Sheng Tai Xue Bao 2001, 12, 355–358. [Google Scholar] [PubMed]
- Antoniacci, L.; Bugiani, R.; Rossi, R.; Calzolari, A.; Alessandrini, A.; Gozzi, R.; Spinelli, F.; Cellini, A.; Mauri, S.; Donati, I. Validation of New Zealand Psa forecasting model in Emilia Romagna Region, Italy. Acta Hortic. 2019, 1243, 71–78. [Google Scholar] [CrossRef]
- Choi, S.; Jayaraman, J.; Segonzac, C.; Park, H.J.; Park, H.; Han, S.W.; Sohn, K.H. Pseudomonas syringae pv. actinidiae type III effectors localized at multiple cellular compartments activate or suppress innate immune responses in Nicotiana benthamiana. Front. Plant Sci. 2017, 8, 2157. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, P.; Scortichini, M. Frost promotes the pathogenicity of Pseudomonas syringae pv. actinidiae in Actinidia chinensis and A. deliciosa plants. Plant Pathol. 2014, 63, 12–19. [Google Scholar] [CrossRef]
- Kiwifruit Vine Health, K. Psa-V Best Practice Guide; Kiwifruit Vine Health (KVH): Mount Maunganui, New Zealand, 2018. [Google Scholar]
- Chiabrando, V.; Giacalone, G. Kiwifruit under plastic covering: Impact on fruit quality and on orchard microclimate. J. Food Nutr. Agric. 2018, 1, 1–6. [Google Scholar] [CrossRef]
- Black, M.Z.; Casonato, S.; Bent, S.J. Opportunities for environmental modification to control Pseudomonas syringae pv. actinidiae in kiwifruit. Acta Hortic. 2015, 1105, 253–260. [Google Scholar] [CrossRef]
- Currie, M.; Martin, P.; Blattmann, M.; Gordon, B.; Patterson, K. Final Report on Growing Tolerant Cultivars in a Psa Environment, VI1296. A Report Prepared for ZESPRI Group Limited; Plant & Food Research: Auckland, New Zealand, 2014. [Google Scholar]
- Gaskin, R.; Manktelow, D.; Cook, S.m.; May, B.; Horgan, D.; van Leeuwen, R. Novel Technologies to Deliver Protectant Sprays to Strung Canopies. A Report Prepared for ZESPRI Group Limited; Plant Protection Chemistry NZ: Rotorua, New Zealand, 2016. [Google Scholar]
- Vanneste, J.l.L.; Moffat, B.; Oldham, J. Survival of Pseudomonas syringae pv actinidiae on Cryptomeria japonica a nonhost plant used as shelter belts in kiwifruit orchards. N. Z. Plant Prot. 2012, 65, 1–7. [Google Scholar] [CrossRef]
- Pattemore, D.E.; Goodwin, R.M.; McBrydie, H.M.; Hoyte, S.M.; Vanneste, J.L. Evidence of the role of honey bees (Apis mellifera) as vectors of the bacterial plant pathogen Pseudomonas syringae. Australas. Plant Pathol. 2014, 43, 571–575. [Google Scholar] [CrossRef]
- Gupta, N.; Debnath, S.; Sharma, S.; Sharma, P.; Purohit, J. Role of nutrients in controlling the plant diseases in sustainable agriculture. In Agriculturally Important Microbes for Sustainable Agriculture: Applications in Crop Production and Protection; Meena, V.S., Mishra, P.K., Bisht, J.K., Pattanayak, A., Eds.; Springer: Singapore, 2017; Volume 2, pp. 217–262. [Google Scholar]
- Cesco, S.; Tolotti, A.; Nadalini, S.; Rizzi, S.; Valentinuzzi, F.; Mimmo, T.; Porfido, C.; Allegretta, I.; Giovannini, O.; Perazzolli, M.; et al. Plasmopara viticola infection affects mineral elements allocation and distribution in Vitis vinifera leaves. Sci. Rep. 2020, 10, 18759. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Zeng, Y.; Liu, P. Metabolic profiling reveals local and systemic responses of kiwifruit to Pseudomonas syringae pv. actinidiae. Plant Direct 2020, 4, e00297. [Google Scholar] [CrossRef] [PubMed]
- Groen, S.C.; Whiteman, N.K. The evolution of ethylene signaling in plant chemical ecology. J. Chem. Ecol. 2014, 40, 700–716. [Google Scholar] [CrossRef] [PubMed]
- Nunes da Silva, M.; Fernandes, A.P.; Vasconcelos, M.W.; Valente, L.C.M.; Carvalho, S.M.P. Influence of the nitrogen source on the tolerance of Actinidia chinensis to Pseudomonas syringae pv. actinidiae. Acta Hortic. 2022, 1332, 103–110. [Google Scholar] [CrossRef]
- Holmes, A. Effect of Soil Nutrition and Composition on the Susceptibility of Hayward and Hort16A to Pseudomonas syringae pv. Actinidiae. ZESPRI Innovation Project V11270 Final Report; ZESPRI: Tauranga, New Zealand, 2012. [Google Scholar]
- Gupta, N.; Bajpai, M.S.; Majumdar, R.; Mishra, P.K. Response of iodine on antioxidant levels of Glycine max L. grown under Cd stress. Adv. Biol. Res. 2015, 9, 40–48. [Google Scholar] [CrossRef]
- Gu, G.; Yang, S.; Yin, X.; Long, Y.; Ma, Y.; Li, R.; Wang, G. Sulfur induces resistance against canker caused by Pseudomonas syringae pv. actinidae via phenolic components Increase and morphological structure modification in the kiwifruit stems. Int. J. Mol. Sci. 2021, 22, 12185. [Google Scholar] [CrossRef]
- Zhang, Z.; Long, Y.; Yin, X.; Yang, S. Sulfur-induced resistance against Pseudomonas syringae pv. actinidiae via triggering salicylic acid signaling pathway in kiwifruit. Int. J. Mol. Sci. 2021, 22, 12710. [Google Scholar] [CrossRef]
- Cellini, A.; Fiorentini, L.; Buriani, G.; Yu, J.; Donati, I.; Cornish, D.A.; Novak, B.; Costa, G.; Vanneste, J.L.; Spinelli, F. Elicitors of the salicylic acid pathway reduce incidence of bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidae. Ann. Appl. Biol. 2014, 165, 441–453. [Google Scholar] [CrossRef]
- Wurms, K.V.; Hardaker, A.J.; Ah Chee, A.; Bowen, J.; Phipps, J.; Taylor, J.; Jensen, D.; Cooney, J.; Wohlers, M.; Reglinski, T. Phytohormone and putative defense gene expression differentiates the response of ‘Hayward’ kiwifruit to Psa and Pfm infections. Front. Plant Sci. 2017, 8, 1366. [Google Scholar] [CrossRef]
- Michelotti, V.; Lamontanara, A.; Buriani, G.; Orrù, L.; Cellini, A.; Donati, I.; Vanneste, J.L.; Cattivelli, L.; Tacconi, G.; Spinelli, F. Comparative transcriptome analysis of the interaction between Actinidia chinensis var. chinensis and Pseudomonas syringae pv. actinidiae in absence and presence of acibenzolar-S-methyl. BMC Genom. 2018, 19, 585. [Google Scholar] [CrossRef]
- Wang, T.; Wang, G.; Jia, Z.-H.; Pan, D.-L.; Zhang, J.-Y.; Guo, Z.-R. Transcriptome analysis of kiwifruit in response to Pseudomonas syringae pv. actinidiae infection. Int. J. Mol. Sci. 2018, 19, 373. [Google Scholar] [CrossRef]
- Wang, T.; Jia, Z.H.; Zhang, J.Y.; Liu, M.; Guo, Z.R.; Wang, G. Identification and analysis of NBS-LRR genes in Actinidia chinensis genome. Plants 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Tahir, J.; Hoyte, S.; Bassett, H.; Brendolise, C.; Chatterjee, A.; Templeton, K.; Deng, C.; Crowhurst, R.; Montefiori, M.; Morgan, E.; et al. Multiple quantitative trait loci contribute to resistance to bacterial canker incited by Pseudomonas syringae pv. actinidiae in kiwifruit (Actinidia chinensis). Hortic. Res. 2019, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Nunes da Silva, M.; Carvalho, S.M.P.; Rodrigues, A.M.; Gómez-Cadenas, A.; António, C.; Vasconcelos, M.W. Defence-related pathways, phytohormones and primary metabolism are key players in kiwifruit plant tolerance to Pseudomonas syringae pv. actinidiae. Plant Cell Environ. 2022, 45, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Wurms, K.; Gould, E.; Chee, A.A.; Taylor, J.; Curran, B.; Reglinski, T. Elicitor induction of defence genes and reduction of bacterial canker in kiwifruit. N. Z. Plant Prot. 2017, 70, 272–284. [Google Scholar] [CrossRef][Green Version]
- Song, Y.; Sun, L.; Lin, M.; Chen, J.; Qi, X.; Hu, C.; Fang, J. Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae pv. actinidiae during early infection. PLoS ONE 2019, 14, e0211913. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Liu, Y.; Zhang, D.; Ni, M.; Jia, B.; Heng, W.; Fang, Z.; Zhu, L.-W.; Liu, P. Transcriptomic and proteomic profiling reveal the key role of AcMYB16 in the response of Pseudomonas syringae pv. actinidiae in kiwifruit. Front. Plant Sci. 2021, 12, 756330. [Google Scholar] [CrossRef]
- Jing, Z.; Liu, Z. Genome-wide identification of WRKY transcription factors in kiwifruit (Actinidia spp.) and analysis of WRKY expression in responses to biotic and abiotic stresses. Genes Genom. 2018, 40, 429–446. [Google Scholar] [CrossRef]
- Michelotti, V.; Urbinati, G.; Gentile, A.; Lucioli, S.; Caboni, E.; Tacconi, G. Preliminary results on the development of a genome editing protocol in Actinidia chinensis var. chinensis as Psa resistance approach. Acta Hortic. 2022, 1332, 111–116. [Google Scholar] [CrossRef]
- Miao, L.; Genjia, T.; Yao, L.; Lian, X. Resistance mechanism of kiwifruit cultivars to Pseudomonas syringae pv. actinidiae. Acta Phytopathol. Sin. 2005, 32, 37–42. [Google Scholar]
- Petriccione, M.; Di Cecco, I.; Arena, S.; Scaloni, A.; Scortichini, M. Proteomic changes in Actinidia chinensis shoot during systemic infection with a pandemic Pseudomonas syringae pv. actinidiae strain. J. Proteom. 2013, 78, 461–476. [Google Scholar] [CrossRef]
- Petriccione, M.; Mastrobuoni, F.; Zampella, L.; Scortichini, M. Reference gene selection for normalization of RT-qPCR gene expression data from Actinidia deliciosa leaves infected with Pseudomonas syringae pv. actinidiae. Sci. Rep. 2015, 5, 16961. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Nunes da Silva, M.; Vasconcelos, M.W.; Pinto, V.; Balestra, G.M.; Mazzaglia, A.; Gomez-Cadenas, A.; Carvalho, S.M.P. Role of methyl jasmonate and salicylic acid in kiwifruit plants further subjected to Psa infection: Biochemical and genetic responses. Plant Physiol. Biochem. 2021, 162, 258–266. [Google Scholar] [CrossRef]
- Reglinski, T.; Vanneste, J.; Wurms, K.; Gould, E.; Spinelli, F.; Rikkerink, E. Using fundamental knowledge of induced resistance to develop control strategies for bacterial canker of kiwifruit caused by Pseudomonas syringae pv. actinidiae. Front. Plant Sci. 2013, 4, 24. [Google Scholar] [CrossRef]
- Liu, L.; Sonbol, F.-M.; Huot, B.; Gu, Y.; Withers, J.; Mwimba, M.; Yao, J.; He, S.Y.; Dong, X. Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat. Commun. 2016, 7, 13099. [Google Scholar] [CrossRef]
- Reglinski, T.; Wurms, K.; Vanneste, J.; Ah Chee, A.; Yu, J.; Oldham, J.; Cornish, D.; Cooney, J.; Jensen, D.; Trower, T. Transient changes in defence gene expression and phytohormone cInduced by acibenzolar-S-methyl in glasshouse and orchard grown kiwifruit. Front. Agron. 2022, 3, 831172. [Google Scholar] [CrossRef]
- Stroud, E.A.; Rikkerink, E.H.A.; Jayaraman, J.; Templeton, M.D. Actigard™ induces a defence response to limit Pseudomonas syringae pv. actinidiae in Actinidia chinensis var. chinensis ‘Hort16A’ tissue culture plants. Sci. Hortic. 2022, 295, 110806. [Google Scholar] [CrossRef]
- Sun, L.-M.; Fang, J.-B.; Zhang, M.; Qi, X.-J.; Lin, M.-M.; Chen, J.-Y. Molecular cloning and functional analysis of the NPR1 homolog in kiwifruit (Actinidia eriantha). Front. Plant Sci. 2020, 11, 551201. [Google Scholar] [CrossRef]
- Melotto, M.; Underwood, W.; Koczan, J.; Nomura, K.; He, S.Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126, 969–980. [Google Scholar] [CrossRef]
- Ton, J.; Flors, V.; Mauch-Mani, B. The multifaceted role of ABA in disease resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Munemasa, S.; Uraji, M.; Nakamura, Y.; Mori, I.C.; Murata, Y. Involvement of endogenous abscisic acid in methyl jasmonate-induced stomatal closure in Arabidopsis. Plant Physiol. 2011, 156, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Cellini, A.; Fiorentini, L.; Dharmaraj, K.; Vanneste, J.L.; Cristescu, S.M.; Harren, F.J.M.; Costa, G.; Spinelli, F. Evidences of Ethylene as a Virulence Factor for Pseudomonas syringae pv. actinidiae and Field Measures to Limit the Kiwifruit Plant Bacterial Canker. In Proceedings of the IXth International Conference on the Plant Hormone Ethylene, Rotorua, New Zealand, 19–23 March 2012. [Google Scholar]
- Collina, M.; Donati, I.; Bertacchini, E.; Brunelli, A.; Spinelli, F. Greenhouse assays on the control of the bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). J. Berry Res. 2016, 6, 407–415. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, J. Chloroplasts at the crossroad of photosynthesis, pathogen infection and plant defense. Int. J. Mol. Sci. 2018, 19, E3900. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Li, L.; Li, D.; Zhang, Q.; Guo, Y.; Wang, S.; Zhong, C.; Huang, H. Whole transcriptome sequencing of Pseudomonas syringae pv. actinidiae-infected kiwifruit plants reveals species-specific interaction between long non-coding RNA and coding genes. Sci. Rep. 2017, 7, 4910. [Google Scholar] [CrossRef]
- Kaji, R.; Yariuchi, R.; Fujii, Y.; Taniguchi, S.; Uji, Y.; Suzuki, G.; Kashihara, K.; Kisaki, G.; Suezawa, K.; Ohtani, M. Expression analysis of defense-related genes in wild kiwifruit (Actinidia rufa) tolerant to bacterial canker. J. Gen. Plant Pathol. 2021, 87, 361–365. [Google Scholar] [CrossRef]
- Shelp, B.J.; Aghdam, M.S.; Flaherty, E.J. γ-Aminobutyrate (GABA) regulated plant defense: Mechanisms and opportunities. Plants 2021, 10, 1939. [Google Scholar] [CrossRef]
- Miao, L.; Genjia, T.; Yao, L.; Kejian, D.; Zhulong, C.; Yun, L. Relationships between the contents of phenolics, soluble proteins in plants of kiwifruit cultivars and their resistance to kiwifruit bacterial canker by Pseudomonas syringae pv. actinidiae. Plant Prot. 2009, 35, 37–41. [Google Scholar]
- Cheng, C.-H. Inheritance of resistance to Pseudomonas syringae pv. actinidiae and genetic correlations with fruit characters in a diploid Actinidia chinensis (kiwifruit) population. Euphytica 2014, 198, 305–315. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).