Enhanced U-Net for Spleen Segmentation in CT Scans: Integrating Multi-Slice Context and Grad-CAM Interpretability
Abstract
1. Introduction
2. Related Work
3. Materials and Methods
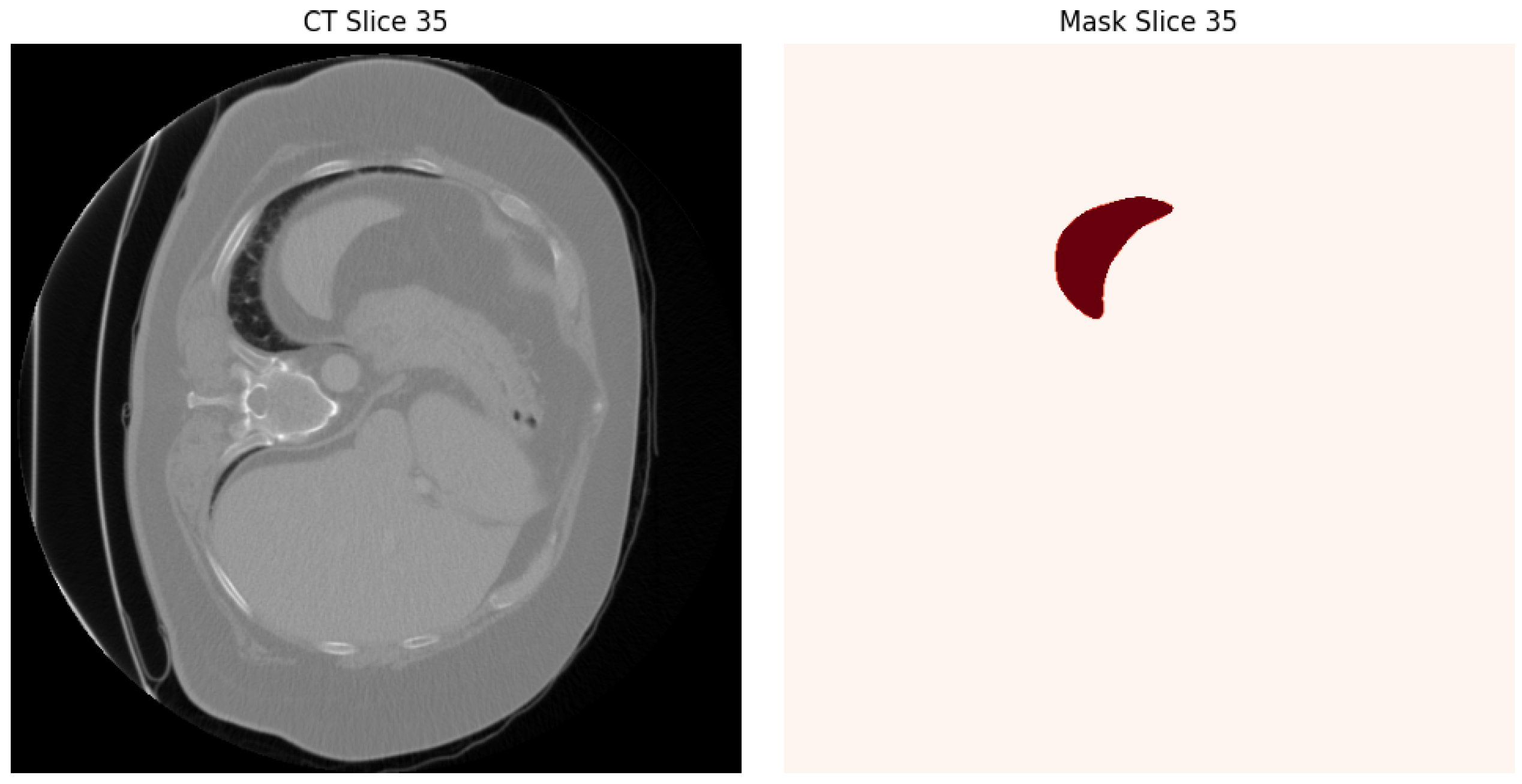
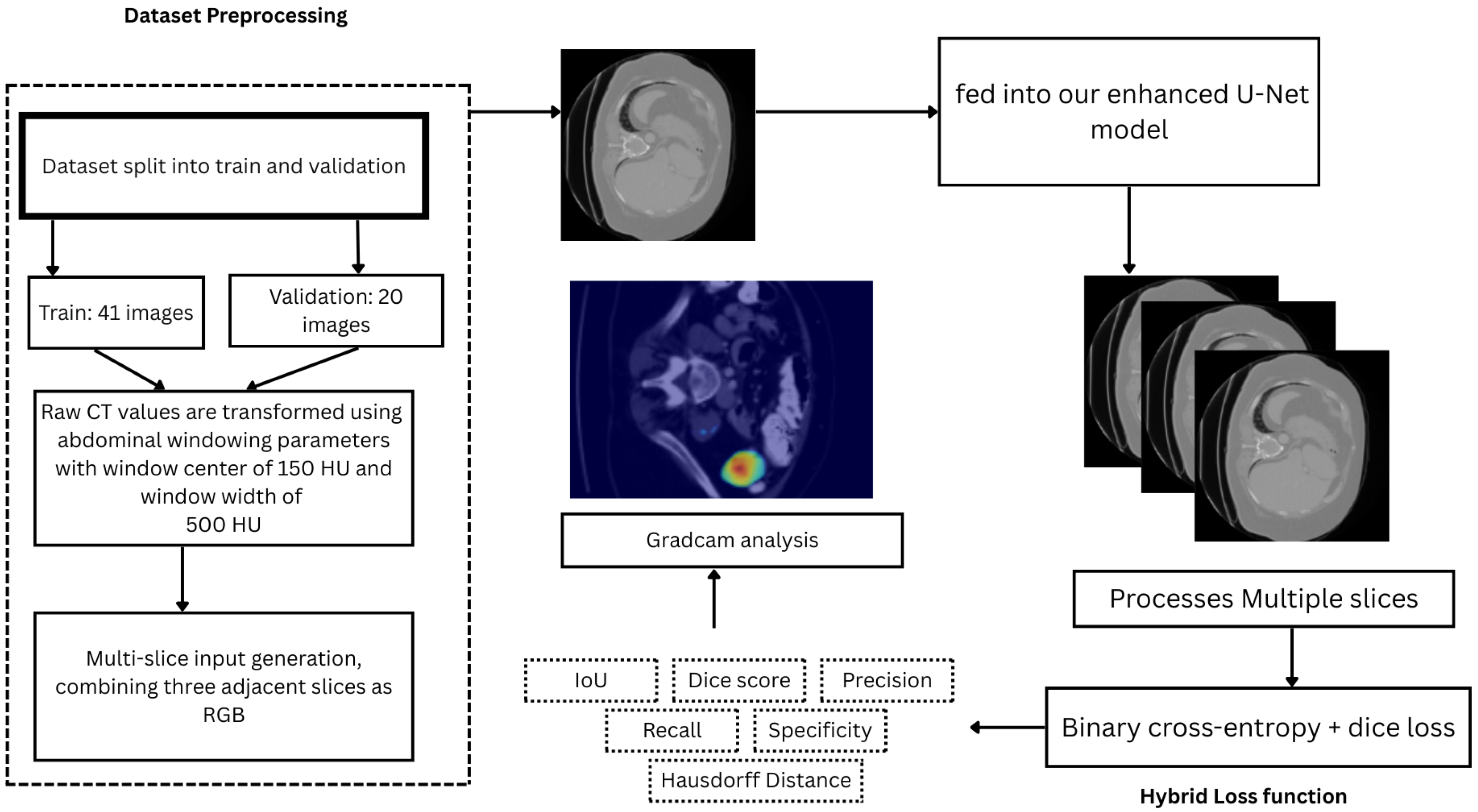
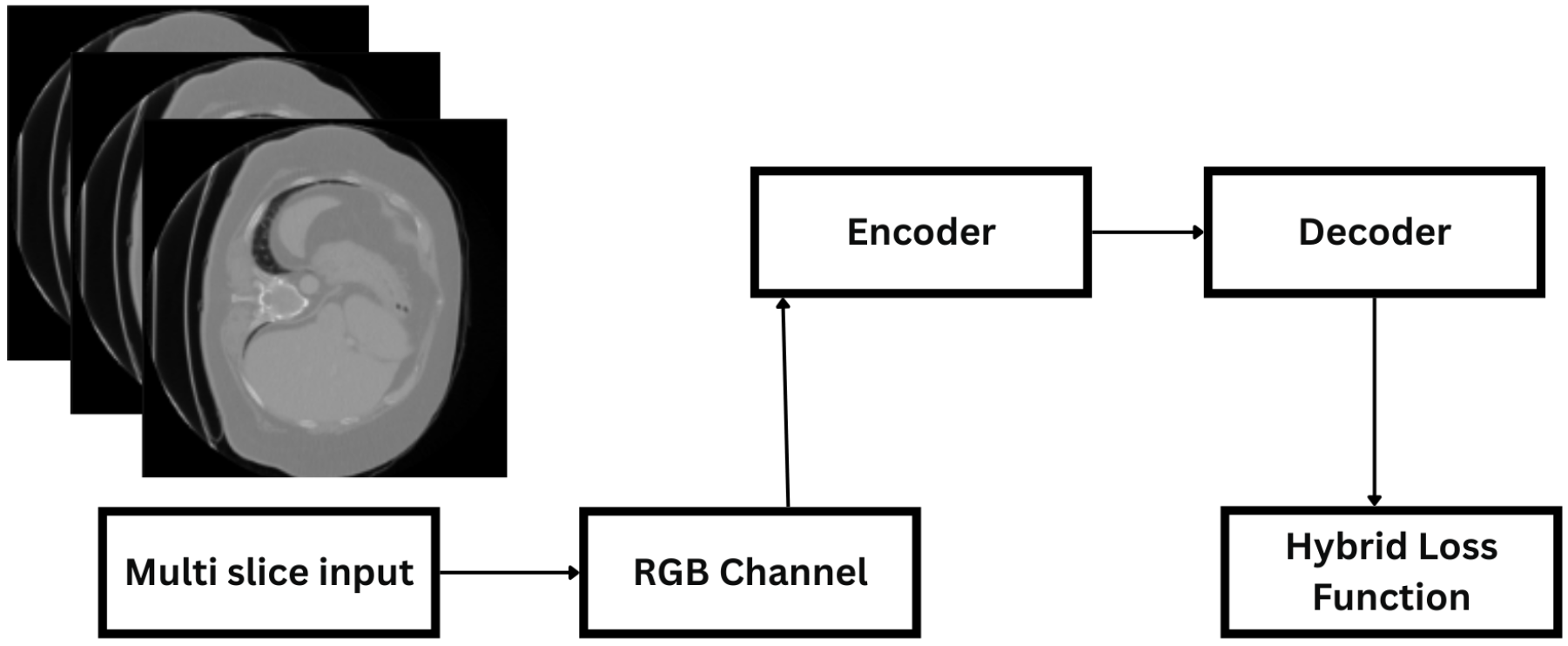
3.1. Dataset and Preprocessing
3.2. System Architecture
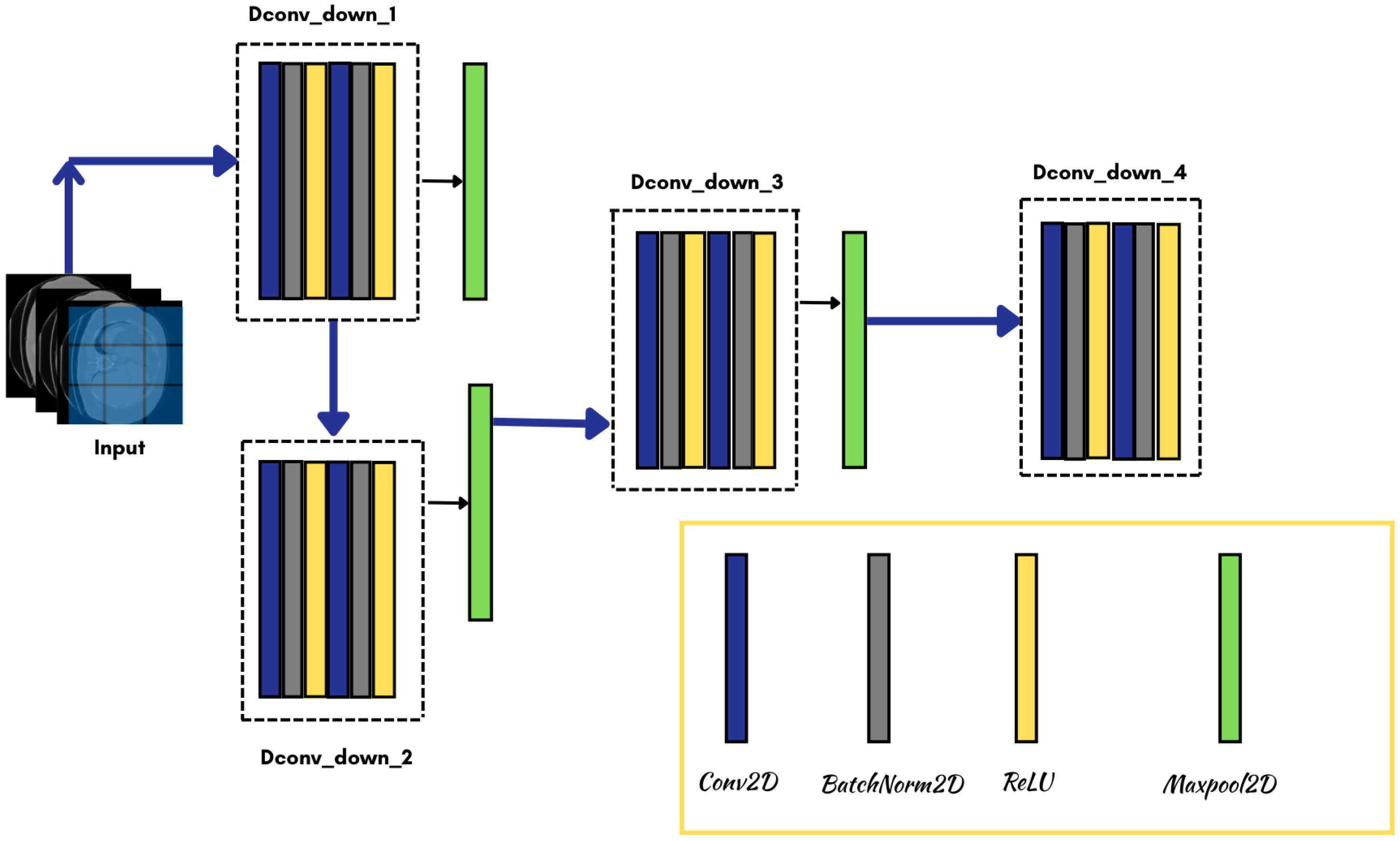
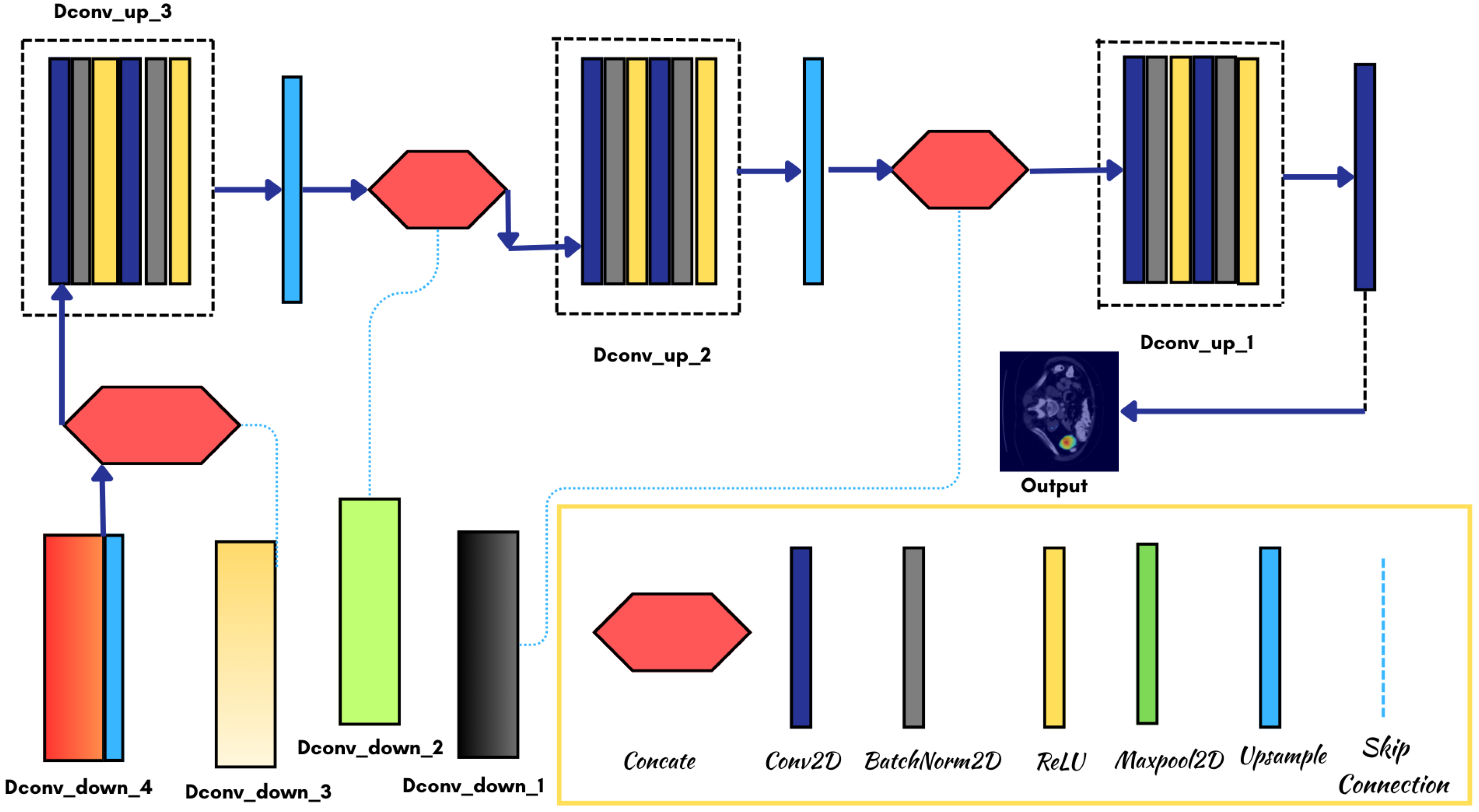
3.3. Model Architecture
4. Results
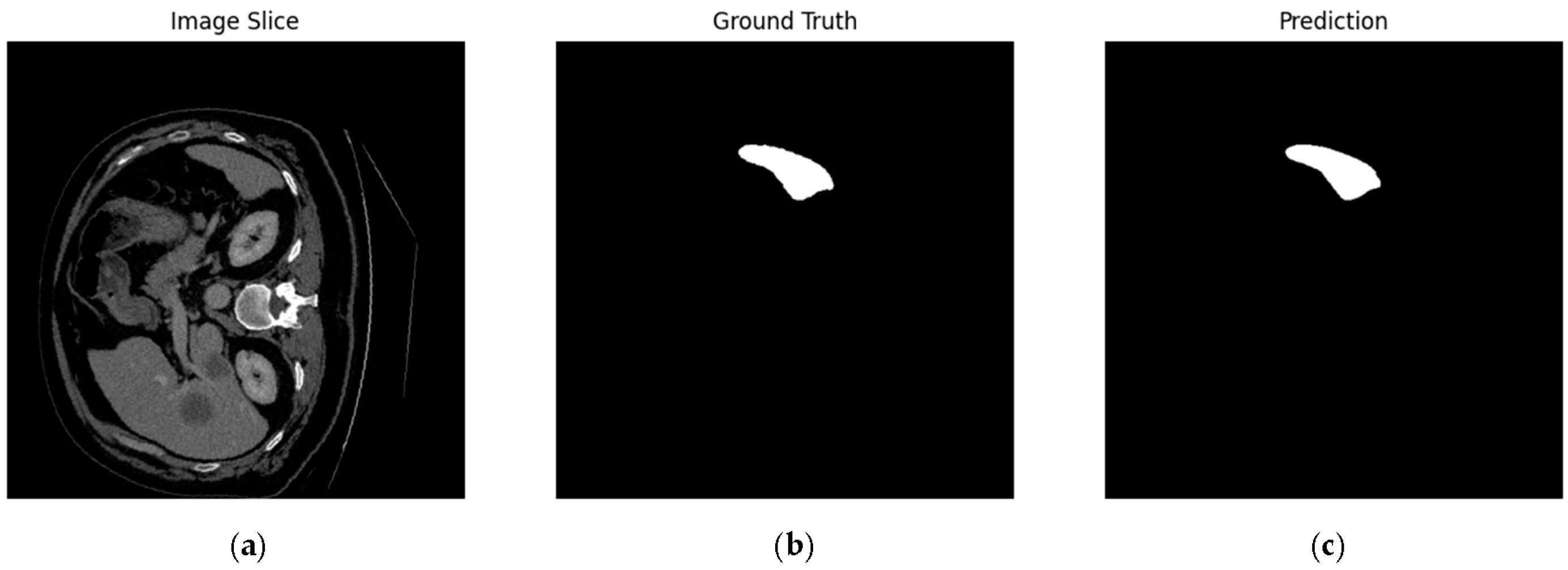
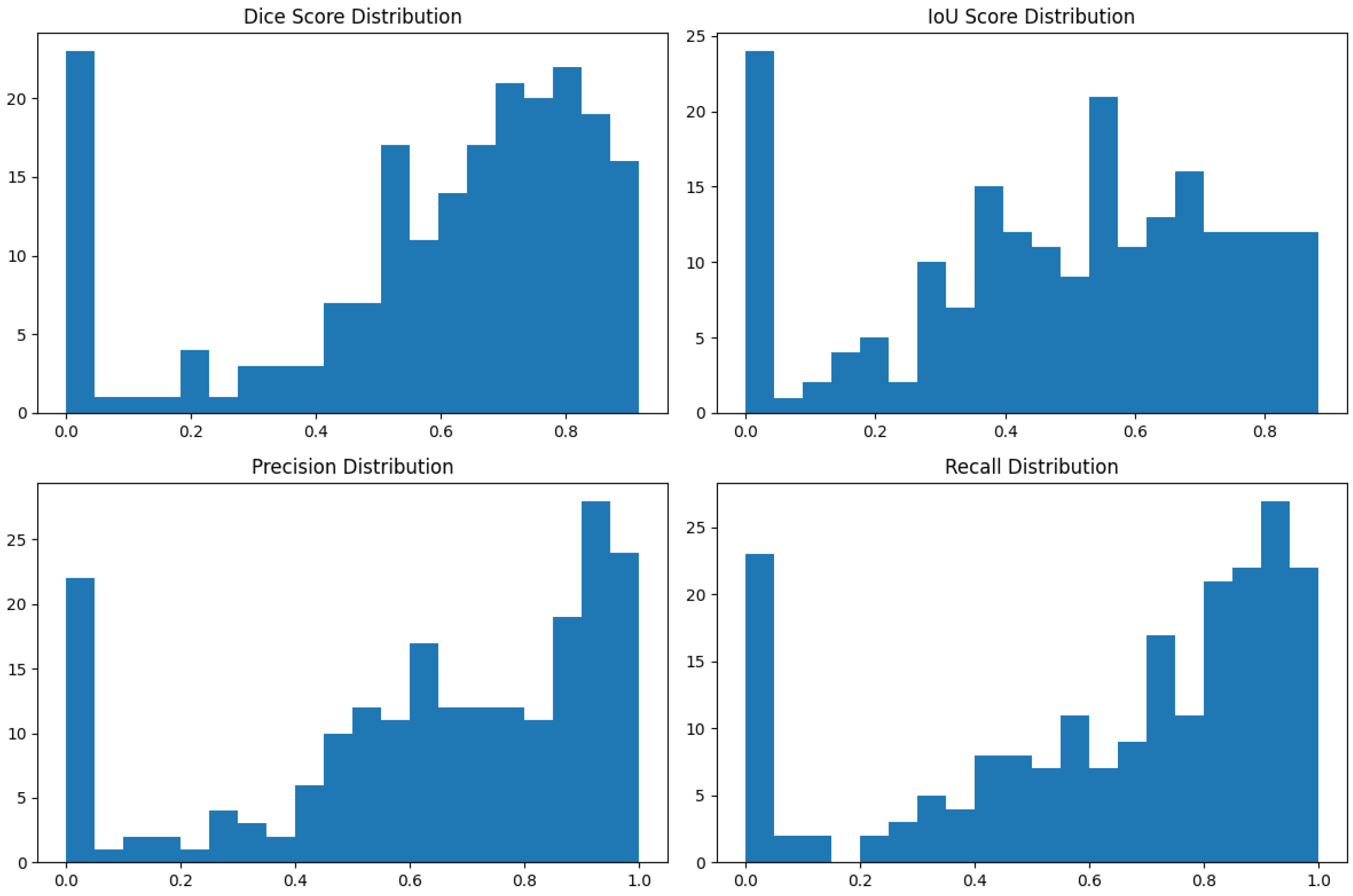
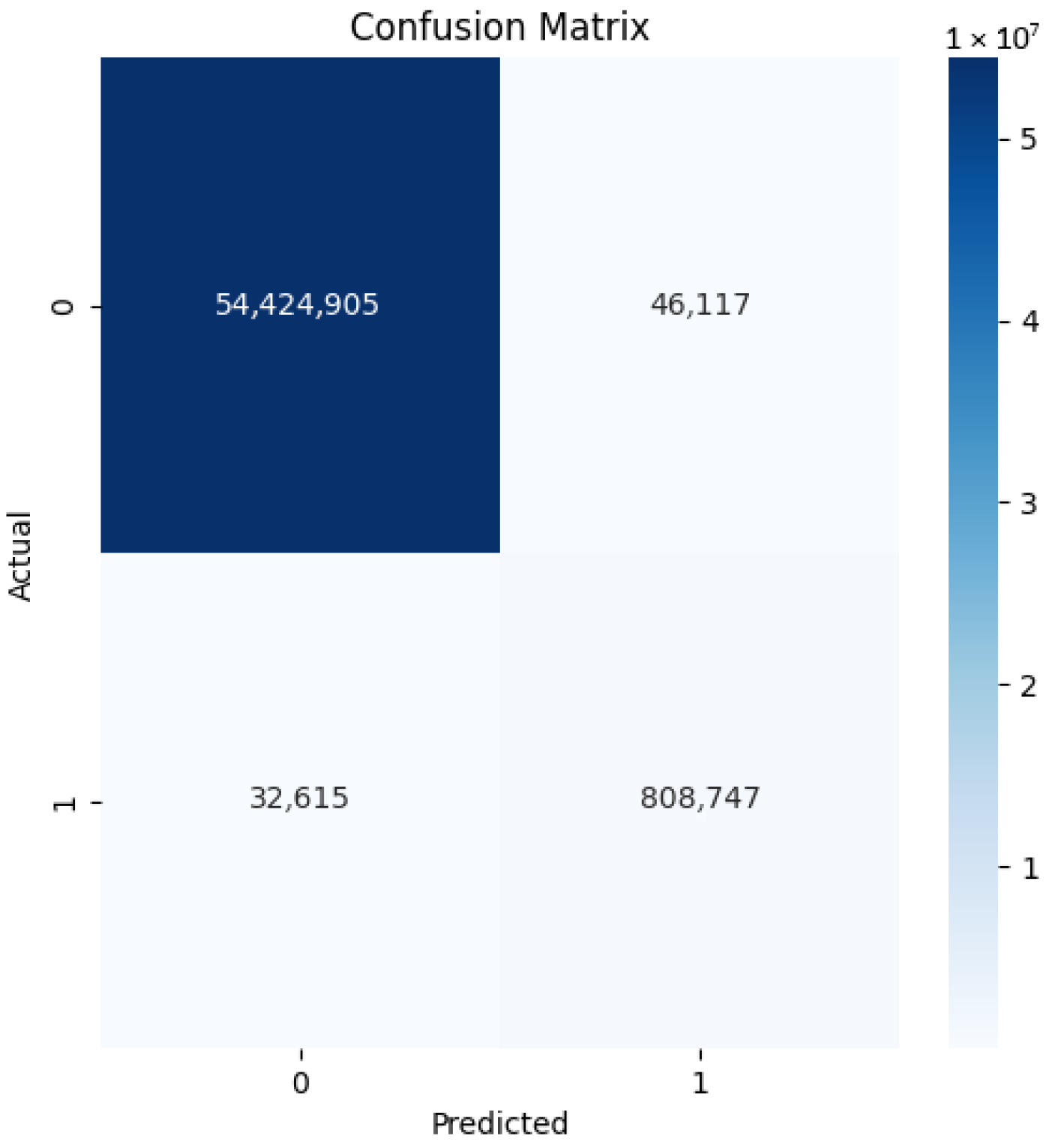
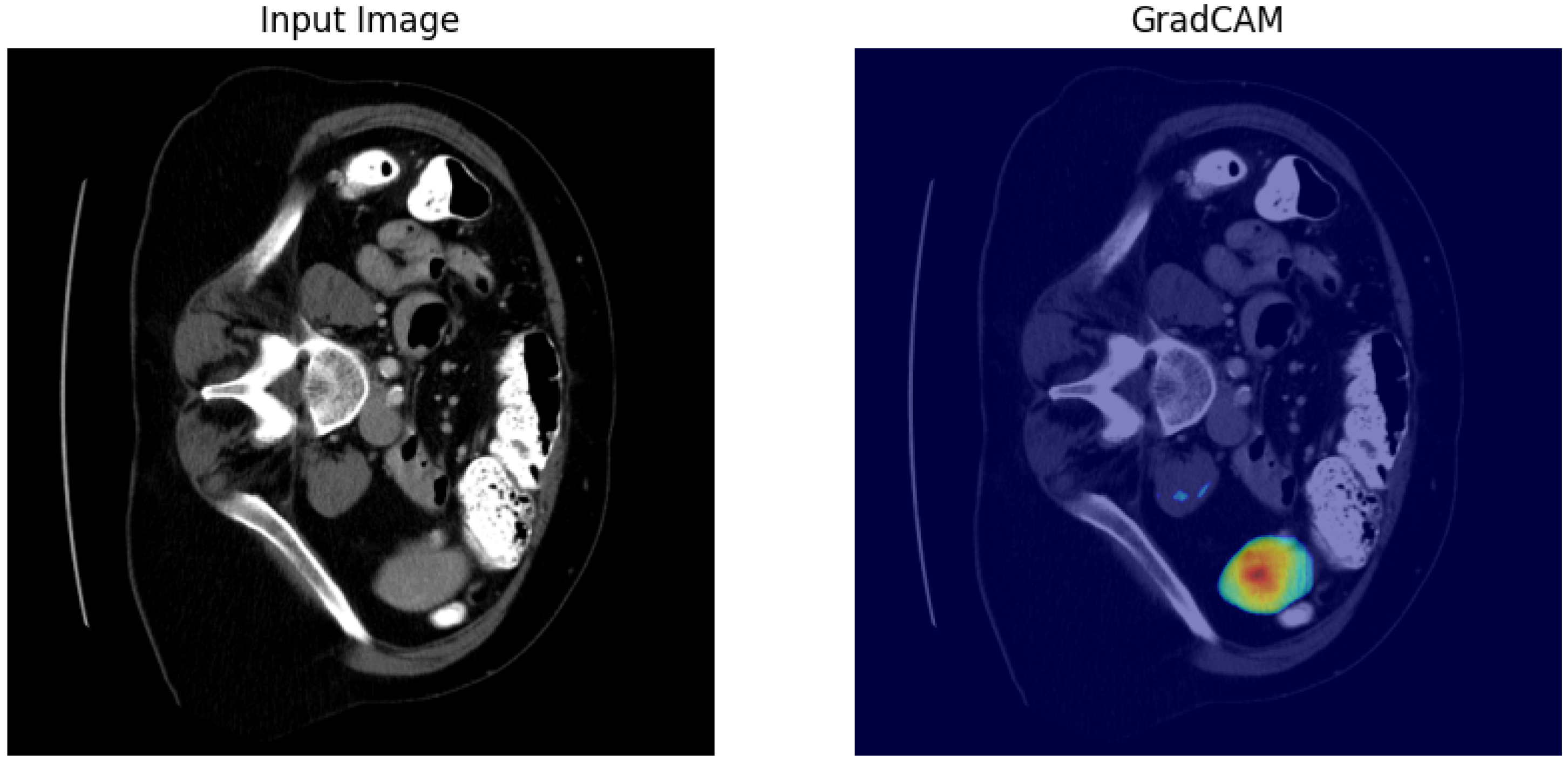
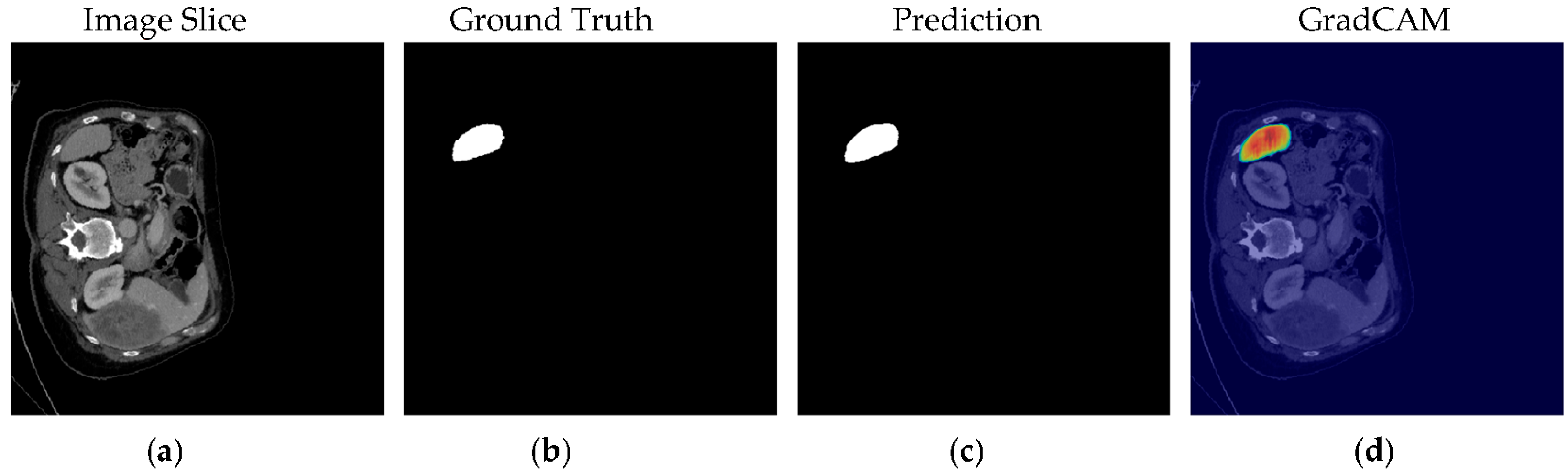
4.1. Our Model’s Results
4.2. Comparison with State-of-the-Art Models
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Gibson, E.; Giganti, F.; Hu, Y.; Bonmati, E.; Bandula, S.; Gurusamy, K.; Davidson, B.; Pereira, S.P.; Clarkson, M.J.; Barratt, D.C. Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans. Med. Imaging 2018, 37, 1822–1834. [Google Scholar] [CrossRef]
- Mebius, R.E.; Kraal, G. Structure and function of the spleen. Nat. Rev. Immunol. 2005, 5, 606–616. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A nested U-Net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Granada, Spain, 20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 3–11. [Google Scholar] [CrossRef]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 19th International Conference, MICCAI 2016, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 424–432. [Google Scholar] [CrossRef]
- Wolz, R.; Chu, C.; Misawa, K.; Fujiwara, M.; Mori, K.; Rueckert, D. Automated abdominal multi-organ segmentation with subject-specific atlas generation. IEEE Trans. Med. Imaging 2013, 32, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Linguraru, M.G.; Hori, M.; Summers, R.M.; Tomiyama, N.; Sato, Y. Abdominal multi-organ segmentation from CT images using conditional shape-location priors. Med. Image Anal. 2015, 26, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.-C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Hierarchical 3D fully convolutional networks for multi-organ segmentation. arXiv 2017, arXiv:1704.06382. [Google Scholar] [CrossRef]
- Zheng, G.; Zheng, G. Multi-stream 3D FCN with Multi-scale Deep Supervision for Multi-modality Isointense Infant Brain MR Image Segmentation. arXiv 2017, arXiv:1711.10212. [Google Scholar] [CrossRef]
- Tang, Y.; Wang, X.; Harrison, A.P.; Lu, L.; Xiao, J.; Summers, R.M. Attention-guided curriculum learning for weakly supervised classification and localization of thoracic diseases on chest radiographs. In Machine Learning in Medical Image Analysis, Proceedings of the 9th International Workshop, MLMI 2018, Granada, Spain, 16 September 2018; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Seo, H.; Huang, C.; Bassenne, M.; Xiao, R.; Xing, L. Modified U-Net (mU-Net) with incorporation of object-dependent high level features for improved liver and liver-tumor segmentation in CT images. IEEE Trans. Med. Imaging 2020, 39, 1316–1325. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. TransUNet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar] [CrossRef]
- Soberanis-Mukul, R.D.; Navab, N.; Albarqouni, S. An Uncertainty-Driven GCN Refinement Strategy for Organ Segmentation. arXiv 2020, arXiv:2012.03352. [Google Scholar] [CrossRef]
- Nath, V.; Yang, D.; Hatamizadeh, A.; Abidin, A.A.; Myronenko, A.; Roth, H.; Xu, D. The Power of Proxy Data and Proxy Networks for Hyper-Parameter Optimization in Medical Image Segmentation. arXiv 2021, arXiv:2107.05471. [Google Scholar] [CrossRef]
- El Jurdi, R.; Petitjean, C.; Honeine, P.; Cheplygina, V.; Abdallah, F. A surprisingly effective perimeter-based loss for medical image segmentation. In Proceedings of the Fourth Conference on Medical Imaging with Deep Learning, Lübeck, Germany, 7–9 July 2021; pp. 158–167. Available online: https://proceedings.mlr.press/v143/el-jurdi21a.html (accessed on 23 August 2025).
- Meddeb, A.; Kossen, T.; Bressem, K.K.; Hamm, B.; Nagel, S.N. Evaluation of a Deep Learning Algorithm for Automated Spleen Segmentation in Patients with Conditions Directly or Indirectly Affecting the Spleen. Tomography 2021, 7, 950–960. [Google Scholar] [CrossRef]
- Smith, A.G.; Kutnár, D.; Vogelius, I.R.; Darkner, S.; Petersen, J. Localise to segment: Crop to improve organ at risk segmentation accuracy. arXiv 2023, arXiv:2304.04606. [Google Scholar] [CrossRef]
- Jha, D.; Tomar, N.K.; Biswas, K.; Durak, G.; Antalek, M.; Zhang, Z.; Wang, B.; Rahman, M.M.; Pan, H.; Medetalibeyoglu, A.; et al. MDNet: Multi-Decoder Network for Abdominal CT Organs Segmentation. arXiv 2024, arXiv:2405.06166. [Google Scholar] [CrossRef]
- Shen, C.; Li, W.; Shi, Y.; Wang, X. Interactive 3D Medical Image Segmentation with SAM 2. arXiv 2024, arXiv:2408.02635. [Google Scholar] [CrossRef]
- Yuan, Z.; Stojanovski, D.; Li, L.; Gomez, A.; Jogeesvaran, H.; Puyol-Antón, E.; Inusa, B.; King, A.P. DeepSPV: A Deep Learning Pipeline for 3D Spleen Volume Estimation from 2D Ultrasound Images. arXiv 2024, arXiv:2411.11190. [Google Scholar] [CrossRef]
- Vu, M.H.; Tronchin, L.; Nyholm, T.; Löfstedt, T. Using Synthetic Images to Augment Small Medical Image Datasets. arXiv 2025, arXiv:2503.00962. [Google Scholar] [CrossRef]
- Samir, J.; Ramadass, K.; Saunders, A.M.; Krishnan, A.R.; Remedios, L.W.; McMaster, E.M.; Landman, B.A. The medical segmentation decathlon without a doctorate. In Proceedings of the Medical Imaging 2025: Clinical and Biomedical Imaging, San Diego, CA, USA, 16–21 February 2025; Volume 13410, p. 134101K. [Google Scholar] [CrossRef]
- Linguraru, M.G.; Sandberg, J.K.; Jones, E.C.; Summers, R.M. Automated segmentation and quantification of liver and spleen from CT images using normalized probabilistic atlases and enhancement estimation. Med. Phys. 2010, 37, 2771–2783. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Seff, A.; Cherry, K.M.; Hoffman, J.; Wang, S.; Liu, J.; Turkbey, E.; Summers, R.M. DeepOrgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Medical Image Computing and Computer-Assisted Intervention, Proceedings of the 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.P.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.-W.; Wu, J. UNet 3+: A full-scale connected UNet for medical image segmentation. In Proceedings of the IEEE International Conference on Acoustics, Speech and Signal Processing, Barcelona, Spain, 4–8 May 2020; pp. 1055–1059. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the Fourth International Conference on 3D Vision, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Jorge Cardoso, M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the Third International Workshop, DLMIA 2017, and 7th International Workshop, ML-CDS 2017, Quebec, QC, Canada, 14 September 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 240–248. [Google Scholar] [CrossRef]
- Calisto, M.B.; Lai-Yuen, S.K. AdaEn-Net: An ensemble of adaptive 2D–3D fully convolutional networks for medical image segmentation. Neural Netw. 2021, 126, 76–94. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2921–2929. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar] [CrossRef]
- Jalali, Y.; Fateh, M.; Rezvani, M.; Abolghasemi, V.; Anisi, M.H. ResBCDU-Net: A deep learning framework for lung CT image segmentation. Sensors 2021, 21, 268. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar] [CrossRef]
- Vu, M.H.; Grimbergen, G.; Nyholm, T.; Löfstedt, T. Evaluation of multislice inputs to convolutional neural networks for medical image segmentation. Med. Phys. 2020, 47, 6216–6231. [Google Scholar] [CrossRef]
- Sørensen, T. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. In Kongelige Danske Videnskabernes Selskab; Munksgaard: Copenhagen, Denmark, 1948; Volume 5, pp. 1–34. Available online: https://books.google.com.bd/books?id=rpS8GAAACAAJ (accessed on 23 August 2025).
- Fawcett, T. An introduction to ROC analysis. Pattern Recognit. Lett. 2006, 27, 861–874. [Google Scholar] [CrossRef]
- Huttenlocher, D.P.; Klanderman, G.A.; Rucklidge, W.J. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 850–863. [Google Scholar] [CrossRef]
- Maier, O.; Menze, B.H.; von der Goltz, J.; Hänsch, R.; Handels, H.; Hoelter, T.; Jakab, A.; Kalavari, V.; Lancaster, J.L.; Marti-Bonmati, L.; et al. ISLES 2015—A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 2017, 35, 250–269. [Google Scholar] [CrossRef]
| Ref. | Methods | Architecture | Dataset | Dice | Research Gap |
|---|---|---|---|---|---|
| [7] | Multi-atlas w/shape priors | Statistical atlas | 38 CT | 0.89 | Computationally intensive |
| [8] | Shape model, deformable reg. | Active shape model | 86 cases | 0.84 | Limited robustness |
| [9] | 3D FCN | 5-layer 3D CNN | BTCV (30) | 0.90 | High memory needs |
| [10] | Multi-scale 3D FCN | 7-layer CNN | LiTS (131) | 0.91 | High GPU memory |
| [11] | Attention U-Net | 6-layer U-Net + attn. | Private (200) | 0.87 | Overfitting risk |
| [5] | UNet++ nested | 5-layer nested U-Net | Med. Decathlon | 0.92 | Slow convergence |
| [12] | nnU-Net auto-config | 6-layer adaptive U-Net | Med. Decathlon | 0.915 | Resource-heavy |
| [13] | Modified U-Net | 5-layer U-Net | TCIA (150) | 0.89 | Boundary issues |
| [14] | Transformer-U-Net | 6-layer CNN-Trans. | Synapse | 0.913 | Low interpretability |
| [15] | Uncertainty-driven GCN refinement | 2D U-Net + GCN | MSD (61 CT) | – | Lesion size imbalance |
| [16] | Proxy data for hyperparameter optimization | CNN-based optimization | MSD proxy | – | Expensive doing optimization |
| [17] | Perimeter-based loss function | U-Net with perimeter loss | MSD (41 CT) | – | Expensive contour losses |
| [18] | Automated segmentation for abnormal spleens | Enhanced 3D U-Net | MSD (61) + in-house | 0.897 | Small dataset, no cross-validation |
| [19] | Two-stage localization + segmentation | Localization + segmentation networks | MSD multi-organ | – | Poor performance for small organs |
| [20] | Multi-decoder with refinement | MiT-B2 + multi-decoders | MSD (41 CT) | 0.917 | Single decoder poor for heterogeneity |
| [21] | Zero-shot segmentation with SAM 2 | SAM 2 foundation model | MSD CT | – | Gap to supervised methods |
| [22] | Segmentation + VAE for volume estimation | Segmentation network + VAE | MSD (60/40 CT) | – | Limited 2D volume estimation accuracy |
| [23] | Synthetic image augmentation | Conditional StyleGAN2 | MSD (one of ten) | – | Synthetic images not always beneficial |
| [24] | Interactive AI-guided labeling | MONAI Label on 3D Slicer | MSD (34 images) | 0.831 | Lack of prospective studies on active learning |
| Ours | Enhanced U-Net | 4-layer + context + hybrid loss | Med. Decathlon (61) | 0.923 | 2D limits, preprocessing |
| Metric | Mean | Standard Deviation |
|---|---|---|
| Dice Similarity Coefficient | 0.923 | 0.04 |
| Intersection over Union | 0.859 | 0.06 |
| Precision | 0.934 | 0.05 |
| Recall (Sensitivity) | 0.918 | 0.06 |
| Specificity | 0.997 | 0.002 |
| Hausdorff Distance (mm) | 9.47 | 4.36 |
| Average Surface Distance (mm) | 1.21 | 0.83 |
| Volumetric Error (%) | 4.7 | 3.2 |
| Total Parameters (M) | 7.82 | - |
| Trainable Parameters (M) | 7.82 | - |
| Non-Trainable Parameters (M) | 0 | - |
| Case Category | Dice Score | Hausdorff Distance (mm) |
|---|---|---|
| Normal spleen morphology | 0.938 | 8.12 |
| Splenomegaly (>500 mL) | 0.917 | 10.84 |
| Post-trauma cases | 0.901 | 12.37 |
| Thin slice (<3 mm) | 0.932 | 8.76 |
| Thick slice (>5 mm) | 0.911 | 11.29 |
| Portal phase contrast | 0.928 | 9.03 |
| Arterial phase contrast | 0.912 | 10.52 |
| High contrast enhancement | 0.935 | 8.45 |
| Low contrast enhancement | 0.908 | 11.23 |
| Configuration | Dice Score | Improvement |
|---|---|---|
| Baseline U-Net | 0.891 | - |
| + Multi-slice input | 0.911 | +2.0% |
| + Hybrid loss function | 0.912 | +2.1% |
| + Data augmentation | 0.918 | +2.7% |
| + Optimized preprocessing | 0.923 | +3.2% |
| Single vs. Multi-slice | ||
| Single-slice input | 0.903 | - |
| Three-slice input | 0.923 | +2.0% |
| Five-slice input | 0.921 | +1.8% |
| Loss function comparison | ||
| Dice loss only | 0.911 | - |
| BCE loss only | 0.889 | −2.2% |
| Hybrid loss () | 0.923 | +1.2% |
| Method | Dice Score | HD (mm) | Training Strategy |
|---|---|---|---|
| Standard U-Net [4] | 0.891 | 12.34 | Encoder–decoder, skip connections, CE loss, basic aug. |
| UNet++ [5] | 0.920 | 10.12 | Nested skip connections, deep supervision, Dice loss |
| Attention U-Net [27] | 0.905 | 11.67 | Attention gates, Dice+CE loss, augmentation |
| nnU-Net [12] | 0.915 | 10.89 | Adaptive preprocessing, Dice+CE loss, heavy aug. |
| TransUNet [14] | 0.913 | 10.45 | Transformer+U-Net, Dice loss, patch-based |
| Our Enhanced U-Net | 0.923 | 9.47 | Multi-slice, hybrid loss, deep supervision aug. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, S.; Raju, M.A.H.; Evna Jafar, A.; Akter, M.; Suma, I.J.; Uddin, J. Enhanced U-Net for Spleen Segmentation in CT Scans: Integrating Multi-Slice Context and Grad-CAM Interpretability. BioMedInformatics 2025, 5, 56. https://doi.org/10.3390/biomedinformatics5040056
Rahman S, Raju MAH, Evna Jafar A, Akter M, Suma IJ, Uddin J. Enhanced U-Net for Spleen Segmentation in CT Scans: Integrating Multi-Slice Context and Grad-CAM Interpretability. BioMedInformatics. 2025; 5(4):56. https://doi.org/10.3390/biomedinformatics5040056
Chicago/Turabian StyleRahman, Sowad, Md Azad Hossain Raju, Abdullah Evna Jafar, Muslima Akter, Israt Jahan Suma, and Jia Uddin. 2025. "Enhanced U-Net for Spleen Segmentation in CT Scans: Integrating Multi-Slice Context and Grad-CAM Interpretability" BioMedInformatics 5, no. 4: 56. https://doi.org/10.3390/biomedinformatics5040056
APA StyleRahman, S., Raju, M. A. H., Evna Jafar, A., Akter, M., Suma, I. J., & Uddin, J. (2025). Enhanced U-Net for Spleen Segmentation in CT Scans: Integrating Multi-Slice Context and Grad-CAM Interpretability. BioMedInformatics, 5(4), 56. https://doi.org/10.3390/biomedinformatics5040056







