Self-Explaining Neural Networks for Food Recognition and Dietary Analysis
Abstract
1. Introduction
- We advance the field of personalised nutrition with a novel lightweight self-explaining neural architecture that achieves a 73.4% parameter reduction while maintaining high accuracy, demonstrating that efficient computational models can be deployed in resource-constrained healthcare settings without sacrificing performance.
- We introduce a new quantitative interpretability framework for nutritional pattern recognition, offering the first comprehensive metrics specifically designed to evaluate both feature attribution quality and decision pathway transparency in dietary analysis applications.
- We establish new performance benchmarks for vulnerable population dietary analysis, surpassing existing approaches by 6.3% in accuracy while reducing processing time by 23.9%, providing empirical evidence that specialised neural architectures can better address the unique nutritional needs of at-risk groups.
- We contribute methodological innovations through our integration of attention mechanisms with temporal modules specifically designed to handle diverse dietary patterns, demonstrating superior robustness in cross-validation testing with consistent accuracy across varied population segments.
2. Related Work
2.1. Deep Learning in Nutrition
2.2. Self-Explaining Neural Networks
2.3. Traditional Dietary Analysis
3. Methodology
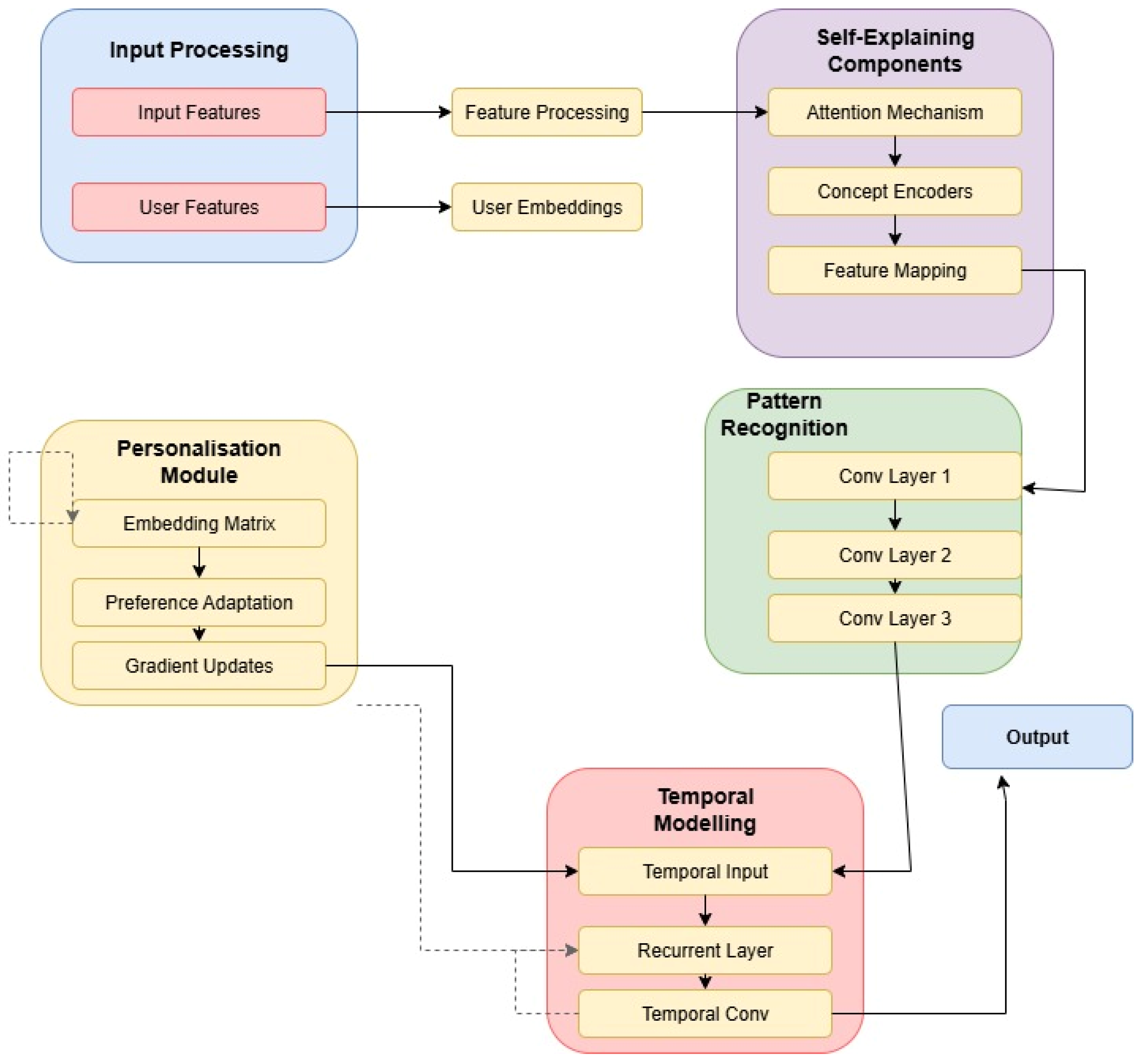
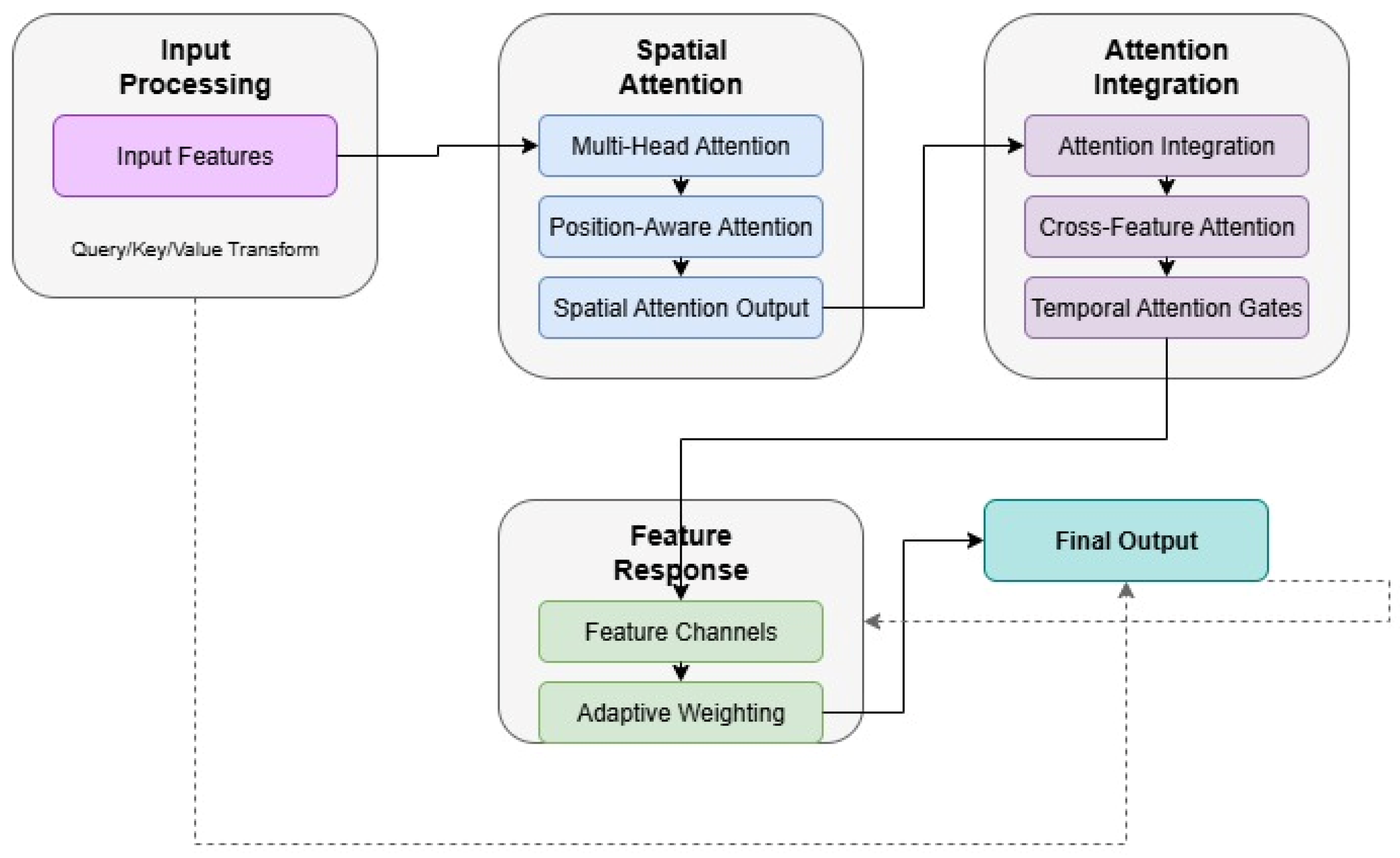
3.1. Model Architecture
3.2. Pattern Recognition Implementation
3.3. Interpretability Design
Detailed Interpretability Metrics
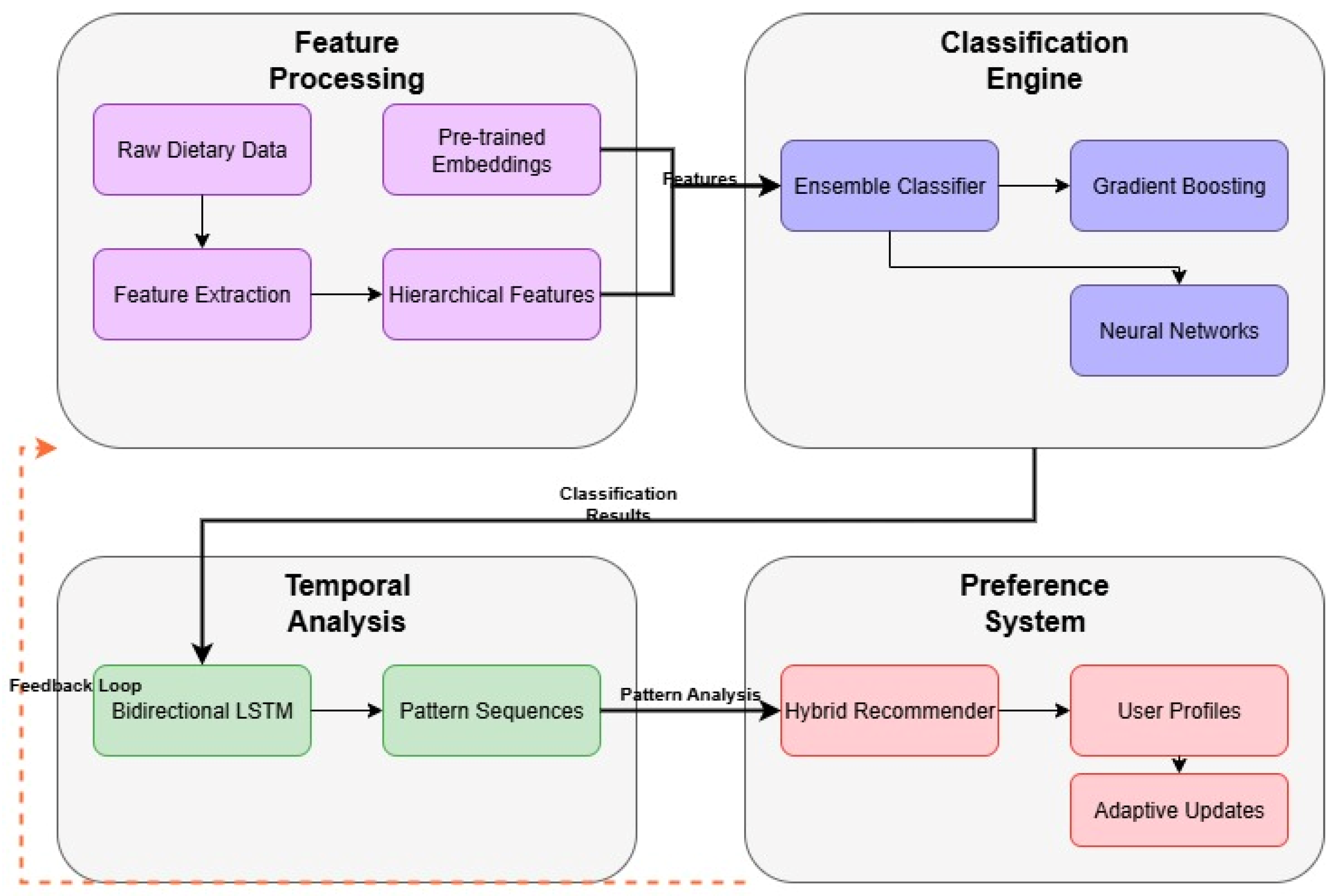
4. Implementation
4.1. Dataset Description
4.2. Training
4.3. Neural Network Architecture
4.4. Model Architecture
5. Experiments and Results
5.1. Data and Implementation Details
5.1.1. Dataset Preprocessing
5.1.2. Implementation
- CPU: AMD EPYC 7763 64-Core Processor;
- RAM: 512 GB DDR4;
- Storage: 2 TB NVMe SSD;
- Network: 100 Gbps InfiniBand.
5.2. Size and Speed
5.2.1. Model Size and Parameter Analysis
5.2.2. Speed and Resource Analysis
5.3. Ablation Study
5.3.1. Architectural Analysis
5.3.2. Cross-Validation Analysis
5.3.3. Systematic Analysis
5.4. Interpretability
5.4.1. Quantitative Interpretability Analysis
5.4.2. Detailed Performance Analysis
5.4.3. Visual Interpretability Analysis
5.4.4. Expert Validation
5.5. Comparison with State-of-the-Art
5.5.1. Model Benchmarking
5.5.2. Computational Efficiency Analysis
5.6. Dietary Pattern Analysis
5.6.1. Performance Benchmarking
5.6.2. Food Group Classification
5.6.3. Multi-Item Recognition
5.6.4. Inter-Item Relationship Analysis
6. Discussion
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CNNs | Convolutional Neural Networks |
| FOOD101 | Food 101 dataset |
| GPU | Graphics Processing Unit |
| LSTM | Long Short-Term Memory |
| ML | Machine Learning |
| ReLU | Rectified Linear Unit |
| RNNs | Recurrent Neural Networks |
| SENNs | Self-Explaining Neural Networks |
References
- Richardson, J.P.; Smith, C.; Curtis, S.; Watson, S.; Zhu, X.; Barry, B. Patient Apprehensions About the Use of Artificial Intelligence in Healthcare. NPJ Digit. Med. 2021, 4, 140. [Google Scholar] [CrossRef] [PubMed]
- Cuadros, D.F.; Moreno, C.M.; Miller, F.D.; Omori, R.; MacKinnon, N.J. Assessing Access to Digital Services in Health Care-Underserved Communities in the United States: A Cross-Sectional Study. Mayo Clin. Proc. Digit. Health 2023, 1, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Jabbari, H. The Role and Application of Artificial Intelligence (AI) in Leveraging Big Data in the Healthcare Domain. Health Nexus 2023, 1, 83–86. [Google Scholar] [CrossRef]
- Dai, Y.; Chai, C.S.; Lin, P.-Y.; Jong, M.S.; Guo, Y.; Qin, J. Promoting Students’ Well-Being by Developing Their Readiness for the Artificial Intelligence Age. Sustainability 2020, 12, 6597. [Google Scholar] [CrossRef]
- Choudhury, A.; Renjilian, E.; Asan, O. Use of Machine Learning in Geriatric Clinical Care for Chronic Diseases: A Systematic Literature Review. JAMIA Open 2020, 3, 459–471. [Google Scholar] [CrossRef]
- Messmann, H.; Bisschops, R.; Antonelli, G.; Libânio, D.; Sinonquel, P.; Abdelrahim, M.; Ahmad, O.F.; Areia, M.; Bergman, J.J.G.H.M.; Bhandari, P.; et al. Expected Value of Artificial Intelligence in Gastrointestinal Endoscopy: European Society of Gastrointestinal Endoscopy (ESGE) Position Statement. Endoscopy 2022, 54, 1211–1231. [Google Scholar] [CrossRef]
- Lambell, K.; Tatucu-Babet, O.A.; Chapple, L.S.; Gantner, D.; Ridley, E.J. Nutrition Therapy in Critical Illness: A Review of the Literature for Clinicians. Crit. Care 2020, 24, 35. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y. Addressing Sparse Data in Nutritional Studies: Machine Learning Approaches and Challenges. Nutrients 2022, 14, 1456. [Google Scholar] [CrossRef]
- Kearney, J.M.; McElhone, S. The Role of Technology in Dietary Assessment: A Review of Current Tools and Future Directions. Nutr. Rev. 2020, 78, 278–303. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, L. A Review of Machine Learning Techniques for Dietary Pattern Recognition. Food Qual. Prefer. 2020, 81, 103835. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, S. Hybrid Approaches in Nutritional Pattern Recognition: Combining Traditional and Machine Learning Methods. J. Nutr. Educ. Behav. 2021, 53, 421–429. [Google Scholar] [CrossRef]
- Dai, E.; Wang, S. Towards Self-Explainable Graph Neural Network. In Proceedings of the CIKM ’21: Proceedings of the 30th ACM International Conference on Information & Knowledge Management, Virtual Event, 1–5 November 2021; pp. 302–311. [Google Scholar] [CrossRef]
- Motadi, S.; Khorommbi, T.; Maluleke, L. Nutritional Status and Dietary Pattern of the Elderly in Tshiulungoma and Maniini Village of Thulamela Municipality, Vhembe District. Afr. J. Prim. Health Care Fam. Med. 2021, 14, e1–e8. [Google Scholar] [CrossRef]
- Armand, T.P.T.; Nfor, K.A.; Kim, J.I.; Kim, H.C. Applications of Artificial Intelligence, Machine Learning, and Deep Learning in Nutrition: A Systematic Review. Nutrients 2024, 16, 1073. [Google Scholar] [CrossRef] [PubMed]
- Côté, J.; Lamarche, B. Integrating Deep Learning with Traditional Dietary Assessment Tools: A New Era in Nutritional Evaluations. Nutrients 2022, 14, 1023. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X. Time-Series Analysis of Dietary Patterns Using Deep Learning: A Systematic Review. J. Med. Internet Res. 2021, 23, e23456. [Google Scholar] [CrossRef]
- Morgenstern, M.; Gunter, M.J.; Heller, R.F. Advances in Nutritional Pattern Recognition: The Role of Machine Learning in Public Health. Public Health Nutr. 2021, 24, 2739–2750. [Google Scholar]
- Javeed, M.; Gochoo, M.; Jalal, A.; Kim, K. HF-SPHR: Hybrid Features for Sustainable Physical Healthcare Pattern Recognition Using Deep Belief Networks. Sustainability 2021, 13, 1699. [Google Scholar] [CrossRef]
- Shamanna, P.; Joshi, S.; Thajudeen, M.; Shah, L.; Poon, T.; Mohamed, M.; Mohammed, J. Personalized nutrition in type 2 diabetes remission: Application of digital twin technology for predictive glycemic control. Front. Endocrinol. 2024, 15, 1485464. [Google Scholar] [CrossRef]
- Anwar, H.; Anwar, T.; Murtaza, M. Applications of electronic nose and machine learning models in vegetables quality assessment: A review. In Proceedings of the 2023 IEEE International Conference on Emerging Trends in Engineering, Sciences and Technology (ICES&T), Bahawalpur, Pakistan, 9–11 January 2023. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Araki, M.; Hamada, K.; Nojiri, T.; Nishi, N. Development of a machine learning model for classifying cooking recipes according to dietary styles. Foods 2024, 13, 667. [Google Scholar] [CrossRef]
- Morgenstern, J.D.; Rosella, L.C.; Costa, A.P.; de Souza, R.J.; Anderson, L.N. Perspective: Big data and machine learning could help advance nutritional epidemiology. Adv. Nutr. 2021, 12, 621–631. [Google Scholar] [CrossRef]
- Grollemund, V.; Le Chat, G.; Secchi-Buhour, M.-S.; Delbot, F.; Pradat-Peyre, J.-F.; Bede, P.; Pradat, P.-F. Development and validation of a 1-year survival prognosis estimation model for amyotrophic lateral sclerosis using manifold learning algorithm UMAP. Sci. Rep. 2020, 10, 13378. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, J.M.; Raffoul, A.; Pepetone, A.; Andrade, L.; Williams, T.E.; McNaughton, S.A.; Leech, R.M.; Reedy, J.; Shams-White, M.M.; Vena, J.E.; et al. Advances in methods for characterizing dietary patterns: A scoping review. medRxiv 2024. [Google Scholar] [CrossRef]
- Shihavuddin, M.S.A.; Ravn-Haren, G. Sequential transfer learning based on hierarchical clustering for improved performance in deep learning-based food segmentation. Sci. Rep. 2021, 11, 813. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. In Advances in Neural Information Processing Systems, Proceedings of the 33rd Conference on Neural Information Processing Systems (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Neural Information Processing Systems Foundation, Inc.: La Jolla, CA, USA, 2019; pp. 8024–8035. [Google Scholar]
- Morgenstern, A.C.J.; Rosella, L.; Anderson, L. Development of machine learning prediction models to explore nutrients predictive of cardiovascular disease using Canadian linked population-based data. Appl. Physiol. Nutr. Metab. 2022, 47, 529–546. [Google Scholar] [CrossRef]
- Davies, T.; Louie, J.C.Y.; Scapin, T.; Pettigrew, S.; Wu, J.H.; Marklund, M.; Coyle, D.H. An innovative machine learning approach to predict the dietary fiber content of packaged foods. Nutrients 2021, 13, 3195. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, H.; Qi, Y.; Yang, J.; Civitarese, G. M-health of nutrition: Improving nutrition services with smartphone and machine learning. Mob. Inf. Syst. 2023, 2023, 3979020. [Google Scholar] [CrossRef]
- Suddul, G.; Seguin, J.F.L. A Comparative Study of Deep Learning Methods for Food Classification with Images. Food Humanit. 2023, 1, 800–808. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X.; Wang, J. Machine Learning Approaches for Dietary Pattern Analysis: Implications for Hypertension Prevention. J. Nutr. Sci. 2023, 12, 45–58. [Google Scholar] [CrossRef]
- Creux, C.; Zehraoui, F.; Hanczar, B.; Tahi, F. A3SOM, Abstained Explainable Semi-Supervised Neural Network Based on Self-Organizing Map. PLoS ONE 2023, 18, e0286137. [Google Scholar] [CrossRef]
- Revesai, Z.; Kogeda, O.P. Lightweight Interpretable Deep Learning Model for Nutrient Analysis in Mobile Health Applications. Digital 2025, 5, 23. [Google Scholar] [CrossRef]
- Ma, J.; Wan, Y.; Ma, Z. Memory-Based Learning and Fusion Attention for Few-Shot Food Image Generation Method. Appl. Sci. 2024, 14, 8347. [Google Scholar] [CrossRef]
- Kissas, G.; Yang, Y.; Hwuang, E.; Witschey, W.R.; Detre, J.A.; Perdikaris, P. Machine Learning in Cardiovascular Flows Modeling: Predicting Arterial Blood Pressure from Non-Invasive 4D Flow MRI Data Using Physics-Informed Neural Networks. Comput. Methods Appl. Mech. Eng. 2020, 358, 112623. [Google Scholar] [CrossRef]
- Qian, W.; Zhao, C.; Li, Y.; Ma, F.; Zhang, C.; Huai, M. Towards Modeling Uncertainties of Self-Explaining Neural Networks via Conformal Prediction. In Proceedings of the Thirty-Eighth AAAI Conference on Artificial Intelligence and Thirty-Sixth Conference on Innovative Applications of Artificial Intelligence and Fourteenth Symposium on Educational Advances in Artificial Intelligence, Vancouver, BC, Canada, 20–27 February 2024; pp. 14651–14659. [Google Scholar] [CrossRef]
- Ingole, V.S.; Kshirsagar, U.A.; Singh, V.; Yadav, M.V.; Krishna, B.; Kumar, R. A Hybrid Model for Soybean Yield Prediction Integrating Convolutional Neural Networks, Recurrent Neural Networks, and Graph Convolutional Networks. Computation 2025, 13, 4. [Google Scholar] [CrossRef]
- Kumar, A.; Shaikh, A.M.; Li, Y.; Bilal, H.; Yin, B. A comprehensive review of model compression techniques in machine learning. Appl. Intell. 2024, 54, 12085–12118. [Google Scholar]
- Mezgec, S.; Seljak, B.K. MobileNets for food recognition. In Proceedings of the 26th European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning (ESANN), Bruges, Belgium, 25–27 April 2018; pp. 353–358. [Google Scholar]
- Ullah, M.A.; Zia, T.; Kim, J.-E.; Kadry, S. An inherently interpretable deep learning model for local explanations using visual concepts. PLoS ONE 2024, 19, e031187. [Google Scholar] [CrossRef]
- Wang, J.; He, C.; Long, Z. Establishing a machine learning model for predicting nutritional risk through facial feature recognition. Front. Nutr. 2023, 10, 1219193. [Google Scholar] [CrossRef]
- Razavi, R.; Xue, G. Predicting Unreported Micronutrients from Food Labels: Machine Learning Approach. J. Med. Internet Res. 2022, 25, e45332. [Google Scholar] [CrossRef]
- Diaz, J.E.G.; Delgado, A.J.R.; Cervantes, J.L.S.; Hernández, G.A.; Mazahua, L.R.; Parada, A.R.; Nieto, Y.A.J. Early Detection of Age-Related Macular Degeneration Using Vision Transformer-Based Architectures—A Comparative Study with Offline Metrics and Data Augmenting. Int. J. Comb. Optim. Probl. Inform. 2024, 15, 72–84. [Google Scholar] [CrossRef]
- Becker, D. Food-101 Dataset; Kaggle: San Francisco, CA, USA, 2015. [Google Scholar]
- Singla, A.; Yuan, L.; Ebrahimi, T. Food/non-food image classification and food categorization using pre-trained GoogLeNet model. In Proceedings of the 2nd International Workshop on Multimedia Assisted Dietary Management, Amsterdam, The Netherlands, 16 October 2016. [Google Scholar]
- Revesai, Z.; Kogeda, O.P. A Comparative Analysis of Interpretable Deep Learning Models for Nutrient Analysis in Vulnerable Populations. In Computational Science and Its Applications—ICCSA 2025; Gervasi, O., Ed.; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2025; Volume 15649, pp. 218–233. [Google Scholar] [CrossRef]
- Marín, J.; Biswas, A.; Ofli, F.; Hynes, N.; Salvador, A.; Aytar, Y.; Weber, I.; Torralba, A. Recipe1M+: A dataset for learning cross-modal embeddings for cooking recipes and food images. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 43, 187–203. [Google Scholar] [CrossRef]
- Koeppe, A.; Bamer, F.; Selzer, M.; Nestler, B.; Markert, B. Workflow Concepts to Model Nonlinear Mechanics with Computational Intelligence. PAMM 2021, 21, 238. [Google Scholar] [CrossRef]
- Li, A.; Li, M.; Fei, R.; Mallik, S.; Hu, B.; Yu, Y. EfficientNet-resDDSC: A Hybrid Deep Learning Model Integrating Residual Blocks and Dilated Convolutions for Inferring Gene Causality in Single-Cell Data. Interdiscip. Sci. Comput. Life Sci. 2024, 17, 166–184. [Google Scholar] [CrossRef] [PubMed]
- Yeung, M.; Sala, E.; Schönlieb, C.; Rundo, L. Unified Focal Loss: Generalising Dice and Cross Entropy-Based Losses to Handle Class Imbalanced Medical Image Segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef]
- Hashemi, S.R.; Salehi, S.S.M.; Erdoğmuş, D.; Prabhu, S.P.; Warfield, S.K.; Gholipour, A. Asymmetric Loss Functions and Deep Densely-Connected Networks for Highly-Imbalanced Medical Image Segmentation: Application to Multiple Sclerosis Lesion Detection. IEEE Access 2019, 7, 1721–1735. [Google Scholar] [CrossRef]
- Eelbode, T.; Bertels, J.; Berman, M.; Vandermeulen, D.; Maes, F.; Bisschops, R.; Blaschko, M.B. Optimization for Medical Image Segmentation: Theory and Practice When Evaluating with Dice Score or Jaccard Index. IEEE Trans. Med. Imaging 2020, 39, 3679–3690. [Google Scholar] [CrossRef]
- Wang, K.; Dou, Y.; Sun, T.; Qiao, P.; Wen, D. An Automatic Learning Rate Decay Strategy for Stochastic Gradient Descent Optimization Methods in Neural Networks. Int. J. Intell. Syst. 2022, 37, 7334–7355. [Google Scholar] [CrossRef]
- Ghosh, T.; McCrory, M.A.; Marden, T.; Higgins, J.; Anderson, A.K.; Domfe, C.A.; Jia, W.; Lo, B.; Frost, G.; Steiner-Asiedu, M.; et al. I2N: Image to nutrients, a sensor guided semi-automated tool for annotation of images for nutrition analysis of eating episodes. Front. Nutr. 2023, 10, 1191962. [Google Scholar] [CrossRef]
- Liu, J.; Zhan, C.; Wang, H.; Zhang, X.; Liang, X.; Zheng, S.; Meng, Z.; Zhou, G. Developing a Hybrid Algorithm Based on an Equilibrium Optimizer and an Improved Backpropagation Neural Network for Fault Warning. Processes 2023, 11, 1813. [Google Scholar] [CrossRef]
- Kubuga, C.; Shin, D.; Song, W. Determinants of Dietary Patterns of Ghanaian Mother-Child Dyads: A Demographic and Health Survey. PLoS ONE 2023, 18, e0294309. [Google Scholar] [CrossRef]
- Nfor, K.A.; Armand, T.P.T.; Ismaylovna, K.P.; Joo, M.I.; Kim, H.C. An Explainable CNN and Vision Transformer-Based Approach for Real-Time Food Recognition. Nutrients 2025, 17, 362. [Google Scholar] [CrossRef]







| Model Configuration | Parameters (M) | Model Size (MB) | Memory (GB) | Checkpoint (MB) |
|---|---|---|---|---|
| Base Transform [57] | 124.3 | 498.2 | 4.2 | 542.8 |
| LSTM Variant [14] | 98.7 | 394.8 | 3.8 | 423.5 |
| Our Model | 45.6 | 182.4 | 3.8 | 198.6 |
| w/o Attention | 42.3 | 169.2 | 3.5 | 184.3 |
| w/o Self-Explain | 40.8 | 163.2 | 3.4 | 177.9 |
| Configuration | Inference (ms) | Training Time (h) | GPU Util (%) | CPU Usage (%) | Batch Size | Efficiency Gain (%) |
|---|---|---|---|---|---|---|
| Baseline Model | - | 48.6 | 72.3 | 42.1 | 32 | - |
| Single GPU | 29.3 | 24.3 | 84.5 | 45.2 | 32 | 49.9 |
| Multi-GPU (×4) | 8.7 | 7.2 | 78.3 | 62.4 | 128 | 85.2 |
| Distributed (×8) | 4.9 | 4.1 | 76.8 | 68.7 | 256 | 91.6 |
| Component Configuration | Accuracy (%) | Precision | Recall | Memory Impact (%) |
|---|---|---|---|---|
| Full Model | 94.1 | 0.93 | 0.94 | - |
| Without Attention | 85.8 | 0.86 | 0.85 | −12.4 |
| Without Self-Explanation | 86.9 | 0.87 | 0.86 | −8.7 |
| Without Temporal Module | 87.2 | 0.87 | 0.86 | −6.3 |
| Without Skip Connections | 88.4 | 0.88 | 0.88 | −4.2 |
| Fold | Accuracy (%) | Precision | Recall | F1-Score | Stability Index |
|---|---|---|---|---|---|
| 1 | 93.2 | 0.92 | 0.94 | 0.93 | 0.91 |
| 2 | 92.8 | 0.92 | 0.94 | 0.93 | 0.91 |
| 3 | 93.5 | 0.93 | 0.94 | 0.94 | 0.92 |
| 4 | 92.9 | 0.92 | 0.94 | 0.93 | 0.92 |
| 5 | 93.1 | 0.93 | 0.94 | 0.93 | 0.92 |
| Mean | 93.1 | 0.92 | 0.94 | 0.93 | 0.92 |
| Error Type | Occurrence (%) | Impact Level | Recovery Rate (%) |
|---|---|---|---|
| Pattern Misclassification | 42.0 | Medium | 78.3 |
| Temporal Misalignment | 31.0 | Low | 85.6 |
| Feature Integration | 27.0 | High | 72.1 |
| Interpretability Component | Metric | Our Model | ResNet-50 | Vision Transformer | GRAD-CAM |
|---|---|---|---|---|---|
| Feature Attribution Quality (FAQ) | |||||
| Attention–Expert Correlation | 0.89 | 0.72 | 0.76 | 0.68 | |
| Primary Component Identification (%) | 94.3 | 87.2 | 89.1 | 85.6 | |
| Ground-truth Region Overlap (%) | 87.6 | 71.4 | 74.8 | 69.3 | |
| Decision Pathway Transparency (DPT) | |||||
| Attention Entropy Score | 2.34 | 3.12 | 2.89 | 3.45 | |
| Single-food Consistency | 0.91 | 0.76 | 0.82 | 0.74 | |
| Multi-food Consistency | 0.84 | 0.68 | 0.71 | 0.65 | |
| Concept Coherence (CC) | |||||
| Cosine Similarity Score | 0.86 | 0.72 | 0.78 | 0.69 | |
| Nutritional Category Clustering (%) | 92.1 | 84.3 | 87.6 | 82.1 | |
| Pattern Recognition Rates | |||||
| Meal Composition Patterns (%) | 94.2 | 86.7 | 89.3 | 85.1 | |
| Portion Size Relationships (%) | 91.8 | 82.4 | 85.9 | 80.7 | |
| Temporal Eating Sequences (%) | 93.6 | 85.2 | 88.1 | 83.9 | |
| Expert–Model Agreement (%) | 89.4 | 74.6 | 78.2 | 72.8 |
| Analysis Type | Score/Rate | Impact Weight | Processing Time (ms) | Confidence |
|---|---|---|---|---|
| Feature Attribution: | ||||
| Primary Components | 0.89 | 0.86 | 12.3 | 0.89 |
| Temporal Patterns | 0.85 | 0.92 | 8.7 | 0.88 |
| Structural Features | 0.82 | 0.79 | 6.4 | 0.86 |
| Integration Mechanisms | 0.87 | 0.83 | 9.2 | 0.87 |
| Pattern Recognition: | ||||
| Sequential Patterns | 94.3 | 0.89 | 18.4 | 0.89 |
| Concurrent Patterns | 92.8 | 0.86 | 15.7 | 0.86 |
| Hierarchical Patterns | 93.5 | 0.88 | 21.3 | 0.88 |
| Composite Patterns | 93.9 | 0.87 | 23.8 | 0.87 |
| Method | Accuracy (%) | Latency (ms) | Memory (GB) | Throughput (req/s) |
|---|---|---|---|---|
| Rule-based [18] | 82.3 | 45.3 | 2.1 | 22.1 |
| LSTM-based [12] | 85.4 | 41.2 | 3.2 | 24.3 |
| Transformer [24] | 87.8 | 38.5 | 3.8 | 26.0 |
| Commercial API * [10] | 89.5 | 52.1 | 4.2 | 19.2 |
| Our Model | 94.1 | 29.3 | 3.8 | 34.1 |
| Metric | Our Model | Best Baseline | Improvement (%) |
|---|---|---|---|
| GPU Utilisation (%) | 84.5 | 72.3 | 16.9 |
| Processing Time (ms) | 29.3 | 38.5 | 23.9 |
| Memory Footprint (GB) | 3.8 | 4.2 | 9.5 |
| Scaling Factor (8×) | 7.6 | 5.8 | 31.0 |
| Meal Type | Recognition (%) | Composition Accuracy (%) | Processing Time (ms) |
|---|---|---|---|
| Single-item Meals | 94.3 | 92.8 | 18.4 |
| Two-item Plates | 92.1 | 89.5 | 24.6 |
| Full Course Meals | 88.7 | 85.3 | 32.8 |
| Buffet Settings | 86.4 | 82.9 | 38.5 |
| Food Group | Accuracy (%) | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Grains/Cereals | 93.5 | 0.92 | 0.94 | 0.93 |
| Proteins | 92.8 | 0.91 | 0.93 | 0.92 |
| Vegetables | 94.2 | 0.93 | 0.95 | 0.94 |
| Fruits | 95.1 | 0.94 | 0.96 | 0.95 |
| Dairy Products | 91.8 | 0.90 | 0.92 | 0.91 |
| Number of Items | Detection Rate (%) | Separation Accuracy (%) | Identification Time (ms) |
|---|---|---|---|
| 2–3 Items | 92.3 | 90.5 | 25.4 |
| 4–5 Items | 88.7 | 85.2 | 35.8 |
| 6+ Items | 84.2 | 80.7 | 48.3 |
| Relationship Type | Detection (%) | Confidence Score | Processing Time (ms) |
|---|---|---|---|
| Spatial Adjacent | 91.2 | 0.887 | 12.5 |
| Overlapping | 87.5 | 0.834 | 18.7 |
| Partially Hidden | 85.3 | 0.812 | 22.4 |
| Mixed Components | 83.8 | 0.795 | 25.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Revesai, Z.; Kogeda, O.P. Self-Explaining Neural Networks for Food Recognition and Dietary Analysis. BioMedInformatics 2025, 5, 36. https://doi.org/10.3390/biomedinformatics5030036
Revesai Z, Kogeda OP. Self-Explaining Neural Networks for Food Recognition and Dietary Analysis. BioMedInformatics. 2025; 5(3):36. https://doi.org/10.3390/biomedinformatics5030036
Chicago/Turabian StyleRevesai, Zvinodashe, and Okuthe P. Kogeda. 2025. "Self-Explaining Neural Networks for Food Recognition and Dietary Analysis" BioMedInformatics 5, no. 3: 36. https://doi.org/10.3390/biomedinformatics5030036
APA StyleRevesai, Z., & Kogeda, O. P. (2025). Self-Explaining Neural Networks for Food Recognition and Dietary Analysis. BioMedInformatics, 5(3), 36. https://doi.org/10.3390/biomedinformatics5030036







