Abstract
Background: Large Language Models (LLMs) have demonstrated strong performances in clinical question-answering (QA) benchmarks, yet their effectiveness in addressing real-world consumer medical queries remains underexplored. This study evaluates the capabilities and limitations of LLMs in answering consumer health questions using the MedRedQA dataset, which consists of medical questions and answers by verified experts from the AskDocs subreddit. Methods: Five LLMs-GPT-4o mini, Llama 3.1-70B, Mistral-123B, Mistral-7B, and Gemini-Flash were assessed using a cross-evaluation framework. Each model generated responses to consumer queries and their outputs were evaluated by every model by comparing them with expert responses. Human evaluation was used to assess the reliability of models as evaluators. Results: GPT-4o mini achieved the highest alignment with expert responses according to four out of the five models’ judges, while Mistral-7B scored the lowest according to three out of five models’ judges. Overall, model responses show low alignment with expert responses. Conclusions: Current small or medium sized LLMs struggle to provide accurate answers to consumer health questions and must be significantly improved.
1. Introduction
In healthcare consultations, clinicians raise questions related to patient care but only find answers to half of those questions due to limited time or the belief that an answer may not exist [1]. Medical QA systems have the potential to address these problems by giving fast responses to clinicians’ questions. These systems are designed to provide accurate and relevant answers to medical queries by leveraging natural language processing techniques. Traditional systems in this domain typically utilize information retrieval techniques to draw responses from structured medical databases or relevant documents. These systems often involve classifying question type, such as Yes/No or factual questions [2] and then employ semantic matching and extraction methods to generate concise responses from the matched documents. More recently, the rise in LLMs such as GPT-4 [3] has facilitated new opportunities in the healthcare domain such as medical record summarization [4] as well as medical QA [5]. However, most research focuses on clinical and multiple-choice benchmarks, leaving open-ended, consumer-based medical QA relatively underexplored.
This study addresses this gap by evaluating the effectiveness of five prominent LLMs in answering consumer-based medical questions from MedRedQA [6], a large-scale dataset of, real-world consumer queries and expert responses extracted from the AskDocs [7] subreddit. The LLMs assessed are GPT-4o mini [8], Llama 3.1 (70B) [9], Mistral-123B [10], Mistral-7B [11], and Gemini-Flash [12]. Employing a cross-evaluation approach, each model’s responses were not only evaluated by itself but also by the other models to minimize bias in the assessment process. To guide this study, two primary research questions are formulated:
RQ1: How effectively do current LLMs perform in answering consumer-based medical questions in MedRedQA?
RQ2: How reliable are different LLMs as evaluators of their own and other models’ responses to medical questions, and does evaluator reliability vary significantly across models?
RQ1 seeks to determine the extent to which LLMs can generate responses aligned with expert answers in the dataset. RQ2 seeks to determine whether LLMs can act as reliable evaluators and whether the choice of evaluator can affect the evaluation results. Given the computational cost and resource constraints associated with using larger models, this study focuses on smaller, more cost-effective models. This practical focus ensures that the findings of the study remain accessible and relevant to future researchers exploring cost-effective methods for medical QA research.
The primary contributions of this paper are summarized as follows:
- Evaluation of Five LLMs in answering Consumer Health Queries: We systematically compare GPT-4o mini, Llama 3.1 (70B), Mistral-123B, Mistral-7B, and Gemini-Flash on a real-world dataset of consumer health questions.
- Cross-Model Evaluation Method: We use a cross-evaluation framework, assessing each model’s ability to judge both its own outputs and those of other models, and compare these findings with human judgments.
- Dataset Limitations and Model Insights: We highlight limitations in MedRedQA and derive some useful insights regarding model performance from analyzing the results.
- Discussion of Practical and Ethical Implications: We examine real-world issues such as model safety mechanisms, misinformation risks, and the challenges of regulatory compliance in healthcare contexts.
The rest of the article is organized as follows. Section 2 (Related Work) provides an overview of existing research on LLMs in medical QA. Section 3 (Methods) describes the selection of models, the rationale for choosing the MedRedQA dataset, the prompt generation strategies, and the evaluation techniques utilized in the study. Section 4 (Results) presents the evaluation outcomes. Section 5 (Discussion) analyzes the performance of each model and discusses the implications of the cross-evaluation findings, including considerations about model sizes and evaluator reliability. Section 6 (Limitations) acknowledges the constraints faced during the study, such as dataset challenges and evaluation complexities. Finally, Section 7 (Conclusions and Future Work) summarizes the key insights and outlines potential directions for enhancing LLM performance in consumer health question answering.
2. Related Work
The integration of artificial intelligence (AI) and knowledge graphs, including ontologies [13], into educational and healthcare domains provides innovative avenues to enhance communication and knowledge access. AI-driven systems use knowledge graphs to organize and interlink vast arrays of information, enabling both educators [14,15] and healthcare providers to access tailored, contextually relevant data [16]. When integrated with chatbots, these systems can facilitate interactive and personalized learning experiences in education, offering students immediate, accurate responses to their inquiries. In healthcare, chatbots powered by AI and ontologies can assist in patient triage, symptom checking [17], and patient education, ensuring that users receive up-to-date medical information efficiently [18]. By leveraging these technologies, educational and healthcare chatbots can move beyond simple transactional interactions to deliver sophisticated, nuanced assistance that supports both learning and clinical decision-making processes [19].
The advent of LLMs such as GPT-4 has introduced transformative possibilities for many use cases in education [20,21], operational support [22], and healthcare. These models, for example, can address the significant documentation burden in EHRs by automating text summarization, allowing clinicians to review condensed, relevant summaries rather than lengthy clinical notes [4]. Several studies have also been conducted to explore the potential of AI and LLMs in medical support and QA [23,24,25].
Current LLMs have been extensively evaluated on clinical QA benchmarks, such as MedQA and PubMedQA [26], where questions are typically structured in a multiple-choice format to assess clinical accuracy and factual recall. However, multiple-choice QA does not fully capture the complexity of real-world medical inquiries, as it limits responses to predefined options and restricts the model’s ability to provide nuanced, explanatory answers. To address these limitations, these datasets have transformed into open-ended question formats, allowing models to handle more elaborate responses that better reflect the complexities of clinical scenarios [5,27]. Additionally, LLMs have been evaluated on qualities beyond factual accuracy, such as safety, bias, and language understanding [28], to better align with the complexities encountered in real-world medical interactions.
Open-ended clinical QA benchmarks, however, are focused on structured, professional queries rather than consumer-based questions. Consumer queries typically lack specific medical terminology [29], use informal language, and may pose open-ended inquiries with limited or ambiguous detail. This difference from clinical-style questions presents a unique challenge for LLMs, which must interpret and respond to questions in a way that accommodates the informal, varied nature of consumer inquiries. Previous work in the consumer medical QA domain includes the CHiQA system [30], which focuses on reliable information retrieval from consumer-friendly sources such as MedlinePlus [31] to address common health questions. By using trusted patient-oriented sources, this system bridges the gap between consumer queries and trustworthy medical content, though it still encounters challenges in matching informal consumer language with precise medical information.
Additionally, research on improving consumer medical QA demonstrates the difficulty consumers face when formulating specific questions that align with their informational needs [32]. Work by Nguyen addresses this issue by proposing improved biomedical representational learning and statistical keyword modeling. These improvements aid in retrieving medical answers even when consumer questions are vague or contain informal language [32]. Another study conducted experiments with a QA system to retrieve answers for real consumer medication queries [33]. By curating a dataset containing genuine consumer questions about medications with corresponding expert verified answers, researchers observed that the QA system struggled with retrieving the correct answers. They also highlighted the need for better contextual understanding in consumer medication QA.
Several existing studies have also assessed LLM responses to questions posted on the AskDocs subreddit. For example, in [34] the authors compared physician and ChatGPT 3.5 (ChatGPT, n.d.) responses to 195 randomly selected questions from the subreddit, finding that healthcare professionals preferred ChatGPT’s responses in 78.6% of 585 evaluations. Other studies have examined LLM responses in specialized fields, such as Otolaryngology [35], where ChatGPT’s responses to 15 domain-specific questions were rated with an accuracy of 3.76 out of 5, and Cancer [36], where physicians evaluated 200 cancer-related questions and rated LLM responses to be of higher quality.
While these studies are valuable in understanding LLM capabilities in answering consumer health queries, there remains a lack of evaluation of LLMs on a large-scale dataset of consumer health questions. The MedRedQA dataset addresses this gap by providing a large collection of consumer-based medical questions and expert answers extracted from the AskDocs subreddit. This dataset includes a wide range of layperson queries on medical topics, offering an opportunity to evaluate LLMs on a large number of real-world, consumer-oriented healthcare questions in non-clinical settings. Physicians’ answers in the dataset (extracted from AskDocs) are used as ground truths in this study. This is justified by the verification of the physicians’ credentials by the subreddit’s moderators. Table 1 summarizes key prior work and highlights how this study contributes to research in consumer-based medical QA by evaluating LLMs on MedRedQA.

Table 1.
Comparative Overview of Key Studies in LLM based Medical QA.
3. Methods
The focus of this study is the evaluation of LLM responses to consumer-based medical questions. The following sections describe the scope and methods used in this study.
3.1. Purpose and Scope
This study addresses the gap in understanding the effectiveness of LLMs in answering consumer-based medical questions. Existing research has predominantly focused on multiple-choice medical QA datasets or smaller, sample-based studies on questions extracted from AskDocs. This study evaluates LLMs on MedRedQA—a dataset consisting of a large number of real-world, consumer-oriented medical inquiries extracted from AskDocs.
3.2. Model Selection
The following five LLMs were selected for this study based on their demonstrated effectiveness in natural language processing tasks. These models provide a diverse selection from both open-source and proprietary sources, capturing a broad view of current LLM capabilities for consumer medical QA.
- GPT-4o mini: A compact variant of the GPT-4o model that prioritizes efficiency while maintaining strong reasoning capabilities.
- Llama 3.1: 70B: Part of the Llama model family, this 70-billion parameter model performs competitively with other compact models and demonstrates robust language understanding across a range of benchmarks.
- Gemini-Flash: This model is part of the Gemini 1.5 series and is developed as a more cost-effective and faster alternative to Gemini-1.5 Pro [12].
- Mistral 7B and Mistral 123B: Known for outperforming larger models on several benchmarks, the Mistral family offers powerful small-scale models that demonstrate competitive performance for their size.
3.3. Dataset Selection
This section describes some of the datasets commonly used in medical QA research and why the MedRedQA dataset is selected for this study over other datasets. Table 2 gives an overview of these datasets.

Table 2.
Overview of Datasets in Medical QA.
3.3.1. Existing Datasets in Medical QA
Several datasets have been used traditionally to evaluate the capabilities of LLMs in medical QA tasks. In multiple choice QA, the most prominent ones include MedQA, PubMedQA, and MMLU-Clinical Knowledge.
MedQA: This dataset is collected from US medical licensing exams (USMLE) study materials and includes questions designed to test structured medical knowledge. The dataset is commonly used to evaluate LLMs’ clinical knowledge, with models such as GPT-4 and Med-Gemini [23] achieving accuracies above 90% [23,25]. It uses a multiple-choice format, which is useful for testing medical knowledge but is less relevant for real-world, open-ended medical queries.
PubMedQA: It consists of clinical research questions extracted from the PubMed database, where answers can take the form of abstracts, yes/no responses, or specific medical conclusions. While the benchmark allows for both short-form and long-form responses, it remains structured around formal medical research rather than the informal queries typical of consumer healthcare.
MMLU-Clinical Knowledge: This benchmark tests a model’s knowledge across multiple domains, including medicine, using a similar multiple-choice format. Like MedQA, this dataset focuses on assessing clinical and factual knowledge.
To address the limitations of multiple-choice formats, some datasets have been transformed or created to require open-ended, detailed answers. These include MedQA-Open, MedQA-CS, and MedQuAD:
MedQA-Open: It is a modified version of MedQA, adapted to require models to generate open-ended answers. While it allows for detailed responses, the questions remain based on medical licensing exams, limiting their relevance to consumer-based, informal queries.
MedQA-CS: This benchmark focuses on clinical skills, modeled after the medical education’s Objective Structured Clinical Examinations (OSCEs). This dataset evaluates LLMs through two tasks: LLM-as-medical-student and LLM-as-clinical-examiner, both reflecting formal clinical scenarios. It provides an assessment of LLMs in settings that are closer to real-world clinical scenarios. However, the benchmark is less relevant for consumer-based medical QA tasks due to its focus on professional clinical settings.
MedQuAD: It contains question-answer pairs extracted from the National Institutes of Health (NIH) website. The dataset includes detailed, structured answers based on expert medical content. However, like other datasets, it does not reflect the informal nature of consumer healthcare questions.
These datasets are crucial for evaluating how well LLMs can handle open-ended questions in clinical scenarios. However, their reliance on formal medical cases or clinical exam formats makes them less suitable for assessing how models respond to consumer-facing queries, which often lack medical precision or structure.
3.3.2. MedRedQA
This dataset includes 51,000 pairs of consumer medical questions and expert answers extracted from the AskDocs subreddit. AskDocs allows consumers to post health-related questions, and only verified medical professionals provide answers. The dataset has two parts, one consisting of samples where expert responses include citations to PubMed articles and the other without any citations. The second part of the dataset is used in this study. The test set of this dataset contains 5099 samples. Each sample contains a title, the body, the response by the medical expert, the response score, and the occupation of the expert. The dataset includes responses that received the highest upvotes (response score), reflecting a consensus on the relevance and quality of the answers.
MedRedQA is used for this study because it provides a large set of real consumer healthcare queries which LLMs can be evaluated on. The informal nature of these questions presents a unique challenge for LLMs and provides a benchmark to evaluate the ability of models to provide accurate answers to medical queries in non-clinical settings.
3.4. Prompt Generation
Two distinct prompts were used in this study, one for the response to user questions, and one for the evaluation of LLM responses for agreement with physician responses. The two prompts are shown in Table 3.

Table 3.
Prompts Used for generating answers (RQ1) and evaluating answers (RQ2).
For both cases, different prompts are tested to make sure that the model responds as required, with answers that are precise and that do not have additional commentary. For the RQ1 prompt, after obtaining the acknowledgment from the model, the title and body of the question are provided to the model to generate the answer. The title is included because it can often contain information that is important to answer the question. For the RQ2 prompt, after obtaining the acknowledgment from the model, the title, body, and the expert and model answers are provided. The acknowledgment message from the models is obtained only once and the same one is used when evaluating each of the samples.
The Mistral-7B model is provided with the same acknowledgment message as the Mistral-123B model, instead of its own, because the model struggles with understanding the instructions properly. Specifically, the model started to create its own example and respond with “Agree” or “Disagree”. The acknowledgment from the Mistral-123B model is used to make sure that the model responds correctly when provided with the two responses to compare. Despite explicit instructions in the prompt that the output should be “Agree” or “Disagree”, responses from the Mistral models are of the form “Agree” or “Disagree” and responses from the Gemini-Flash model are of the form “Agree\n” or “Disagree\n”. These responses are treated as “Agree” or “Disagree”, respectively. Responses other than these are classified as “other”.
3.5. Evaluation of Model Responses
Responses generated by LLMs are evaluated through a cross-model approach, where each model evaluates its own output and is cross-evaluated by all other models to reduce bias. In the evaluation process models are instructed to classify responses as either “Agree” or “Disagree” based on their similarity to expert-provided answers. This approach was chosen over traditional metrics such as ROUGE [40] and BERT-SCORE [41] because these metrics primarily measure surface-level lexical similarities, which may not reflect deeper semantic alignment. Even when the wording between two answers differs, the core information can still be highly aligned, which LLM-based evaluation may be able to better capture.
LLMs have been successfully used as evaluators in multiple research contexts [42], including pairwise comparisons, where they assess responses based on factors such as helpfulness, fluency, and factual accuracy [43]. Other studies have demonstrated the effectiveness of LLMs as evaluators in comparing expert responses with LLM generated responses in the medical domain [5]. These prior successes make LLMs suitable evaluators for this study.
LLM generated answers are categorized as either “Agree” or “Disagree”. While some evaluation methodologies include a “Neutral” category to account for responses that are neither fully correct nor incorrect, the category is not included in this evaluation because preliminary experiments showed that most of the answers were classified as “Neutral”. This could be because model responses do not perfectly match expert answers, which makes the models classify most samples as Neutrals. Using a binary evaluation instead forces the models to offer clearer distinctions, improving the utility of the results. Furthermore, this evaluation assumes the accuracy of expert answers as only verified individuals can respond to questions.
3.6. Assessment of Model Reliability as Evaluators
To judge the effectiveness of each model as an evaluator, a sample of 50 expert-model response pairs was selected for manual review, yielding 250 total evaluation pairs across the five models. For each pair of samples, the human evaluator compared the expert response with the model response and classified it as “Agree” if they both contained the same information and “Disagree” if they contained different information. Specifically, an LLM response is labeled as “Agree” if it captures the core recommendation or factual claim of the expert’s answer without contradicting or omitting critical information. Minor stylistic differences or additional non-contradictory disclaimers (e.g., “I am not a doctor”) do not affect the label. However, if an LLM response leaves out an important piece of the expert’s direct advice (such as telling a user to seek immediate medical help) or states the opposite, it is labeled as “Disagree”. Questions and their corresponding expert answers were reviewed manually and only those samples were chosen where it would be straightforward to determine whether a model-generated response matches the expert response. Specifically, the selection criteria included:
- Short and direct expert responses: For example, if a consumer asked, “Should I be concerned about this symptom?” and the expert responded with a direct confirmation or denial with a “Yes” or “No” in the response, or a strongly implied affirmative or negative stance. These samples were included as it would be easy to verify whether the model response matches the expert response or not.
- Responses requesting additional information: Some expert responses requested more details rather than answering the question. These samples were included as they provided a clear decision rule: model responses that recognized the need for further details were labeled as “Agree” and those that did not were labeled as “Disagree”.
- No long responses: Longer expert responses–i.e., those that did not have a definitive “Yes” or “No” response–were not included as determining whether a model response matches the expert response might have required domain knowledge.
Each of the five models’ evaluation output (“Agree” or “Disagree”) for the 250 pairs was compared with the human evaluation output. The percentage of responses where the model evaluation output was the same as the human evaluation output is reported. To minimize bias, the model evaluator’s initial classifications were not visible during manual evaluation.
4. Results
Evaluation results are shown in Figure 1. GPT-4o mini responses achieved the highest percentage agreement with expert answers as evaluated by four out of the five model judges. Results also show that Mistral-7B tends to give higher agreement scores as an evaluator across the board, possibly indicating a bias toward lenient evaluations. On the other hand, the Gemini-Flash and Mistral-123B models tend to provide lower agreement scores, suggesting that these models are more critical evaluators. It should also be noted that the agreement and disagreement scores do not sum up to 100 except when the evaluator is GPT-4o mini. This is because there is a small percentage of responses for which the models either do not provide answers or provide answers other than “Agree” or “Disagree”. Since the number of such responses is very small, it does not affect the results by a significant amount.
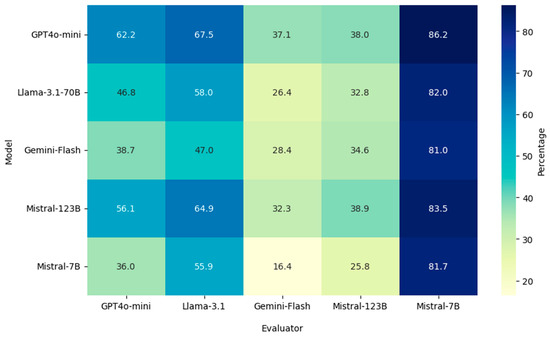
Figure 1.
Cross Model Evaluation Results of LLM Responses.
Table 4 shows the results of the manual evaluation. As shown in the table, evaluation by the Gemini-Flash model achieved the highest alignment with human evaluations, scoring 77.2%, while Mistral-7B showed the lowest alignment at 51.6%. Table 5 shows agreement scores for each model’s responses using Gemini-Flash as the evaluator, with GPT-4o mini achieving the highest score (37.1%).

Table 4.
Percentage Alignment between Model and Human evaluations for subset of samples.

Table 5.
Percentage Agreement between Model and Physician responses with Gemini-Flash as Evaluator.
5. Discussion
This section discusses key findings from the results, including variations in model evaluation behavior, the impact of safety mechanisms, and the broader implications for using LLMs in consumer health QA.
Safety Mechanisms in Gemini-Flash: Experiments revealed that the Gemini-Flash model sometimes refused to answer questions entirely due to its built-in safety mechanisms, instead returning a “safety error”. Moreover, the model often responded by saying that “it was an AI model and could not offer medical advice” and in one example which involved surgery, the model refused to provide an answer saying instead that it was a “dangerous procedure” and recommended seeking professional medical help. This resulted in a lot of disagreement outputs, and contributed to the low agreement score for the model as compared to other models.
The safety mechanisms in Gemini-Flash highlight two important real-world considerations: medical questions can include sensitive topics and avoiding them entirely may limit a model’s usefulness to users seeking information. At the same time, providing responses that resemble professional medical advice could lead to risks, such as encouraging self-diagnosis or replacing expert consultation. This underscores the challenges of using LLMs in the real-world for consumer-facing medical queries, where the goal of providing accurate medical information must be weighed against the potential risks of misdiagnosis or overreliance on AI-driven advice.
Discrepancies Across Evaluation Scores: The results show variations in model behavior as evaluators, where Mistral-7B shows a tendency toward lenient assessments resulting in scores greater than 80% for all models. On the other hand, the Gemini-Flash and Mistral-123B models are more critical, providing accuracy scores lower than 40% across all models. Evaluation results for the models as judges indicate that the Mistral-7B model is the worst performing evaluator, which explains its high agreement scores across all evaluations. Gemini-Flash performs the best as an evaluator (Table 4) but has the second lowest average agreement score (37.2%). This difference suggests that, although Gemini-Flash’s safety mechanisms lead to lower agreement scores due to frequent refusals to answer, these same mechanisms do not affect its ability to evaluate the responses of other models.
Model Sizes: Mistral-7B achieves the lowest agreement score and the lowest accuracy as an evaluator. This might suggest that closed models (GPT4o-mini, Gemini-Flash) are closer in size to Llama 3.1-70B and Mistral-123B than Mistral-7B, as similarly sized models would be expected to exhibit comparable performance levels. The significantly lower performance of Mistral-7B may be attributed to its smaller size, which could limit its ability to answer consumer based medical questions accurately. However, whether specialized fine-tune models smaller in size exhibit the same patterns is planned for future work.
Low Average Agreement Scores: Low average agreement scores for each model across all five model evaluators, as shown in Table 6, emphasize the difficulties current LLMs face in providing accurate answers to consumer-based medical questions. Excluding the Mistral-7B model due to its relatively poor evaluator performance, we observed that even the highest-performing model, GPT4o-mini, achieved only 51.2% accuracy. This finding highlights the challenges mid or small sized LLMs encounter when attempting to address medical inquiries posed by consumers. Future work should consider fine-tuning LLMs specifically on consumer-based medical QA datasets. Specialized models fine-tuned on medical data have been shown to improve performance on medical datasets, for example, Med-Gemini currently achieves the highest accuracy (91.1%) on MedQA [23]. Fine-tuning may also improve the accuracy of model responses to consumer medical queries.

Table 6.
Average Agreement Scores Across All Model Evaluators.
Another promising approach is retrieval-augmented generation (RAG). RAG combines LLMs with information retrieval techniques to pull relevant data from external medical sources, such as MedlinePlus, before generating an answer. This approach allows models to use information outside their learned parameters to generate responses to questions. RAG has shown promising results in improving the accuracy of LLMs for medical QA [44]. Consumer-facing QA systems may also benefit by augmenting LLMs with external medical knowledge before responding to queries. The results of this study also show that LLMs should be evaluated on broader datasets to better understand the limitations and capabilities of models in this domain in the real-world.
6. Limitations
This section outlines the limitations of this study, specifically focusing on challenges related to the MedRedQA dataset and the evaluation process used.
Incomplete Questions: One limitation of this study lies in the nature of the MedRedQA dataset, which includes instances where expert responses prompt the user for additional information or clarity. These situations often involve requests for supplementary details or, at times, visual inputs, such as clear images, which are not included in the dataset due to privacy restrictions. As a result, model responses are sometimes misaligned with expert answers, especially when interpreting cases where an image would provide critical context.
This limitation also extends to cases where expert answers hinge on situational or time-specific knowledge. For example, questions related to health protocols during the COVID-19 pandemic may lack explicit mention of the pandemic context, yet experts assume this context in their responses. Models, however, may not interpret these questions correctly without this contextual indicator, potentially leading to inaccurate or irrelevant responses. An example is questions such as whether it is safe to bring elderly individuals to hospitals during the COVID period; without a clear indication in the question, models may miss the situational implications present in the expert responses.
Future dataset development should aim to incorporate richer metadata for each question. This could include:
- Structured Annotations: Labeling questions that require external information (e.g., images) to flag cases where responding to the question may not be possible due to missing contextual data.
- Follow-up Exchanges: Capturing multi-turn interactions where users provide additional details requested by experts, making the dataset more representative of real-world consultations.
- Multiple Expert Responses: Multiple physician responses per question could enhance the evaluation of LLM responses by providing a greater number of responses to evaluate the LLM responses against.
Using LLMs as evaluators: Using LLMs as evaluators can address limitations with ROUGE and BERT-SCORE but introduces challenges of its own. An LLM may be biased towards evaluating its own response as correct, a limitation that this study tries to address by using a cross-evaluation method where each LLM evaluates both its own responses and responses of other LLMs. LLM-based evaluation is also quicker and cost-effective for larger datasets compared to evaluation by medical experts, which is time-consuming and expensive at scale. However, while LLMs can semantically compare responses, they are not licensed medical professionals, and their assessments may not be as reliable as those of experts.
When used in cross-evaluation, models which are trained on similar data and share similar knowledge gaps or biases may propagate these weaknesses, affecting the reliability of results.
Improvements to the evaluation method used could include using contextualized prompts which contain medical guidelines or short reference passages so that judgments hinge on established medical consensus rather than the models’ internal knowledge alone. Another improvement could be to fine-tune LLMs specifically for evaluation tasks rather than using each model in a zero-shot manner without any fine-tuning.
Sample Size and Number of Human Evaluators: Another limitation is that we relied on a single human evaluator to assess a small, hand-picked set of relatively straightforward questions, which may not fully reflect the model’s true capabilities in more complex or ambiguous cases. Because only one human was involved, we could not measure inter-rater reliability using standard metrics (e.g., Cohen’s kappa) or verify the consistency of these judgments. Future work should include a larger and more diverse selection of questions-covering a broader range of medical topics and complexities-as well as multiple evaluators, ideally with varied backgrounds or medical expertise. Such an approach would not only provide a more robust measurement of each model’s evaluator performance but also increase overall confidence in the reliability and generalizability of the findings.
Small or Mid-sized LLMs: This study uses small or mid-sized LLMs for cost efficiency purposes. This limits the generalizability of our findings to state-of-the-art, large-scale LLMs.
Credibility of Physician Responses: The study assumes that physician responses are credible based on the verification performed by the subreddit’s moderators. However, this verification might not necessarily imply that the answers by the physicians are always correct.
7. Conclusions and Future Work
LLMs have enormous potential in transforming the medical QA domain. In the clinical QA domain, they could help physicians quickly receive answers to their queries and consequently improve the quality of care that they offer to their patients. In the consumer QA domain, they could help consumers receive the preliminary guidance that they often seek before deciding to go for a hospital visit. However, current LLMs are not yet capable of being deployed for real-world consumer medical questions, answering applications in a zero-shot manner.
As the results of this study show, LLMs struggle in providing accurate answers to consumer health questions. There is a need to extensively evaluate LLMs on a broader range of consumer QA benchmarks that reflect real-world healthcare scenarios to accurately assess their reliability in providing answers to consumer questions. The findings of this study highlight both the potential and the current limitations of using LLMs for consumer health QA. While models like GPT-4o mini show promise, the overall low agreement scores indicate that significant improvements are needed before LLMs can be reliably deployed in real-world healthcare settings.
One immediate avenue for improvement is the fine-tuning of LLMs on datasets specifically curated for consumer health questions. As shown in other domains, specialized fine-tuning can substantially enhance a model’s performance by allowing it to better understand the nuances and linguistic patterns typical of the target domain. Developing models that are trained on large-scale, diverse datasets like MedRedQA could help bridge the gap between current capabilities and the requirements for effective consumer health assistance. Furthermore, expanding LLMs to handle multimodal inputs, such as images or voice recordings, presents an opportunity to better mimic the versatility of human practitioners. Enabling models to process and interpret medical images, for instance, could enhance their ability to provide comprehensive answers when textual information alone is insufficient.
Integrating LLMs with medical knowledge bases and evidence-based guidelines presents another opportunity to improve accuracy. RAG techniques, which combine LLMs with information retrieval systems, allow models to access up-to-date and authoritative medical information during response generation. By fetching relevant data from trusted sources like MedlinePlus or PubMed, models can provide more accurate and contextually appropriate answers, reducing the risk of misinformation.
Addressing the challenge of incomplete or ambiguous questions is crucial. Future research could explore methods for LLMs to handle incomplete information more effectively, such as by generating clarifying questions or recognizing when additional data are needed. Developing models capable of understanding implicit context—like situational factors during a pandemic—would also enhance their applicability in real-world scenarios. The study underscores the need for more comprehensive evaluation methods to assess LLM performance accurately. Future work should involve larger and more diverse sample sizes for manual evaluation, such as including questions other than those that have simple, definitive answers, as well as more human evaluators. Additionally, establishing standardized benchmarks and metrics for consumer health QA can provide a clearer picture of model capabilities and areas needing improvement.
As AI technologies advance within the healthcare sector, adhering to regulatory requirements becomes essential. Future work should consider compliance with health information regulations like HIPAA in the United States or GDPR in Europe when handling sensitive medical data. Using LLMs for consumer healthcare applications also raises ethical concerns regarding misinformation risks. LLMs exhibit a tendency for hallucinations, producing misleading or incorrect information. Even small inaccuracies can have serious consequences for the consumer, influencing their health decisions and potentially delaying necessary medical care. Ensuring regulatory compliance regarding handling consumer data and providing consumers with AI generated medical information will be critical for real-world deployment. Improving the interpretability of LLMs can enhance trust and facilitate their adoption in healthcare. Future research could focus on developing tools and techniques that allow users and practitioners to understand how models arrive at their conclusions. Interpretability can aid in identifying errors, biases, and areas where the model may lack sufficient knowledge.
Author Contributions
M.A.: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Writing—Original Draft, and Visualization. Y.S.: Conceptualization, Methodology, Writing—Review and Editing, Investigation, Validation. I.D.: Writing—Review and Editing, Project administration, Supervision, Funding acquisition, and Resources. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Patient consent was waived as the study does not involve human subjects and uses a publicly available dataset.
Data Availability Statement
The MedRedQA dataset is available at the following link: https://data.csiro.au/collection/csiro:62454 (accessed on 23 February 2025).
Acknowledgments
Gabriel Vald provided valuable assistance towards the development of this study.
Conflicts of Interest
The authors declare no conflict of interest.
Glossary
These are definitions of some specialized terms used in the article.
| Natural Language Processing | A field of AI that enables computers to understand and process human language. |
| Large Language Models (LLMs) | AI models trained on vast amounts of text data to generate human-like language and answer complex queries |
| Retrieval Augmented Generation | A method combining LLMs with external information retrieval to improve response accuracy. |
| Fine-Tuning | The process of training a pre-existing AI model on a specialized dataset to improve its performance in a specific domain |
| Electronic Health Records (EHRs) | Digital Medical Records containing patient history and treatment details. |
| AskDocs subreddit | A reddit forum where verified medical experts answer user questions. |
References
- Del Fiol, G.; Workman, T.E.; Gorman, P.N. Clinical questions raised by clinicians at the point of care: A systematic review. JAMA Intern. Med. 2014, 174, 710–718. [Google Scholar] [CrossRef]
- Sarrouti, M.; Lachkar, A.; Ouatik, S.E.A. Biomedical question types classification using syntactic and rule based approach. In Proceedings of the 2015 7th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (IC3K), Lisbon, Portugal, 12–14 November 2015; IEEE: New York, NY, USA, 2015; Volume 1, pp. 265–272. [Google Scholar]
- Achiam, J.; Adler, S.; Agarwal, S.; Ahmad, L.; Akkaya, I.; Aleman, F.L.; Almeida, D.; Altenschmidt, J.; Altman, S.; Anadkat, S.; et al. Gpt-4 technical report. arXiv 2023, arXiv:2303.08774. [Google Scholar]
- Li, L.; Zhou, J.; Gao, Z.; Hua, W.; Fan, L.; Yu, H.; Hagen, L.; Zhang, Y.; Assimes, T.L.; Hemphill, L.; et al. A scoping review of using large language models (llms) to investigate electronic health records (ehrs). arXiv 2024, arXiv:2405.03066. [Google Scholar]
- Yao, Z.; Zhang, Z.; Tang, C.; Bian, X.; Zhao, Y.; Yang, Z.; Wang, J.; Zhou, H.; Jang, W.S.; Ouyang, F.; et al. Medqa-cs: Benchmarking large language models clinical skills using an ai-sce framework. arXiv 2024, arXiv:2410.01553. [Google Scholar]
- Nguyen, V.; Karimi, S.; Rybinski, M.; Xing, Z. MedRedQA for Medical Consumer Question Answering: Dataset, Tasks, and Neural Baselines. In Proceedings of the 13th International Joint Conference on Natural Language Processing and the 3rd Conference of the Asia-Pacific Chapter of the Association for Computational Linguistics, Bali, Indonesia, 1–4 November 2023; Volume 1: Long Papers, pp. 629–648. [Google Scholar]
- Reddit. AskDocs. 2024. Available online: https://www.reddit.com/r/AskDocs/ (accessed on 27 October 2024).
- OpenAI. GPT-4o Mini: Advancing Cost-Efficient Intelligence. 2024. Available online: https://openai.com/index/gpt-4o-mini-advancing-cost-efficient-intelligence/ (accessed on 27 October 2024).
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Vaughan, A.; Yang, A.; et al. The llama 3 herd of models. arXiv 2024, arXiv:2407.21783. [Google Scholar]
- Mistral, A.I. News on Mistral Large. 2024. Available online: https://mistral.ai/news/mistral-large-2407/ (accessed on 27 October 2024).
- Jiang, A.Q.; Sablayrolles, A.; Mensch, A.; Bamford, C.; Chaplot, D.S.; Casas, D.D.L.; Bressand, F.; Lengyel, G.; Lample, G.; Saulnier, L.; et al. Mistral 7B. arXiv 2023, arXiv:2310.06825. [Google Scholar]
- Team, G.; Georgiev, P.; Lei, V.I.; Burnell, R.; Bai, L.; Gulati, A.; Tanzer, G.; Vincent, D.; Pan, Z.; Wang, S.; et al. Gemini 1.5: Unlocking multimodal understanding across millions of tokens of context. arXiv 2024, arXiv:2403.05530. [Google Scholar]
- Baydaroğlu, Ö.; Yeşilköy, S.; Sermet, Y.; Demir, I. A comprehensive review of ontologies in the hydrology towards guiding next generation artificial intelligence applications. J. Environ. Inform. 2023, 42, 90–107. [Google Scholar] [CrossRef]
- Sajja, R.; Sermet, Y.; Cikmaz, M.; Cwiertny, D.; Demir, I. Artificial intelligence-enabled intelligent assistant for personalized and adaptive learning in higher education. Information 2024, 15, 596. [Google Scholar] [CrossRef]
- Sajja, R.; Sermet, Y.; Demir, I. End-to-End Deployment of the Educational AI Hub for Personalized Learning and Engagement: A Case Study on Environmental Science Education. EarthArxiv 2024, 7566. [Google Scholar] [CrossRef]
- Chi, N.C.; Nakad, L.; Fu, Y.K.; Demir, I.; Gilbertson-White, S.; Herr, K.; Demiris, G.; Burnside, L. Tailored Strategies and Shared Decision-Making for Caregivers Managing Patients’ Pain: A Web App. Innov. Aging 2020, 4 (Suppl. S1), 153. [Google Scholar] [CrossRef]
- Chi, N.C.; Shanahan, A.; Nguyen, K.; Demir, I.; Fu, Y.K.; Herr, K. Usability Testing of the Pace App to Support Family Caregivers in Managing Pain for People with Dementia. Innov. Aging 2023, 7 (Suppl. S1), 857. [Google Scholar] [CrossRef]
- Sermet, Y.; Demir, I. A semantic web framework for automated smart assistants: A case study for public health. Big Data Cogn. Comput. 2021, 5, 57. [Google Scholar] [CrossRef]
- Pursnani, V.; Sermet, Y.; Kurt, M.; Demir, I. Performance of ChatGPT on the US fundamentals of engineering exam: Comprehensive assessment of proficiency and potential implications for professional environmental engineering practice. Comput. Educ. Artif. Intell. 2023, 5, 100183. [Google Scholar] [CrossRef]
- Sajja, R.; Sermet, Y.; Cwiertny, D.; Demir, I. Platform-independent and curriculum-oriented intelligent assistant for higher education. Int. J. Educ. Technol. High. Educ. 2023, 20, 42. [Google Scholar] [CrossRef]
- Sajja, R.; Sermet, Y.; Cwiertny, D.; Demir, I. Integrating AI and Learning Analytics for Data-Driven Pedagogical Decisions and Personalized Interventions in Education. arXiv 2023, arXiv:2312.09548. [Google Scholar]
- Samuel, D.J.; Sermet, M.Y.; Mount, J.; Vald, G.; Cwiertny, D.; Demir, I. Application of Large Language Models in Developing Conversational Agents for Water Quality Education, Communication and Operations. EarthArxiv 2024, 7056. [Google Scholar] [CrossRef]
- Saab, K.; Tu, T.; Weng, W.H.; Tanno, R.; Stutz, D.; Wulczyn, E.; Zhang, F.; Strother, T.; Park, C.; Vedadi, E.; et al. Capabilities of gemini models in medicine. arXiv 2024, arXiv:2404.18416. [Google Scholar]
- Zhang, M.; Zhu, L.; Lin, S.Y.; Herr, K.; Chi, C.L.; Demir, I.; Dunn Lopez, K.; Chi, N.C. Using artificial intelligence to improve pain assessment and pain management: A scoping review. J. Am. Med. Inform. Assoc. 2023, 30, 570–587. [Google Scholar] [CrossRef]
- Nori, H.; Lee, Y.T.; Zhang, S.; Carignan, D.; Edgar, R.; Fusi, N.; King, N.; Larson, J.; Li, Y.; Liu, W.; et al. Can generalist foundation models outcompete special-purpose tuning? case study in medicine. arXiv 2023, arXiv:2311.16452. [Google Scholar]
- Jin, Q.; Dhingra, B.; Liu, Z.; Cohen, W.W.; Lu, X. Pubmedqa: A dataset for biomedical research question answering. arXiv 2019, arXiv:1909.06146. [Google Scholar]
- Nachane, S.S.; Gramopadhye, O.; Chanda, P.; Ramakrishnan, G.; Jadhav, K.S.; Nandwani, Y.; Dinesh, R.; Joshi, S. Few shot chain-of-thought driven reasoning to prompt LLMs for open ended medical question answering. arXiv 2024, arXiv:2403.04890. [Google Scholar]
- Kanithi, P.K.; Christophe, C.; Pimentel, M.A.; Raha, T.; Saadi, N.; Javed, H.; Maslenkova, S.; Hayat, N.; Rajan, R.; Khan, S. Medic: Towards a comprehensive framework for evaluating llms in clinical applications. arXiv 2024, arXiv:2409.07314. [Google Scholar]
- Welivita, A.; Pu, P. A survey of consumer health question answering systems. Ai Mag. 2023, 44, 482–507. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Mrabet, Y.; Ben Abacha, A. Consumer health information and question answering: Helping consumers find answers to their health-related information. J. Am. Med. Inform. Assoc. 2020, 27, 194–201. [Google Scholar] [CrossRef]
- MedlinePlus. U.S. National Library of Medicine, National Institutes of Health. Available online: https://medlineplus.gov/ (accessed on 27 October 2024).
- Nguyen, V. Consumer Medical Question Answering: Challenges and Approaches. Doctoral Dissertation, The Australian National University, Canberra, Australia, 2024. ANU Open Research Repository. [Google Scholar]
- Abacha, A.B.; Mrabet, Y.; Sharp, M.; Goodwin, T.R.; Shooshan, S.E.; Demner-Fushman, D. Bridging the gap between consumers’ medication questions and trusted answers. In MEDINFO 2019: Health and Wellbeing e-Networks for All; IOS Press: Amsterdam, The Netherlands, 2019; pp. 25–29. [Google Scholar]
- Ayers, J.W.; Poliak, A.; Dredze, M.; Leas, E.C.; Zhu, Z.; Kelley, J.B.; Faix, D.J.; Goodman, A.M.; Longhurst, C.A.; Hogarth, M.; et al. Comparing physician and artificial intelligence chatbot responses to patient questions posted to a public social media forum. JAMA Intern. Med. 2023, 183, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Carnino, J.M.; Pellegrini, W.R.; Willis, M.; Cohen, M.B.; Paz-Lansberg, M.; Davis, E.M.; Grillone, G.A.; Levi, J.R. Assessing ChatGPT’s Responses to Otolaryngology Patient Questions. Ann. Otol. Rhinol. Laryngol. 2024, 133, 658–664. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Parsa, R.; Hope, A.; Hannon, B.; Mak, E.; Eng, L.; Liu, F.-F.; Fallah-Rad, N.; Heesters, A.M.; Raman, S. Physician and Artificial Intelligence Chatbot Responses to Cancer Questions from Social Media. JAMA Oncol. 2024, 10, 956–960. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Pan, E.; Oufattole, N.; Weng, W.H.; Fang, H.; Szolovits, P. What disease does this patient have? A large-scale open domain question answering dataset from medical exams. Appl. Sci. 2021, 11, 6421. [Google Scholar] [CrossRef]
- Hendrycks, D.; Burns, C.; Basart, S.; Zou, A.; Mazeika, M.; Song, D.; Steinhardt, J. Measuring massive multitask language understanding. arXiv 2020, arXiv:2009.03300. [Google Scholar]
- Ben Abacha, A.; Demner-Fushman, D. A question-entailment approach to question answering. BMC Bioinform. 2019, 20, 511. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y. Rouge: A package for automatic evaluation of summaries. In Text Summarization Branches Out; Association for Computational Linguistics: Barcelona, Spain, 2004; pp. 74–81. [Google Scholar]
- Zhang, T.; Kishore, V.; Wu, F.; Weinberger, K.Q.; Artzi, Y. Bertscore: Evaluating text generation with bert. arXiv 2019, arXiv:1904.09675. [Google Scholar]
- Zhang, X.; Li, Y.; Wang, J.; Sun, B.; Ma, W.; Sun, P.; Zhang, M. Large language models as evaluators for recommendation explanations. In Proceedings of the 18th ACM Conference on Recommender Systems, Bari, Italy, 14–18 October 2024; pp. 33–42. [Google Scholar]
- Chern, S.; Chern, E.; Neubig, G.; Liu, P. Can large language models be trusted for evaluation? Scalable meta-evaluation of llms as evaluators via agent debate. arXiv 2024, arXiv:2401.16788. [Google Scholar]
- Xiong, G.; Jin, Q.; Lu, Z.; Zhang, A. Benchmarking retrieval-augmented generation for medicine. arXiv 2024, arXiv:2402.13178. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).