1. Introduction
Traditionally, pharmacokinetic (PK) parameters in human therapeutic drug monitoring (TDM) have been estimated using in vitro and in vivo methods. Pharmacokinetic data are frequently utilized in pharmacokinetic/pharmacodynamic (PKPD) studies to establish the relationship between drug exposure and response, such as the area under the concentration–time curve (AUC). However, when sparse data methods are employed, population PK/PD models (popPKPD) are suitable and commonly employed for understanding the exposure–response relationship [
1,
2].
Machine learning methods have emerged as powerful tools in pharmacokinetics methodology, marking a new trend. They enable the management of intricate relationships within large datasets and the analysis of high-dimensional data in clinical practice. The recent integration of artificial intelligence (AI) has further propelled the utilization of ML for drug-dose predictions. ML demonstrates remarkable computational efficiency and holds substantial potential in the realm of drug development [
3].
Although ML is less commonly utilized for drug PK predictions compared to population PK modeling, there are examples in the literature where ML has been successfully employed for forecasting PK data [
4,
5,
6]. For instance, Keutzer et al. [
7] conducted a study to evaluate the performance of various ML algorithms in predicting Rifampicin PK and compared them to population PK modeling. The authors trained lasso regression models, gradient boosting machines, XGBoost models, and random forest models to predict plasma concentration–time series and the area under the concentration-versus-time curve from 0 to 24 h (AUC0-24 h) after repeated dosing. The results showed that the predictive performance of the models improved as the number of plasma concentrations per patient increased, highlighting the impact of data availability on model accuracy. Similarly, in a study involving adults with nephrotic syndrome and membranous nephropathy, Yuan et al. [
8] investigated the use of ML models to predict tacrolimus (TAC) blood concentration in real-world settings. The XGBoost model exhibited good predictive ability for TAC blood concentration. Yet another example is the utilization of neural networks, which are well known for their ability to perform automated predictive analytics, to enhance temporal prediction metrics for patient response time courses. The author of Lu et al. [
9] employed neural networks to analyze longitudinal platelet response data from 665 patients who received T-DM1. The dataset includes patients from multiple clinical studies. By leveraging the power of neural networks, the aim was to improve the accuracy of predicting patient responses over time.
Therefore, the application of ML methods in PK has gained substantial interest in the field of clinical pharmacology in recent years. Examples include the use of ML techniques to predict drug exposure, such as TAC and mycophenolic acid, to improve the individual clearance predictions of renally cleared drugs in adult or neonate kidney transplant recipients [
10,
11,
12]. Consequently, these ML approaches have opened up new possibilities in therapeutic drug monitoring (TDM). ML models have the potential to revolutionize drug development, enabling more efficient and cost-effective prediction of PK parameters and informing decision-making in the early stages of drug development [
13,
14]. However, it is vital to acknowledge the challenges associated with this approach. One key challenge is the requirement for high-quality input data since inaccurate or incomplete data can lead to unreliable predictions. Additionally, the use of ML models in drug development raises concerns about interpretability and transparency, as these models are often seen as “black boxes” that are difficult to understand and validate [
15].
TAC is an immunosuppressant calcineurin inhibitor (CNI) commonly used in solid organ transplants to mitigate the risk of rejection. However, its usage is limited due to various factors, including a narrow therapeutic window and a highly variable pharmacological profile encompassing both PK and PD. In addition, studies have shown that only
to
of kidney transplant recipients treated with an initial weight-based tacrolimus dose were within the target concentration of the first steady-state TAC [
16,
17,
18]. Thus, TAC concentrations in the early post-transplant period are usually not measured at a steady-state, which can take up to 3 weeks for transplant recipients to reach the target concentration range, increasing the risks of rejection, acute tubular necrosis, and other complications in the early stages after renal transplantation. However, TAC concentrations decrease over time [
19]. TAC is known for its intricate pharmacokinetics, which involve liver-mediated autoinduction of elimination, concentration-dependent clearance with circadian rhythms, and dose-dependent bioavailability [
20,
21,
22,
23]. TAC is commercialized under different brand names. One of the first TAC formulations developed and approved by regulatory agencies was Prograf, which is given twice daily. However, other formulations were developed to reduce pharmacokinetic variation in blood levels and facilitate compliance, such as prolonged-release TAC formulations like Advagraf, which is administered once daily [
24]. Consequently, these pharmacological differences increase the complexity and time required in the modeling process for TAC. TDM serves as a fundamental approach in mitigating these challenges by allowing for individualized dosing of TAC, reducing toxicity risks, and minimizing the likelihood of rejection. In clinical practice, monitoring blood concentrations, adjusting treatment plans, and administering personalized TAC dosages are essential to achieve optimal therapeutic outcomes [
25].
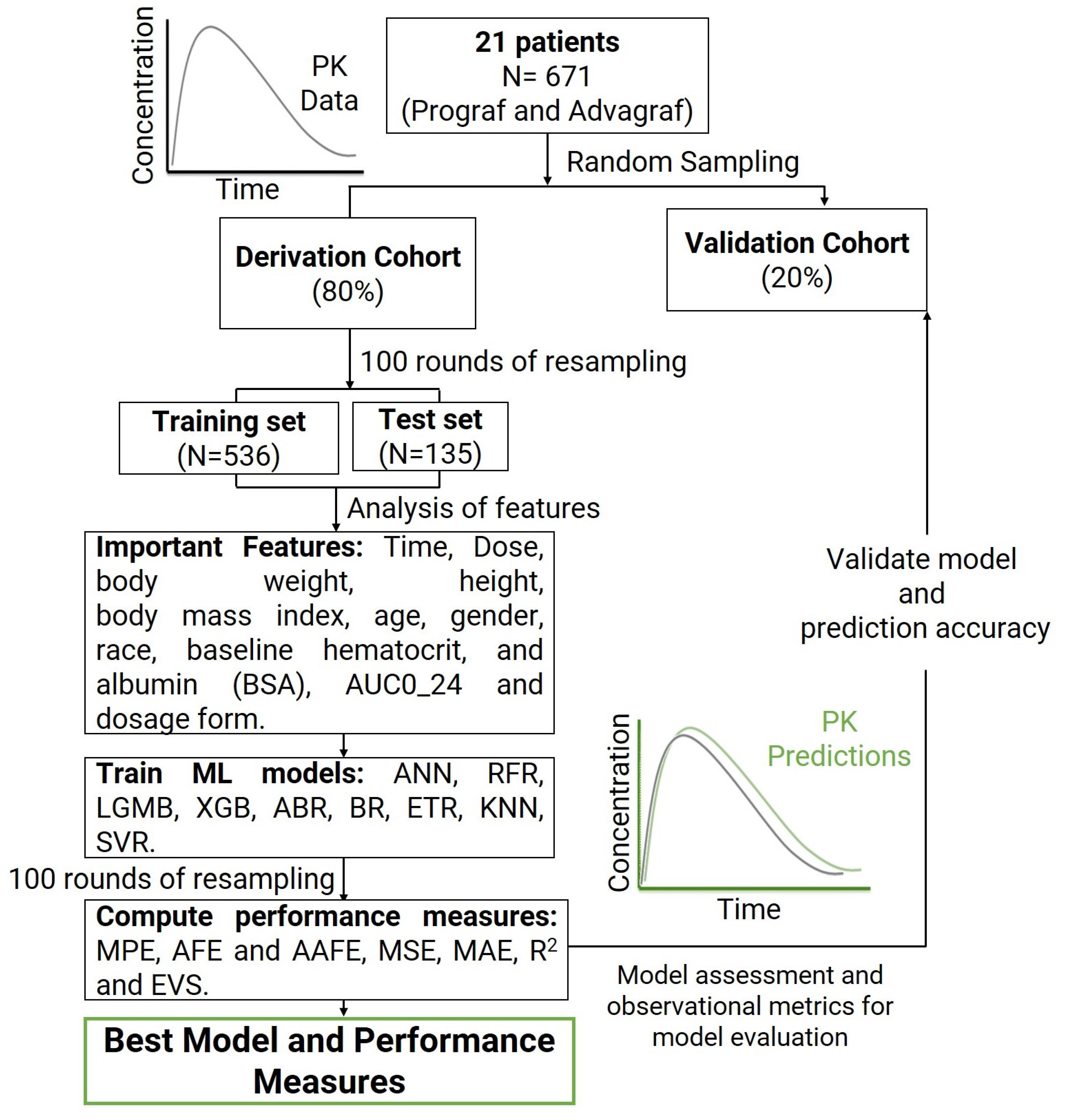
In this context, the main objectives of this research are: (i) to implement ML methods for accurately and precisely predicting the plasma concentration of tacrolimus over time for individual TAC formulations (Prograf and Advagraf individually); (ii) to analyze the capabilities of the ML models in achieving accurate PK predictions; (iii) to evaluate the external predictability of the models using an independent dataset; and (iv) to apply ML models to enhance the effectiveness of personalized medicine (PM) and provide clinicians with rationale initial dosage recommendations that maximize the likelihood of achieving the desired tacrolimus concentrations after the initial dose. Consequently, this research aims to contribute to advancing individualized treatment strategies and improving therapeutic outcomes. To the best of our knowledge, this is the first study to employ ML models for predicting TAC steady-state trough concentration. The data were sourced from a retrospective study of stable TAC plasma concentrations over time in the pediatric population with kidney pediatric transplants who received administration of Prograf and Advagraf [
26].
The rest of this paper is structured as follows.
Section 2 presents the materials and methods used.
Section 3 describes the obtained results, while
Section 4 provides a comprehensive discussion. Finally,
Section 5 draws conclusions and outlines potential lines of future research.
4. Discussion
Our study’s findings indicate that most of the ML models used for TAC prediction demonstrated a high accuracy. The models that achieved better results for AFE and AAFE values were the ETR, BR, RFR, KNN, XGB, and LGMB models.
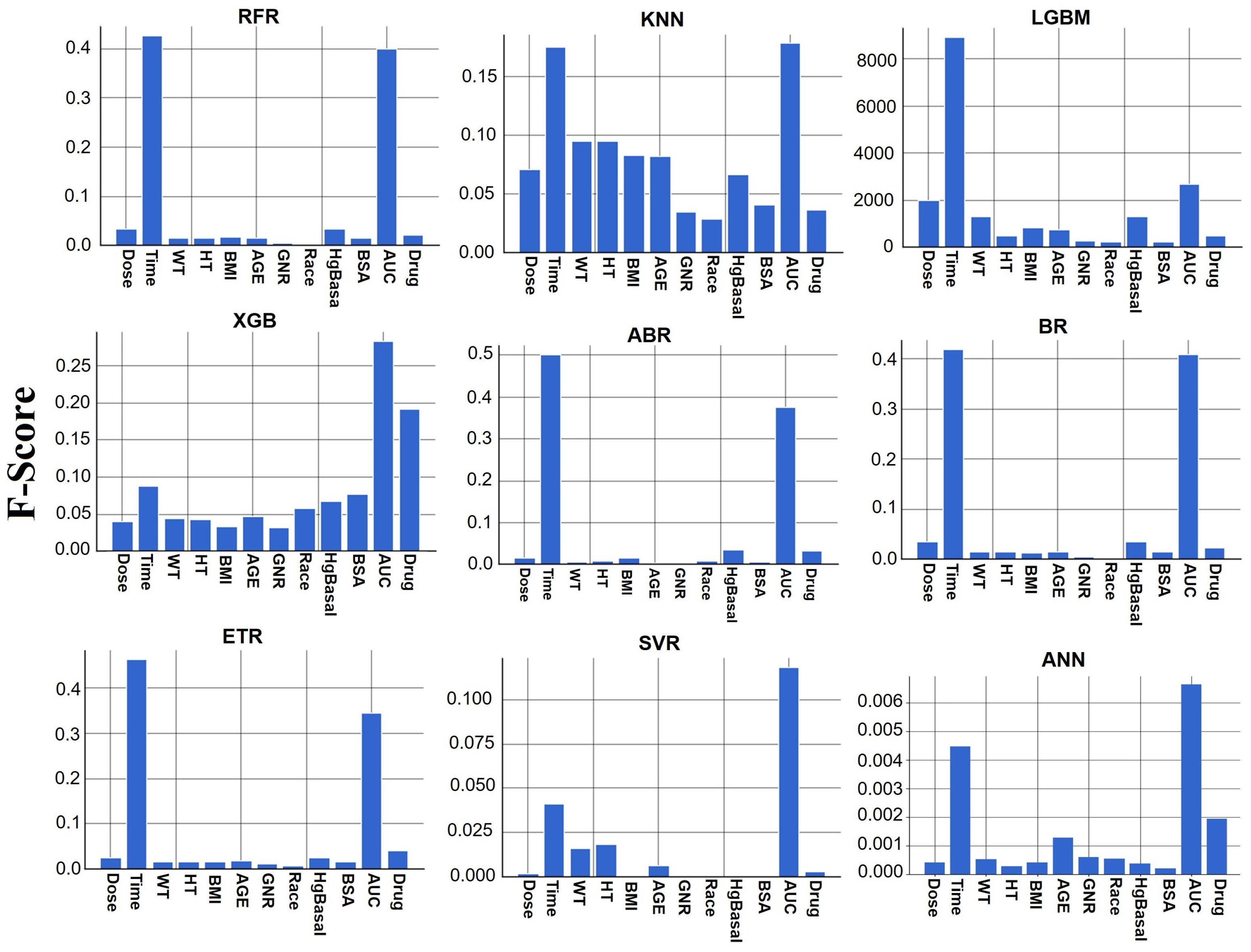
The ETR model, which implements a meta-estimator involving randomized decision trees and averaging, achieved slightly better performance. This advantage could be attributed to its ability to control overfitting and improve predictive accuracy by using multiple subsamples of the dataset. This finding emphasizes the importance of considering the characteristics of different ML models and their potential advantages in specific scenarios.
The top three ML models for TAC concentration prediction in this study were ETR, BR, and RFR, while XGB and LGMB also demonstrated good accuracy. This is similar to the findings of other research on TAC predictions in adults [
8,
33].
The results show that there were no significant accuracy differences between the top three or five best models, which suggests that these models perform comparably well.
Overall, the successful performance of ML models in predicting TAC concentrations in pediatric patients suggests that they could be valuable tools in real-world clinical settings. By providing accurate predictions of TAC concentrations, these models can aid in individualized treatment strategies, optimizing dosage regimens, and ultimately improving therapeutic outcomes for pediatric renal transplant recipients.
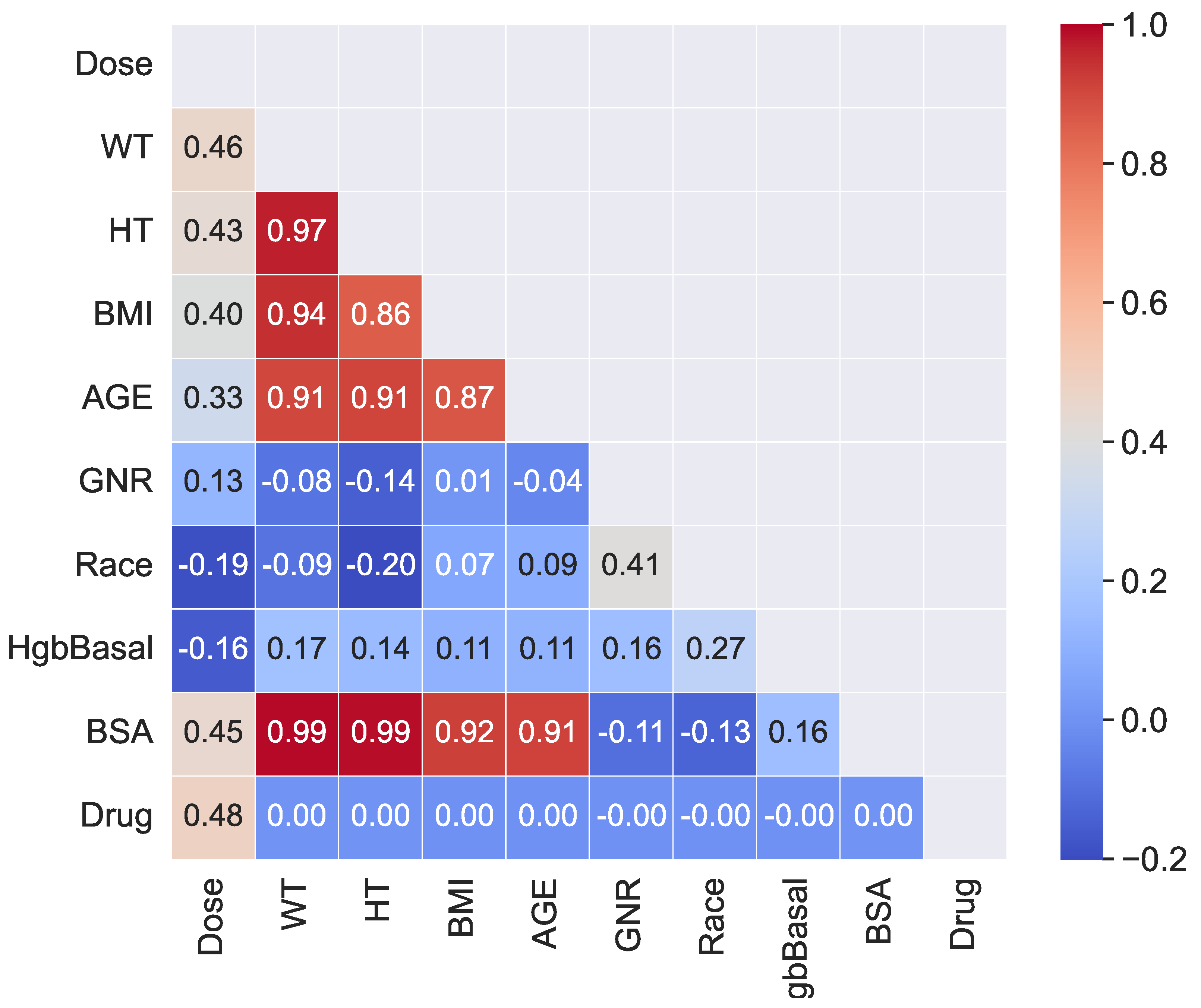
The feature importance analysis for the ETR model revealed that the area under the concentration–time curve (AUC) of TAC blood concentration had a significant effect on TAC blood concentration. This finding aligns with the existing knowledge in the field, as AUC is a critical PK parameter used to assess drug exposure and is considered the preferred measure for TAC exposure in clinical practice [
52,
53]. Interestingly, the importance of AUC was also supported by other ML models used in this study, including RFR, LGMB, ABR, and BR. This consistency in feature importance across different models reinforces the significance of AUC as a critical factor in predicting TAC blood concentrations and its relevance in guiding individualized dosing strategies. Furthermore, some of the models considered the importance of the pharmaceutical form of TAC (Prograf vs. Advagraf) in predicting blood concentrations. This is a logical consideration, as the dosing regimens and concentration–time profiles differ between Prograf (twice-daily administration) and Advagraf (once-daily administration). The number of maximum concentration points for each pharmaceutical form is indeed different, which could influence the overall concentration–time profile. Therefore, taking into account the pharmaceutical form as a feature in the models can help capture these differences and improve prediction accuracy.
Validating ML methods for TAC predictions in the presence of other co-administered drugs is crucial for real-world clinical applications. The PK of TAC can be affected by drug–drug interactions, where the presence of other drugs in the patient’s regimen can influence its metabolism, absorption, distribution, and elimination.
In addition, drug interactions may not only affect the PK of TAC but also impact the therapeutic outcomes and safety of the patient. Therefore, the ability of ML models to accurately predict TAC blood concentrations in the presence of co-administered drugs can have significant clinical implications, guiding clinicians in optimizing dosing regimens and minimizing the risk of adverse drug events [
54].
Unfortunately, this dataset does not take into account genomic information. Despite numerous factors that may affect the pharmacokinetics of tacrolimus, genetic factors are quite important and common. TAC is metabolized by two enzymes of the cytochrome P450 family: CYP3A5 and CYP3A4. The effect of CYP3A5 and CYP3A4 genotypes on TAC bioavailability has been demonstrated, and a significant portion of the interindividual variability in its PK is explained by mutations in the CYP3A4 and CYP3A5 enzymes. For example, studies have shown that the mean dose-adjusted blood TAC concentration was significantly higher among CYP3A53 homozygotes compared to carriers of the wild-type allele (CYP3A51) [
55]. In a recent prospective study, a group of kidney transplant patients received a TAC dose either based on the CYP3A5 genotype (the adapted group) or according to the standard regimen (the control group) [
56]. Consequently, additional studies are necessary to determine whether the pharmacogenetic approach could help reduce the necessity for induction therapy and co-immunosuppressors [
55].
ML methods have become a prominent trend in predicting drug concentrations in the blood, and this approach has also been applied to predict TAC blood concentrations in previous research. The majority of these studies utilized artificial neural networks and regression models for their predictions [
8,
11,
57,
58,
59,
60,
61].
However, it is essential to acknowledge that these earlier studies faced certain limitations. Firstly, they often dealt with a relatively limited amount of data, which may impact the generalizability of their models. Additionally, the lack of external validation in many of these studies raises concerns about the robustness and reliability of their findings.
Furthermore, when comparing modeling approaches in PK, there are some key points to consider. PK methods primarily focus on estimating parameters for the structural model, variability, and covariate model parameters within a population, which contributes to mechanistic understanding, biological interpretability of the results, and the ability to simulate in silico experiments from the model. Conversely, ML is primarily geared towards predicting outcomes and ML has the inherent danger of producing results that are not therapeutically meaningful. Consequently, PK/PD analysis provides valuable mechanistic insights into biological processes, whereas ML models, while trained more swiftly, offer fewer mechanistic insights and can be perceived as enigmatic ’black boxes’, making it challenging to extract underlying mechanisms [
6]. This underscores the necessity for ML to have access to substantial training data that can reasonably be assumed to be exchangeable with the test data. Conversely, Bayesian inference excels when dealing with sparse data and a dense model, thereby requiring fewer patients to obtain meaningful results in PK methods [
7].
Because of the numerous issues that PM and ML encounter, research in this field remains in its exploratory phase, underscoring the need for further investigation and validation. The fusion of PK and ML holds the potential to yield precise estimations of drug exposure by simulating rich concentration-versus-time profiles, by exploring and learning the relationships within all the patient covariates [
62] or by using faster models and performing faster analyses [
63]. For instance, the ML approach has been shown to confer advantages over traditional approaches, including increased accuracy and reduced variance [
64]. These innovative approaches represent a significant advancement compared to the prior situation where extensive databases were essential to train an ML algorithm, leaving scarce independent datasets for validation purposes [
7].
As ML methods continue to advance and more data become available, it is hoped that these limitations can be addressed and the potential of ML fully harnessed in drug concentration prediction, benefiting both adult and pediatric populations alike.