Synthetic MRI Generation from CT Scans for Stroke Patients
Abstract
:1. Introduction
2. Materials and Methods
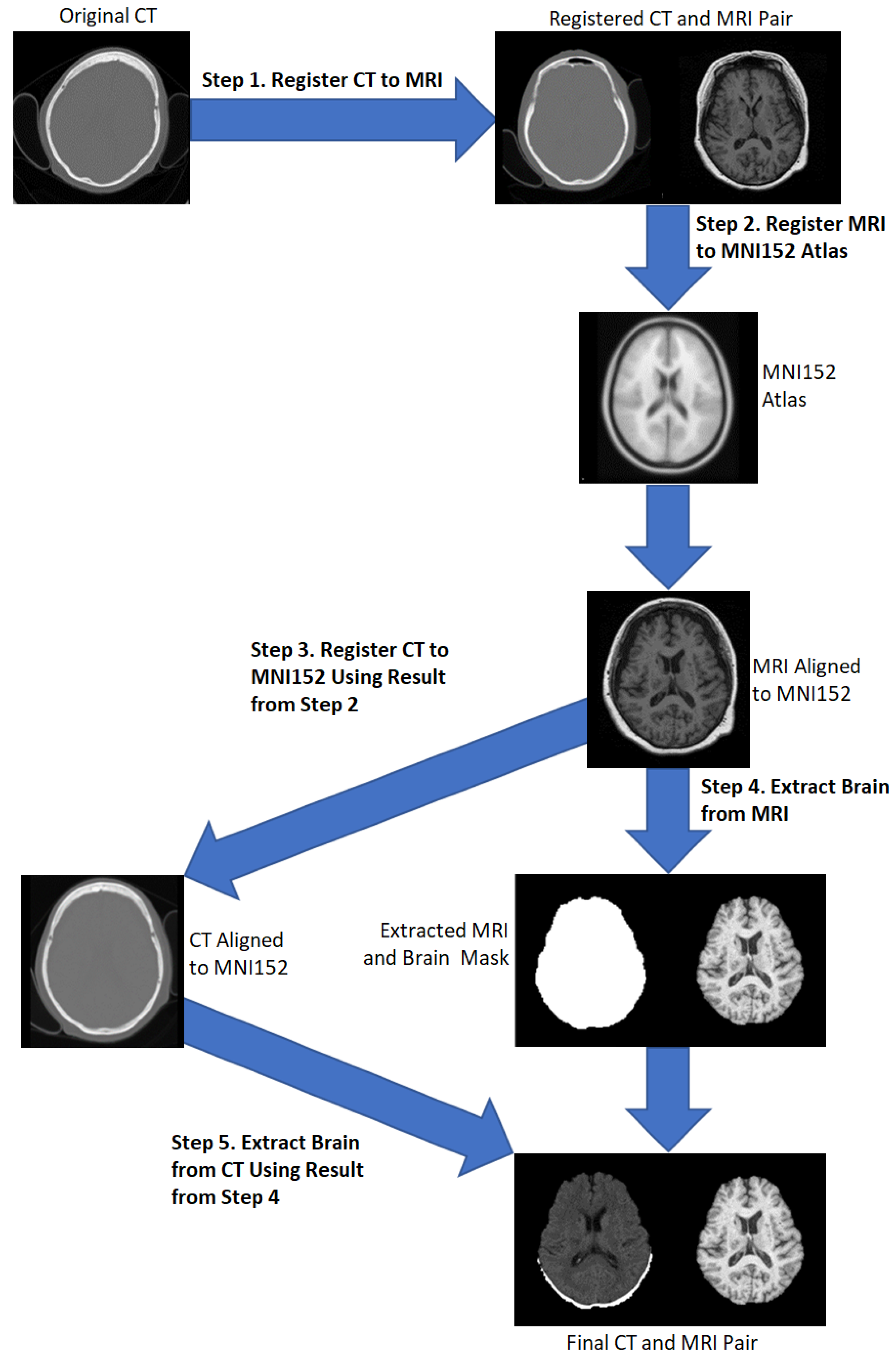
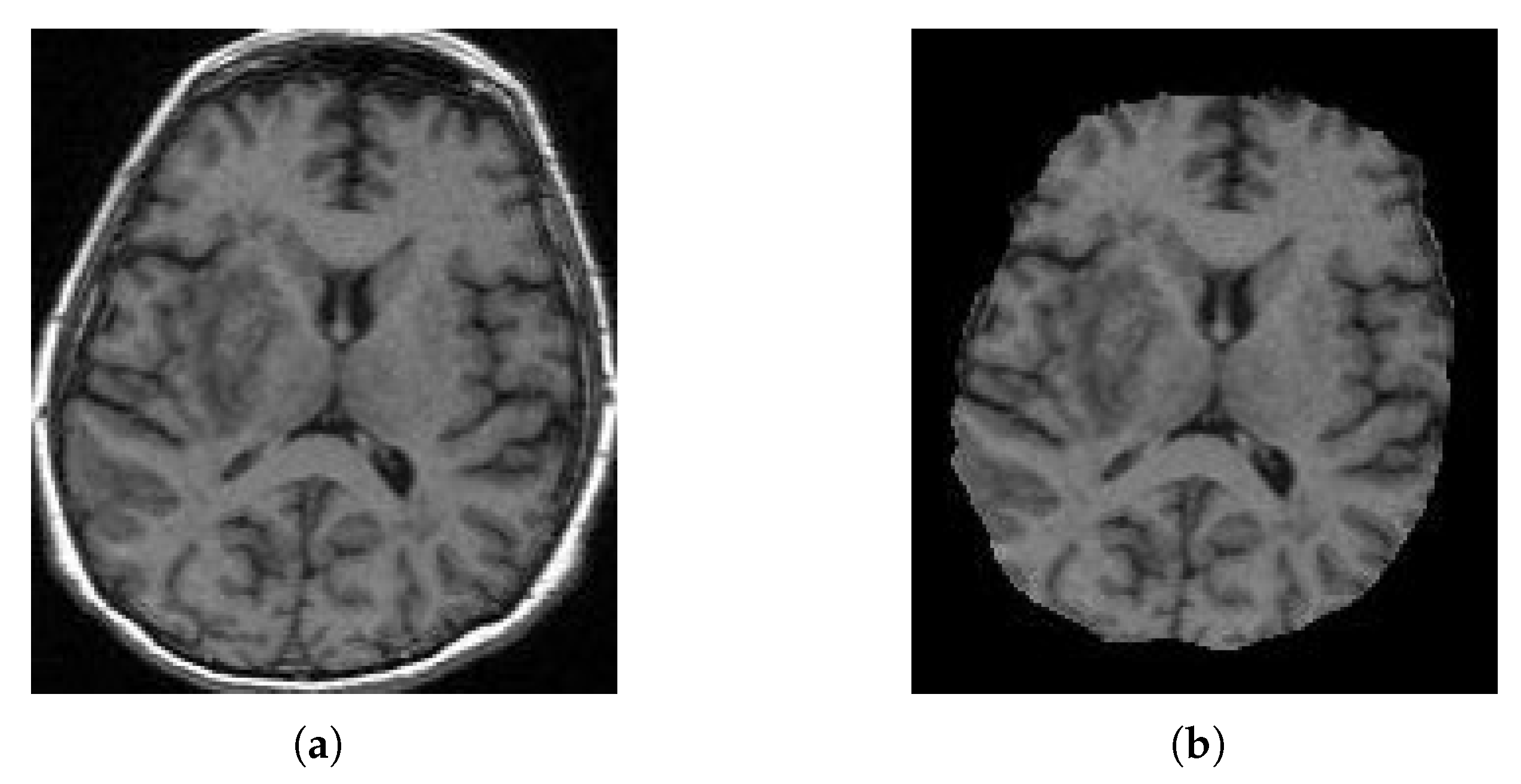
2.1. Preprocessing
- Applying the transformation matrix from step 2 to the resultant CT from step 1.
- Apply the runhdbet function of HD-BET [32] to the resultant MRI from step 2. The resultant extraction brain and brain mask are then saved.
- Using pixelwise multiplication between the resultant CT scan from step 3 and the brain mask from step 4 to extract the brain from the CT.
2.2. Model Architectures
2.3. Training and Evaluation
3. Results
3.1. UNet V1
3.2. UNet V2
3.3. Patch-Based UNet
3.4. 2D UNet
3.5. UNet++
3.6. Attention UNet
3.7. Transformer UNet
3.8. CycleGAN
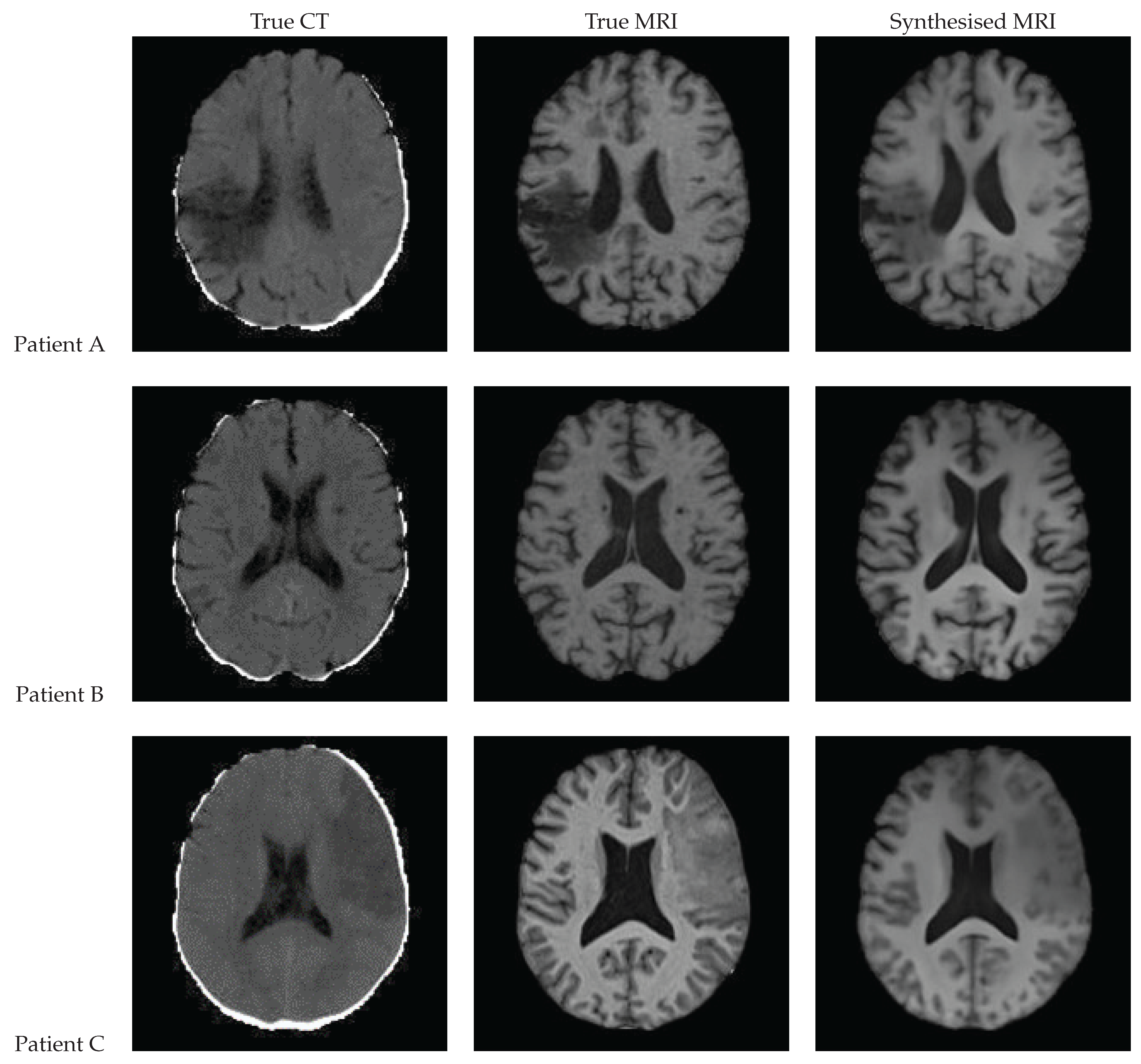
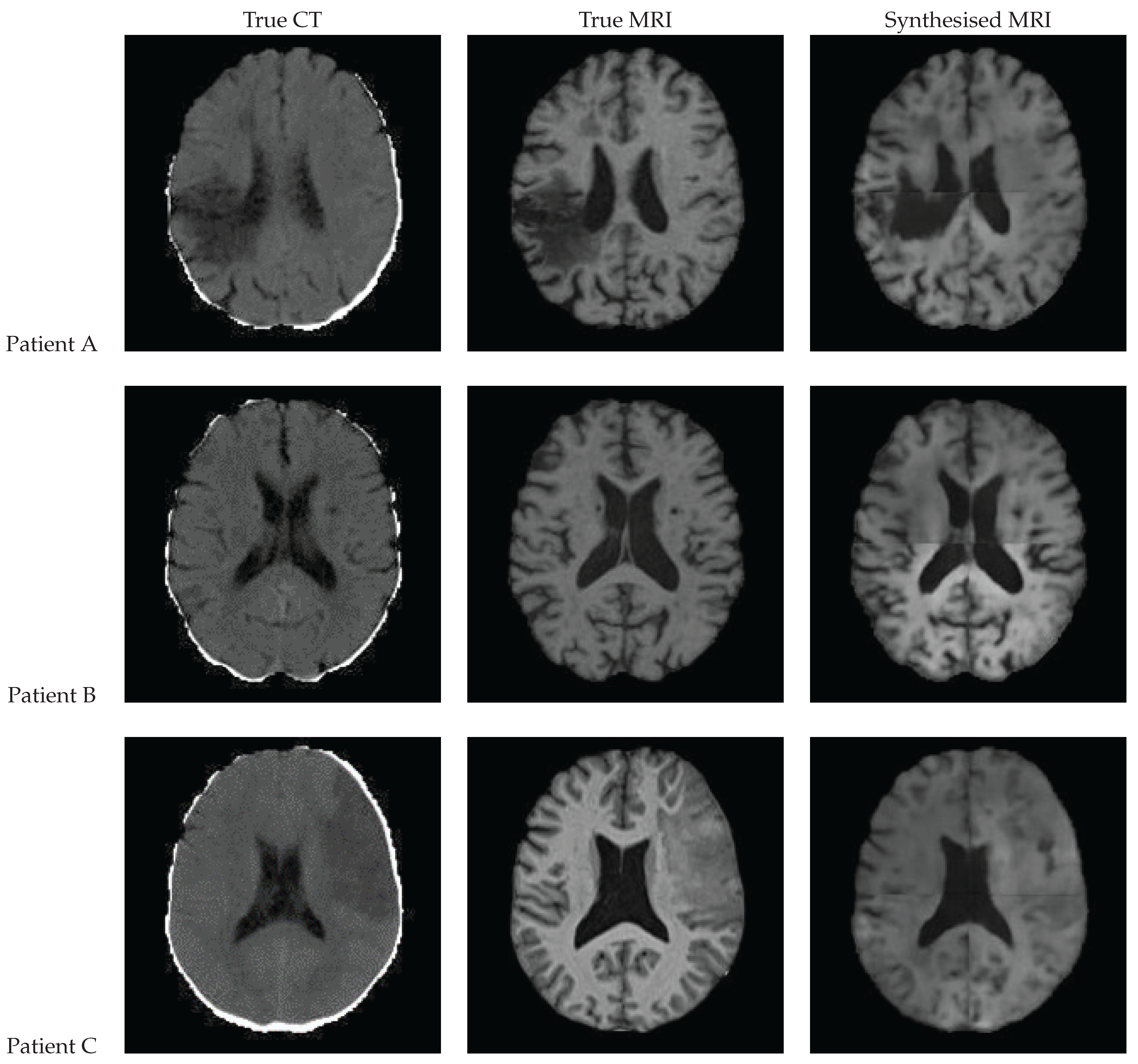
3.9. Qualitative Assessment
3.10. Quantitative Assessment
3.11. Performance at Clinically Relevant Tasks
3.11.1. Registration
3.11.2. Lesion Segmentation
3.11.3. Brain Tissue Segmentation
4. Discussion
4.1. Different Architectures
4.2. Limitations
4.3. Input Data Quality
4.4. Metrics
4.5. Other Datasets
4.6. Further Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CNN | Convolutional Neural Network |
| CSF | Cerebrospinal fluid |
| CT | Computed Tomography |
| DSC | Dice Score |
| GAN | Generative Adversarial Network |
| GM | Grey matter |
| MAE | Mean Absolute Error |
| MRI | Magnetic Resonance Imaging |
| MSE | Mean Squared Error |
| PSNR | Peak Signal-to-Noise Ratio |
| SSIM | Structural Similarity Index Measure |
| WM | White matter |
References
- Chalela, J.A.; Kidwell, C.S.; Nentwich, L.M.; Luby, M.; Butman, J.A.; Demchuk, A.M.; Hill, M.D.; Patronas, N.; Latour, L.; Warach, S. Magnetic resonance imaging and computed tomography in emergency assessment of patients with suspected acute stroke: A prospective comparison. Lancet 2007, 369, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Moreau, F.; Asdaghi, N.; Modi, J.; Goyal, M.; Coutts, S.B. Magnetic Resonance Imaging versus Computed Tomography in Transient Ischemic Attack and Minor Stroke: The More You See the More You Know. Cerebrovasc. Dis. Extra 2013, 3, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Provost, C.; Soudant, M.; Legrand, L.; Ben Hassen, W.; Xie, Y.; Soize, S.; Bourcier, R.; Benzakoun, J.; Edjlali, M.; Boulouis, G.; et al. Magnetic Resonance Imaging or Computed Tomography Before Treatment in Acute Ischemic Stroke. Stroke 2019, 50, 659–664. [Google Scholar] [CrossRef]
- Birenbaum, D.; Bancroft, L.W.; Felsberg, G.J. Imaging in acute stroke. West. J. Emerg. Med. 2011, 12, 67–76. [Google Scholar] [PubMed]
- Wu, J.; Ngo, G.H.; Greve, D.; Li, J.; He, T.; Fischl, B.; Eickhoff, S.B.; Yeo, B.T. Accurate nonlinear mapping between MNI volumetric and FreeSurfer surface coordinate systems. Hum. Brain Mapp. 2018, 39, 3793–3808. [Google Scholar] [CrossRef]
- Talairach, J.; Tournoux, P. Co-Planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System: An Approach to Cerebral Imaging; Thieme Publishers: Stuttgart, Germany, 1988. [Google Scholar]
- Brodmann, K.; Garey, L. Brodmann’s: Localisation in the Cerebral Cortex; Springer: New York, NY, USA, 2007. [Google Scholar]
- Mori, S.; Wakana, S.; van Zijl, P.; Nagae-Poetscher, L. MRI Atlas of Human White Matter; Elsevier Science: Amsterdam, The Netherlands, 2005. [Google Scholar]
- Zachiu, C.; de Senneville, B.D.; Moonen, C.T.W.; Raaymakers, B.W.; Ries, M. Anatomically plausible models and quality assurance criteria for online mono- and multi-modal medical image registration. Phys. Med. Biol. 2018, 63, 155016. [Google Scholar] [CrossRef]
- Liu, L.; Johansson, A.; Cao, Y.; Dow, J.; Lawrence, T.S.; Balter, J.M. Abdominal synthetic CT generation from MR Dixon images using a U-net trained with ‘semi-synthetic’ CT data. Phys. Med. Biol. 2020, 65, 125001. [Google Scholar] [CrossRef]
- Dinkla, A.; Florkow, M.; Maspero, M.; Savenije, M.; Zijlstra, F.; Doornaert, P.; van Stralen, M.; Philippens, M.; van den Berg, C.; Seevinck, P. Dosimetric Evaluation of Synthetic CT for Head and Neck Radiotherapy Generated by a Patch-Based Three-Dimensional Convolutional Neural Network. Med. Phys. 2019, 46, 4095–4104. [Google Scholar] [CrossRef]
- Brou Boni, K.N.D.; Klein, J.; Vanquin, L.; Wagner, A.; Lacornerie, T.; Pasquier, D.; Reynaert, N. MR to CT synthesis with multicenter data in the pelvic area using a conditional generative adversarial network. Phys. Med. Biol. 2020, 65, 075002. [Google Scholar] [CrossRef]
- Han, X. MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef]
- Chen, S.; Peng, Y.; Qin, A.; Liu, Y.; Zhao, C.; Deng, X.; Deraniyagala, R.; Stevens, C.; Ding, X. MR-based synthetic CT image for intensity-modulated proton treatment planning of nasopharyngeal carcinoma patients. Acta Oncol. 2022, 61, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Florkow, M.C.; Willemsen, K.; Zijlstra, F.; Foppen, W.; van der Wal, B.C.H.; van der Voort van Zyp, J.R.N.; Viergever, M.A.; Castelein, R.M.; Weinans, H.; van Stralen, M.; et al. MRI-based synthetic CT shows equivalence to conventional CT for the morphological assessment of the hip joint. J. Orthop. Res. 2022, 40, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lei, Y.; Wang, T.; Kayode, O.; Tian, S.; Liu, T.; Patel, P.; Curran, W.J.; Ren, L.; Yang, X. MRI-based treatment planning for liver stereotactic body radiotherapy: Validation of a deep learning-based synthetic CT generation method. Br. J. Radiol. 2019, 92, 20190067. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Harms, J.; Wang, T.; Liu, Y.; Shu, H.K.; Jani, A.B.; Curran, W.J.; Mao, H.; Liu, T.; Yang, X. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med. Phys. 2019, 46, 3565–3581. [Google Scholar] [CrossRef]
- Kazemifar, S.; McGuire, S.; Timmerman, R.; Wardak, Z.; Nguyen, D.; Park, Y.; Jiang, S.; Owrangi, A. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother. Oncol. 2019, 136, 56–63. [Google Scholar] [CrossRef]
- Qi, M.; Li, Y.; Wu, A.; Jia, Q.; Li, B.; Sun, W.; Dai, Z.; Lu, X.; Zhou, L.; Deng, X.; et al. Multi-sequence MR image-based synthetic CT generation using a generative adversarial network for head and neck MRI-only radiotherapy. Med. Phys. 2020, 47, 1880–1894. [Google Scholar] [CrossRef]
- Kalantar, R.; Messiou, C.; Winfield, J.M.; Renn, A.; Latifoltojar, A.; Downey, K.; Sohaib, A.; Lalondrelle, S.; Koh, D.M.; Blackledge, M.D. CT-Based Pelvic T(1)-Weighted MR Image Synthesis Using UNet, UNet++ and Cycle-Consistent Generative Adversarial Network (Cycle-GAN). Front. Oncol. 2021, 11, 665807. [Google Scholar] [CrossRef]
- Dong, X.; Lei, Y.; Tian, S.; Wang, T.; Patel, P.; Curran, W.J.; Jani, A.B.; Liu, T.; Yang, X. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother. Oncol. 2019, 141, 192–199. [Google Scholar] [CrossRef]
- Hong, K.T.; Cho, Y.; Kang, C.; Ahn, K.S.; Lee, H.; Kim, J.; Hong, S.; Kim, B.H.; Shim, E. Lumbar Spine Computed Tomography to Magnetic Resonance Imaging Synthesis Using Generative Adversarial Network: Visual Turing Test. Diagnostics 2022, 12, 530. [Google Scholar] [CrossRef]
- Dai, X.; Lei, Y.; Wang, T.; Zhou, J.; Roper, J.; McDonald, M.; Beitler, J.J.; Curran, W.J.; Liu, T.; Yang, X. Automated delineation of head and neck organs at risk using synthetic MRI-aided mask scoring regional convolutional neural network. Med. Phys. 2021, 48, 5862–5873. [Google Scholar] [CrossRef]
- Kieselmann, J.P.; Fuller, C.D.; Gurney-Champion, O.J.; Oelfke, U. Cross-modality deep learning: Contouring of MRI data from annotated CT data only. Med. Phys. 2021, 48, 1673–1684. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Qin, W.; Liang, X.; Xu, J.; Xiong, J.; Xie, Y. Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy. Quant. Imaging Med. Surg. 2020, 10, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, W.; Xiong, J.; Xia, J.; Xie, Y. Comparison of Supervised and Unsupervised Deep Learning Methods for Medical Image Synthesis between Computed Tomography and Magnetic Resonance Images. BioMed Res. Int. 2020, 2020, 5193707. [Google Scholar] [CrossRef] [PubMed]
- Feng, E.; Qin, P.; Chai, R.; Zeng, J.; Wang, Q.; Meng, Y.; Wang, P. MRI Generated From CT for Acute Ischemic Stroke Combining Radiomics and Generative Adversarial Networks. IEEE J. Biomed. Health Inform. 2022, 26, 6047–6057. [Google Scholar] [CrossRef] [PubMed]
- McNaughton, J. CT to Synthetic MRI Generation. 2023. Available online: https://github.com/jakemcnaughton/CT-to-Synthetic-MRI-Generation/ (accessed on 3 June 2023).
- Jenkinson, M.; Smith, S. A global optimisation method for robust affine registration of brain images. Med. Image Anal. 2001, 5, 143–156. [Google Scholar] [CrossRef]
- Jenkinson, M. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. NeuroImage 2002, 17, 825–841. [Google Scholar] [CrossRef] [PubMed]
- Greve, D.N.; Fischl, B. Accurate and robust brain image alignment using boundary-based registration. NeuroImage 2009, 48, 63–72. [Google Scholar] [CrossRef]
- Isensee, F.; Schell, M.; Pflueger, I.; Brugnara, G.; Bonekamp, D.; Neuberger, U.; Wick, A.; Schlemmer, H.; Heiland, S.; Wick, W.; et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum. Brain Mapp. 2019, 40, 4952–4964. [Google Scholar] [CrossRef]
- Johnson, H.; Harris, G.; Williams, K. BRAINSFit: Mutual Information Registrations of Whole-Brain 3D Images, Using the Insight Toolkit. Insight J. 2007, 180, 1–10. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Springer: New York, NY, USA, 2014; pp. 277–289. [Google Scholar] [CrossRef]
- Kapur, T.; Pieper, S.; Fedorov, A.; Fillion-Robin, J.C.; Halle, M.; O’Donnell, L.; Lasso, A.; Ungi, T.; Pinter, C.; Finet, J.; et al. Increasing the impact of medical image computing using community-based open-access hackathons: The NA-MIC and 3D Slicer experience. Med. Image Anal. 2016, 33, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Odena, A.; Dumoulin, V.; Olah, C. Deconvolution and Checkerboard Artifacts. Distill 2016, 1, e3. [Google Scholar] [CrossRef]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. In Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Volume 11045, pp. 3–11. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Le Folgoc, L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.u.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Nice, France, 2017; Volume 30. [Google Scholar]
- Iommi, D. 3D-CycleGan-Pytorch-MedImaging. 2021. Available online: https://github.com/davidiommi/3D-CycleGan-Pytorch-MedImaging (accessed on 3 June 2023).
- Brudfors, M.; Chalcroft, L. ATLAS_UNET. 2022. Available online: https://grand-challenge.org/algorithms/atlas_unet-2/ (accessed on 1 April 2023).
- Zhang, Y.; Brady, M.; Smith, S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 2001, 20, 45–57. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.P.; Luck, M.; Honari, S. Distribution Matching Losses Can Hallucinate Features in Medical Image Translation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2018, Granada, Spain, 16–20 September 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer: Cham, Switzerland, 2018; pp. 529–536. [Google Scholar]

















| Study | Region of Interest | Number of Patients | Paired | GAN | CNN | Transformers |
|---|---|---|---|---|---|---|
| [20] | Pelvis | 17 | ✓ | ✓ | ✓ | ✗ |
| [21] | 140 | ✓ | ✓ | ✗ | ✗ | |
| [22] | Lumbar Spine | 285 | ✓ | ✓ | ✗ | ✗ |
| [23] | Head and Neck | 118 | ✓ | ✓ | ✗ | ✗ |
| [24] | 229 * | ✗ | ✓ | ✗ | ✗ | |
| [25] | Brain | 34 | ✓ | ✓ | ✓ | ✗ |
| [26] | 34 | ✓ | ✓ | ✓ | ✗ | |
| [27] | 103 | ✓ | ✓ | ✗ | ✗ | |
| Ours | 181 | ✓ | ✓ | ✓ | ✓ |
| Study | n | Scanner | TR (ms) | TE (ms) | TI (ms) | Flip (∘) | Sequence * |
|---|---|---|---|---|---|---|---|
| 1 | 55 | Avanto 1.5 T | 11 | 4.94 | n/a | 15 | FLASH3D |
| 2 | 47 | Avanto 1.5 T | 13 | 4.76 | n/a | 25 | FLASH3D |
| 3 | 8 | Skyra 3 T | 23 | 2.46 | n/a | 23 | FLASH3D |
| 4 | 18 | Skyra 3 T | 1900 | 2.07 | 900 | 9 | FLASH3D, MPRAGE |
| 5 | 53 | Avanto 1.5 T | 2200 | 2.97 | 900 | 8 | FLASH3D, MPRAGE |
| Model | Number of Epochs | Learning Rate | Patch Based | Input Dimension | Batch Size | Loss Function | Training Environment |
|---|---|---|---|---|---|---|---|
| UNet V1 | 400 | ✗ | 176 × 192 × 176 | 1 | MAE | 1 × 80 GB Tensorflow | |
| UNet V2 | 400 | ✗ | 176 × 192 × 176 | 1 | MAE | 1 × 80 GB Tensorflow | |
| UNet Patch | 400 | ✓ | 96 × 96 × 96 | 4 | MAE | 4 × 32 GB Tensorflow | |
| UNet 2D | 400 | ✗ | 192 × 176 | 16 | MAE | 4 × 32 GB Tensorflow | |
| UNet++ | 400 | ✓ | 96 × 96 × 96 | 4 | MAE | 4 × 32 GB Tensorflow | |
| Attention UNet | 400 | ✓ | 96 × 96 × 96 | 4 | MAE | 4 × 32 GB Tensorflow | |
| Transformer UNet | 400 | ✓ | 96 × 96 × 96 | 1 | MAE | 1 × 80 GB Tensorflow | |
| CycleGAN | 200 | ✓ | 112 × 112 × 112 | 6 | MAE/BCE * | 6 × 32 GB Pytorch |
| Model | MAE ↓ | MSE ↓ | SSIM ↑ | PSNR ↑ | Total SSIM ↑ * |
|---|---|---|---|---|---|
| UNet | |||||
| UNet V2 | |||||
| 2D UNet | |||||
| Patch-Based UNet | |||||
| Attention UNet | |||||
| UNet++ | |||||
| Transformer UNet | |||||
| CycleGAN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
McNaughton, J.; Holdsworth, S.; Chong, B.; Fernandez, J.; Shim, V.; Wang, A. Synthetic MRI Generation from CT Scans for Stroke Patients. BioMedInformatics 2023, 3, 791-816. https://doi.org/10.3390/biomedinformatics3030050
McNaughton J, Holdsworth S, Chong B, Fernandez J, Shim V, Wang A. Synthetic MRI Generation from CT Scans for Stroke Patients. BioMedInformatics. 2023; 3(3):791-816. https://doi.org/10.3390/biomedinformatics3030050
Chicago/Turabian StyleMcNaughton, Jake, Samantha Holdsworth, Benjamin Chong, Justin Fernandez, Vickie Shim, and Alan Wang. 2023. "Synthetic MRI Generation from CT Scans for Stroke Patients" BioMedInformatics 3, no. 3: 791-816. https://doi.org/10.3390/biomedinformatics3030050
APA StyleMcNaughton, J., Holdsworth, S., Chong, B., Fernandez, J., Shim, V., & Wang, A. (2023). Synthetic MRI Generation from CT Scans for Stroke Patients. BioMedInformatics, 3(3), 791-816. https://doi.org/10.3390/biomedinformatics3030050










