1. Introduction
Time series data are widely used in many fields, and various data-driven modeling techniques are developed to represent the dynamic characteristics of systems and forecast the future behavior. The growing research in artificial intelligence has provided powerful machine learning (ML) techniques to contribute to data-driven model development. Real-world data provide several challenges to modeling and forecasting, such as missing values and outliers. Such imperfections in data can reduce the accuracy of ML and the models developed. This necessitates data preprocessing for the imputation of missing values, down- and up-sampling, and data reconciliation. Data preprocessing is a laborious and time-consuming effort since big data are usually stacked on a large scale [
1]. When models are used for forecasting, the accuracy of forecasts improve if the effects of future possible disturbances based on behavior patterns extracted from historical data are incorporated in the forecasts. This paper focuses on these two problems and investigates the benefits of preprocessing the real-world data and the performance of different recurrent neural network (RNN) models for detecting various events that affect blood glucose concentration (BGC) in people with type 1 diabetes (T1D). The behavior patterns detected are used for more accurate predictions of future BGC variations, which can be used for warnings and for increasing the effectiveness of automated insulin delivery (AID) systems.
Time series data captured in daily living of people with chronic conditions have many of these challenges to modeling, detection, and forecasting. Focusing on people with T1D, the medical objective is to forecast the BGC of a person with T1D and prevent the excursion of BGC outside a “desired range” (70–180 mg/dL) to reduce the probability of hypo- and hyperglycemia events. In recent years, the number of people with diabetes has grown rapidly around the world, reaching pandemic levels [
2,
3]. Advances in continuous glucose monitoring (CGM) systems, insulin pump and insulin pen technologies, and in novel insulin formulations has enabled many powerful treatment options [
4,
5,
6,
7,
8,
9]. The current treatment options available to people with T1D range from manual insulin injections to AID. Manual injection (insulin bolus) doses are computed based on the person’s characteristics and the properties of the meal consumed. Current AID systems necessitate the manual entry of meal information to give insulin boluses for mitigating the effects of meal on the BGC. A manual adjustment of the basal insulin dose and increasing the BGC target level and/or consumption of snacks are the options to mitigate the effects of physical activity. Some people may forget to make these manual entries and a system that can nudge them to provide appropriate information can reduce the extreme excursions in BGC. Commercially available AID systems are hybrid closed-loop systems, and they require these manual entries by the user. AID systems, also called artificial pancreas (AP), consist of a CGM, an insulin pump, and a closed-loop control algorithm that manipulates the insulin infusion rate delivered by the pump based on the recent CGM values reported [
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22,
23]. More advanced AID systems that use a multivariable approach [
10,
24,
25,
26] use additional inputs from wearable devices (such as wristbands) to automatically detect the occurrence of physical activity and incorporate this information to the automated control algorithms for a fully automated AID system [
27]. Most AID systems use model predictive control techniques that predict future BGC values in making their insulin dosing decisions. Knowing the habits of the individual AID user improves the control decisions since the prediction accuracy of the future BGC trajectories can explicitly incorporate the future potential disturbances to the BGC, such as meals and physical activities, that will occur with high likelihood during the future BGC prediction window [
24,
26]. Consequently, the detection of meal and physical activity events from historical free-living data of a person with T1D will provide useful information for decision making by both the individual and by the AID system.
CGM systems report subcutaneous glucose concentration to infer BGC with a sampling rate of 5 min. Self-reported meal and physical activity data are often based on diary entries. Physical activity data can also be captured by wearable devices. The variables reported by wearable devices may have artifacts, noise, missing values, and outliers. The data used in this work include only CGM values, insulin dosing information, and diary entries of meals and physical activities.
Analyzing long-term data of people with T1D indicates that individuals tend to repeat daily habitual behaviors.
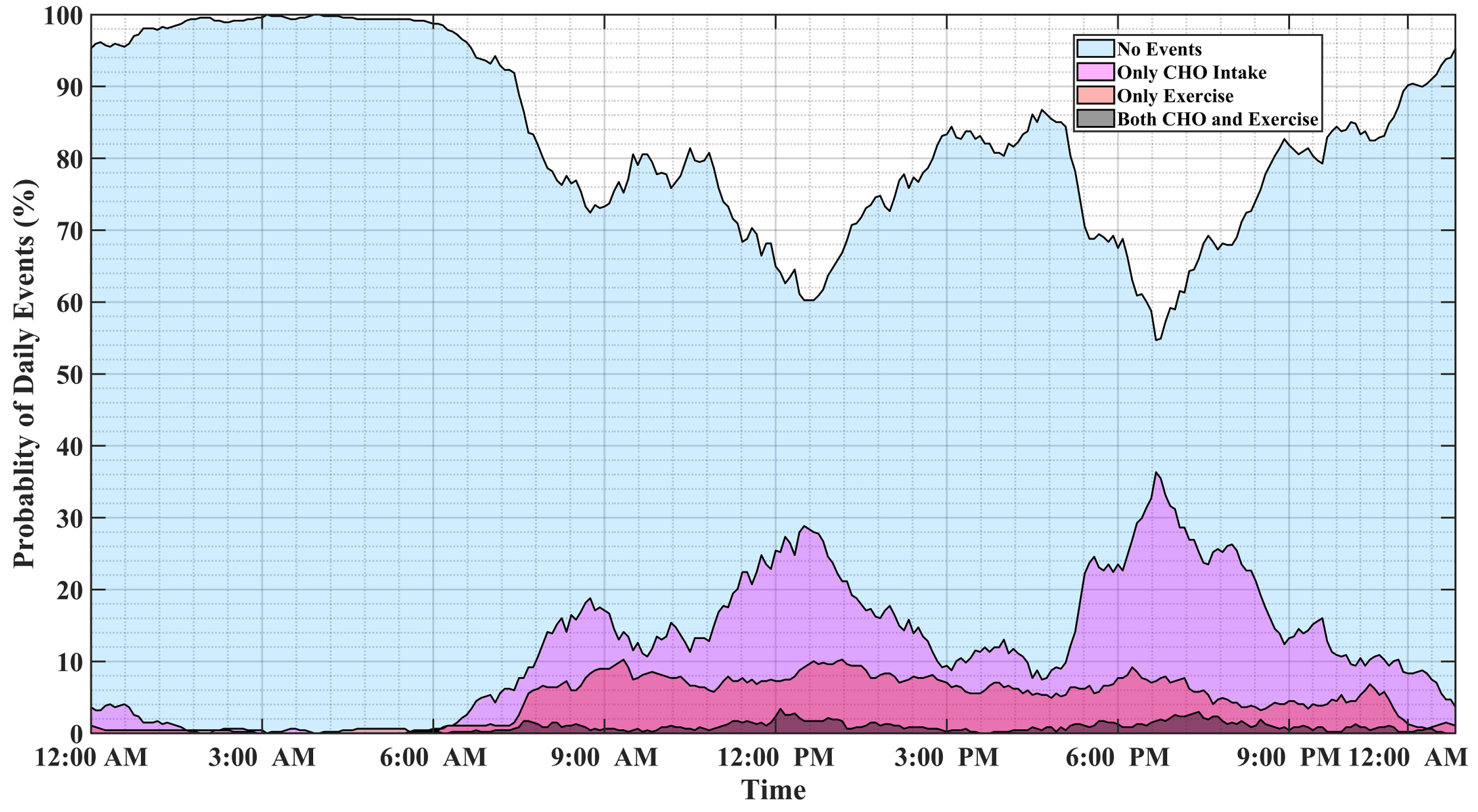
Figure 1 illustrates the probability of physical activity and meal (indicated as carbohydrate intake) events, either simultaneously or disjointly, for 15 months of self-reported CGM, meal, insulin pump, and physical activity data of individuals with T1D. Major factors affecting BGC variations usually occur at specific time windows and conditions, and some combinations of events are mutually exclusive. For example, insulin-bolusing and physical activity are less likely to occur simultaneously or during hypoglycemia episodes, since people do not exercise when their BGC is low. People may have different patterns of behavior during the work week versus weekends or holidays. Predicting the probabilities of exercise, meal consumption, and their concurrent occurrence based on historical data using ML can provide important information on the behavior patterns for making medical therapy decisions in diabetes.
Motivated by the above considerations, this work develops a framework for predicting the probabilities of meal and physical activity events, including their independent and simultaneous occurrences. A framework is built to handle the inconsistencies and complexities of real-world data, including missing data, outlier removal, feature extraction, and data augmentation. Four different recurrent neural network (RNN) models are developed and evaluated for estimating the probability of events causing large variations in BGC. The advent of deep neural networks (NNs) and their advances have paved the way for processing and analyzing various types of information, namely: time-series, spatial, and time-series–spatial data. Long short-term memory (LSTM) NN models are specific sub-categories of recurrent NNs introduced to reduce the computational burden of storing information over extended time intervals [
28,
29]. LSTMs take advantage of nonlinear dynamic modeling without knowing time-dependency information in the data. Moreover, their multi-step-ahead prediction capability makes them an appropriate choice for detecting upcoming events and disturbances that can deteriorate the accuracy of model predictions.
The main contributions of this work are the development of NN models capable of estimating the occurrences of meals and physical activities without requiring additional bio-signals from wearable devices, and the integration of convolution layers with LSTM that enable the NN to accurately estimate the output from glucose–insulin input data. The proposed RNN models can be integrated with the control algorithm of an AID system to enhance its performance by readjusting the conservativeness and aggressiveness of the AID system.
The remainder of this paper is organized as follows: the next section provides a short description of the data collected from people with T1D. The preprocessing step, including outlier removal, data imputation, and feature extraction is presented in
Section 3.
Section 4 presents various RNN configurations used in this study. A case study with real-world data and a discussion of the results are presented in
Section 5 and
Section 6, respectively. Finally,
Section 6 provides the conclusions.
4. Detection and Classification Methods
Detecting the occurrence of events causing large glycemic variations requires solving a supervised classification problem. Hence, all samples required labeling using the information provided in the datasets, specifically using variables “Activity.duration” and “Nutrition.carbohydrate”. In order to determine the index sets of each class, let
N be the total number of samples and
be the sample duration of physical activity at each sampling time
k. Define sets of sample indexes as:
The label indexes defined by (
11) corresponds to classes “Meal and Exercise”, “no Meal but Exercise”, “no Exercise but Meal”, “neither Meal nor Exercise”, respectively.
Four different configurations of the RNN models were studied to assess the accuracy and performance of each in estimating the joint probability of the carbohydrate intake and physical activity. All four models used 24 past samples of the selected feature variables, and event estimations were performed one sample backward. Estimating the co-occurrences of the external disturbances should be performed at least one step backward as the effect of disturbance variables needs to be seen first, before parameter adjustment and event prediction can be made.
Since the imputation of gaps with a high number of consecutive missing values adversely affects the prediction of meal–exercise classes, all remaining samples with missing values after the data imputation step were excluded from parameter optimization. Excluding missing values inside the input tensor can be carried out either by using a placeholder for missing samples and filtering samples through masking layer or by manually removing incomplete samples.
Each recurrent NN models used in this study encompasses a type of LSTM units [
50] (see
Figure 2) to capture the time-dependent patterns in the data. The first NN model consists of a masking layer to filter out unimputed samples, followed by a LSTM layer, two dense layers, and a softmax layer to estimate the probability of each class. The LSTM and dense layers undergo training with dropout and parameter regularization strategies to avoid the drastic growth of hyperparameters. Additionally, the recurrent information stream in the LSTM layer was randomly ignored in the calculation at each run. At each layer of the network, the magnitude of both weights and intercept coefficients was penalized by adding a
regularizer term to the loss function. The rectified linear unit (ReLu) activation function was chosen as a nonlinear component in all layers. The input variables of the regular LSTM network will have the shape of
, which denotes the size of samples, the size of lagged samples, and the number of feature variables, respectively.
The second model encompasses a series of two 1D convolution layers, each one followed by a max pool layer for downsampling feature maps. The output of the second max pool layer was flattened to achieve a time-series extracted feature to feed to to the LSTM layer. A dense layer after LSTM was added to the model and the joint probability of events was estimated by calculating the output of the softmax layer. Like the first RNN model, the ReLU activation function was employed in all layers to capture the nonlinearity in the data. A
regularization method was applied to all hyperparameters of the model. Adding convolution layers with repeated operations to an RNN model paves the way for extracting features for the sequence regression or classification problem. This approach has shown a breakthrough in visual time-series prediction from the sequence of images or videos for various problems, such as activity recognition, textual description, and audio and word sequence prediction [
51,
52]. Time-distributed convolution layers scan and elicit features from each block of the sequence of the data [
53]. Therefore, each sample was reshaped into
, with
blocks at each sample.
The third classifier has a 2D convolutional LSTM (ConvLSTM) layer, one dropout layer, two dense layers, and a softmax layer for the probability estimation of each class from the sequences of data. A two-dimensional ConvLSTM structure was designed to capture both temporal and spatial correlation in the data, moving pictures in particular, by employing a convolution operation in both input-to-state and state-to-state transitions [
50]. In comparison to a regular LSTM cell, ConvLSTMs perform the convolution operation by an internal multiplication of inputs and hidden states into kernel filter matrices (
Figure 2c). Similar to previously discussed models, the
regularization constraint and ReLU activation function were considered in constructing the ConvLSTM model. A two-dimensional ConvLSTM import sample of spatiotemporal data in the format of
, where
and
, stands for the size of the rows and columns of each tensor, and
is the number of channels/features on the data [
54].
Finally, the last model comprises two 1D convolution layers, two max pooling layers, a flatten layer, a bidirectional LSTM (Bi-LSTM) layer, a dense layer, and a soft max layer to predict classes. Bi-LSTM units capture the dependency in the sequence of the data in two directions. Hence, as a comparison to a regular LSTM memory unit, Bi-LSTM requires reversely duplicating the same LSTM unit and employing a merging strategy to calculate the output of the cell [
55]. The use of this approach was primarily observed in speech recognition tasks, where, instead of real-time interpretation, the whole sequence of the data was analyzed and its superior performance over the regular LSTM was justified [
56]. The joint estimation of glycemic events was made one step backward. Therefore, the whole sequence of features were recorded first, and the use of an RNN model with Bi-LSTM units for the detection of unannounced disturbances was quite justifiable. The tensor of input data is similar to LSTM with 1D convolutional layers.
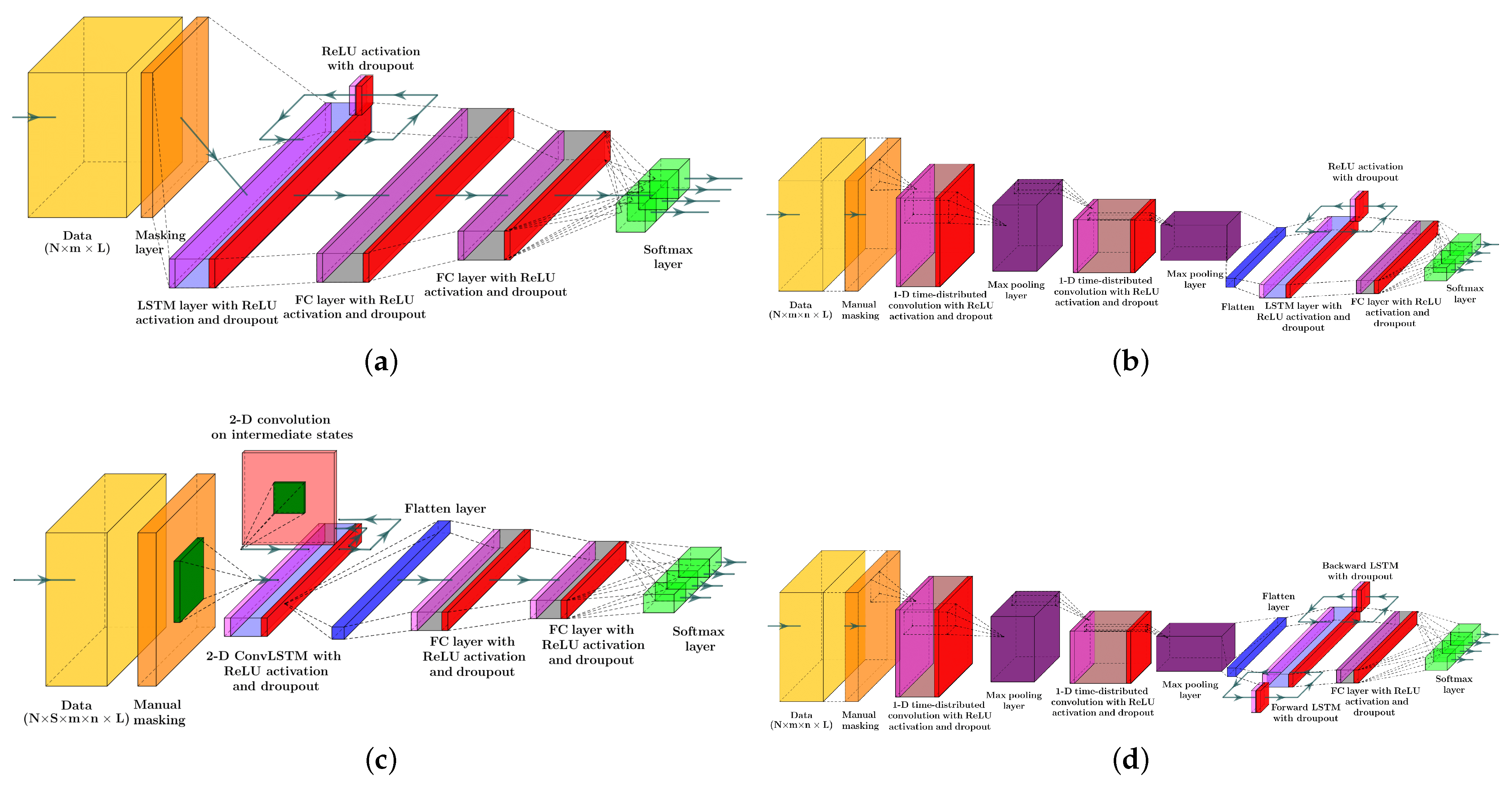
Figure 2 is the schematic diagram of a regular LSTM, a Bi-LSTM, and a ConvLSTM unit.
Figure 3 depicts the structure of the four RNN models to estimate the probability of meal consumption, physical activity, and their concurrent occurrence. The main difference between models (a) and (b) in
Figure 3 is the convolution and max-pooling layers added before the LSTM layer to extract features map from time series data. Although adding convolutional blocks to an RNN model increases the number of learnable parameters, including weights, biases, and kernel filters, calculating temporal feature maps from input data better discriminates the target classes.
6. Discussion of Results
Each classifier was evaluated by testing a 12.5% split of all sensor and insulin pump recordings for each subject, corresponding to 3–12 weeks of data for a subject. The average and the standard deviation of performance indexes are reported in
Table 6. The lowest performance indexes were achieved by 2D ConvLSTM models. Bi-LSTM with 1D convolution layer RNN models achieve the highest accuracy for six subjects out of eleven, and LSTM with 1D convolution RNN for three subjects. Bi-LSTM with 1D convolution layer RNN models outperformed other models for four subjects, with weighted F1 scores ranging from 91.41–96.26%. Similarly, LSTM models with 1D convolution layers achieved the highest weighted F1 score for another four subjects, with score values within 93.65–96.06%. Glycemic events for the rest of the three subjects showed to be better predicted by regular LSTM models, with a weighted F1 score between 93.31–95.18%. This indicates that 1D convolution improves both the accuracy and F1 scores for most of the subjects. Based on the number of adjustable parameters for the four different RNN models used for a specific subject, LSTMs are the most computational demanding blocks in the model. To assess the computational load of developing the various RNN models, we compared the number of learnable parameters (details provided in
Supplementary Materials). These values can be highly informative, as the number of dropouts in each model and the number of learnable parameters at each epoch (iteration) are invariant.
A comparison between 1D conv-LSTM and 1D-Bi-LSTM for one randomly selected subject shows that the number of learnable parameters increases by at least 54%, mainly stemming from an extra embedded LSTM in the bidirectional layer (
Table S1). While comparing adjustable parameters may not be the most accurate way of determining the computational loads for training the models, they provide a good reference to compare the computational burden of different RNN models.
Figure 4 displays a random day selected from the test data to compare the effectiveness of each RNN model in detecting meal and exercise disturbances. Among four possible realizations for the occurrence of events, detecting joint events,
, is more challenging as it usually shows overlaps with
and
. Another reason for the lower detection is the lack of enough information on
, knowing that people would usually rather have a small snack before and after exercise sessions over having a rescue carbohydrate during physical activity. Furthermore, the AID systems used by subjects automatically record only CGM and insulin infusion values, and meal and physical activity sessions need to be manually entered to the device, which is, at times, an action that may be forgotten by the subject. Meal consumption and physical activity are two prominent disturbances that disrupt BGC regulation, but their opposite effect on BGC makes the prediction of
less critical than each of meal intake or only physical activity classes.
The confusion matrices of the classification results for one of the subjects (No. 2) are summarized in
Table 7. As can be observed from
Figure 4 and
Table 7, detecting
(physical activity) is more challenging in comparison to the carbohydrate intake (
) and
(no meal or exercise). One reason for this difficulty is the lack of biosignal information, such as 3D accelerometer, blood volume pulse, and heart rate data. Some erroneous detections, such as confusing meals and exercise, are dangerous, since meals necessitate an insulin bolus while exercise lowers BGC, and the elimination of insulin infusion and/or increase in target BGC are needed. RNNs with LSTM and 1D convolution layers provide the best overall performance in minimizing such confusions: two meals events are classified as exercise (0.003%) and eight exercise events are classified as meals (0.125%).
Two limitations of the study are the quality and accuracy of data collected in free living and the variables that are measured. As stated in the Introduction and Data Preprocessing sections, the missing data in the time series of CGM readings is one limitation that we addressed by developing data preprocessing techniques. The second limitation is the number of variables that are measured. In this data set, there are only CGM and insulin pump data and the voluntary information provided by the patients about meal consumption and exercising. This information is usually incomplete (sometimes people may forget or have no time to enter this information). These events can be captured objectively by other measurements from wearable devices. Such data were not available in this data set and limited the accuracy of the results, especially when the meal and exercise occurred concurrently.
The proportion of correctly detected exercise and meal events to all actual exercise and meal events for all subjects reveals that a series of convolution–max-pooling layers could elicit informative feature maps for classification efficiently. Although augmented features, such as the first and second derivatives of CGM and PIC, enhance the prediction power of the NN models, the secondary feature maps, extracted from all primary features, show to be a better fit for this classification problem. In addition, repeated 1D kernel filters in convolution layers better suit the time-series nature of the data, as opposed to extracting feature maps by utilizing 2D convolution filters on the data.