Abstract
Photosensitization of proteins mediated by chromophores is a mechanism commonly employed by nature and mimicked in a broad array of laboratory research and applications. Nature has evolved specialized complexes of proteins and photosensitizers (PS) that assemble to form photoreceptor proteins (PRP). These are used by many organisms in diverse processes, such as energy conversion, protection against photodamage, etc. The same concept has been used in laboratory settings for many applications, such as the stimulation of neurons or the selective depletion of proteins in a signaling pathway. A key issue in laboratory settings has been the relationship between the photooxidation of proteins and conformational changes in host proteins. For several years, we have been interested in creating non-native PRP using porphyrin PS. In this study, we investigated the self-assembled complex between zinc protoporphyrin IX (ZnPPIX) and bovine β-lactoglobulin (BLG) as a model of non-native PRP. Since BLG undergoes a significant conformational transition near physiological pH, the study was carried out at acidic (pH 5) and alkaline (pH 9) conditions where the two conformations are respectively prevalent. We employed a series of steady-state and time-resolved optical spectroscopies as well as gel electrophoresis to experimentally characterize the photosensitization mechanisms and their effect on the host protein. Our results show that ZnPPIX prompts light-dependent modifications of BLG, which appear to be much more significant at alkaline pH. The modifications seem to be driven by photooxidation of amino acid residues that do not lead to the formation of cross-links or protein fragmentation.
1. Introduction
The use of light to manipulate the folding and functionality of biomolecules has attracted significant attention over the years [1,2]. This approach has many appealing aspects, including the ability to use a wide range of wavelengths to activate photosensitive mechanisms, a large selection of photosensitizers (PS) with various photo-responsive properties, an array of fabrication strategies, and the relatively inexpensive and minimally invasive nature of light delivery [1,3,4,5,6,7,8,9]. A common paradigm found in nature and in nature-mimicking constructs encompasses the following features: (1) PS co-factors are linked (covalently or non-covalently) to biomolecules; (2) optical excitation of the PS triggers a photosensitization event (e.g., photoisomerization); (3) the event causes chemical and or conformational modification of the biomolecule; (4) the conformational change activates or suppresses their functions. In nature, these systems are ubiquitous and often categorized as photoreceptor proteins (PRP), which are utilized by many organisms, from bacteria to mammals [10,11]. PRPs have inspired a variety of non-native uses in fields as diverse as neuroscience [4], targeted theranostics [2], fundamental biophysics [1,12], and fuel cells [13].
In nature, most PRPs utilize reversible mechanisms whereby the photoreceptor returns to its pre-excited state through a chemical cycle [14]. In other cases, such as the visual cycle, the process following photon absorption is irreversible, and the PRP must be recycled through a cellular process [15]. The non-native use of complexes of PS and proteins often contends with irreversible mechanisms without having cellular mechanisms that can restore the complexes. In this case, proteins and PS undergo a photoinduced depletion of the system of interest. This mechanism is well-known in photodynamic therapy (PDT) [16,17,18] and leads to a limitation of the treatment. Irreversible changes are a limitation in other non-native applications of PS:protein complexes, such as chromophore/fluorophore-assisted laser/light inactivation (CALI or FALI) of proteins [19,20], whereby the irradiation of the PS:protein complex produces massive chemical and structural changes in targeted proteins [20]. Irreversible changes are typically caused by the reaction between optically excited transient products, such as singlet oxygen (1O2) or radicals, with the side chains of amino acids or the backbone of polypeptides [21].
Typically, the type of PS dictates the molecular mechanisms that follow the absorption of photons. Porphyrin-type PS (such as protoporphyrin IX, PPIX, and its analogs) are efficient generators of oxidizing species, such as 1O2 and radicals [22], which can oxidize protein residues, in particular Met, Trp, Tyr, His, and Cys [23,24]. Modifications of these residues can lead to a number of effects that include small conformational changes, cross-linking, and even fragmentation of proteins [23,24,25]. It is therefore important to correlate the photoinduced chemical changes to their effects on protein conformation to improve the understanding of the mechanisms that lead from the optical excitation of the PS ligand to the conformational changes of the host protein and design strategies for the generation of non-native PS:protein complexes to develop novel PRP systems or for their deployment in biomedical applications [26].
This work focuses on the complex formed by a particular metal PPIX (ZnPPIX, Scheme S1) and a small protein model represented by bovine β-lactoglobulin. ZnPPIX has been used often for its photosensitizing properties [27,28,29], while BLG is widely utilized as a protein model due to its well-characterized structure and ability to bind small ligands [30,31]. At pH > 4.5 and micromolar concentrations, BLG is postulated to be mostly dimeric [32]. BLG has been shown to undergo a significant structural transition near pH 7.4 (Tanford transition), triggered by the deprotonation of the Glu residue at position 89 [33,34]. Due to its properties, despite being a bovine protein, BLG has been investigated as a potential model carrier for drug delivery [35,36,37,38].
Our group has studied the effects of porphyrins as model PS for the creation of non-native PRP [39,40,41]. Among our earlier investigations were the self-assembly of metal-free porphyrins (cationic and neutral) with BLG [39,40,42]. In recent studies, we have demonstrated that metal-free (mf) and metal-PPIXs (including ZnPPIX) non-covalently bind BLG dimers, likely in the groove at the interface between the two monomers (Scheme S1) [43]. In the same study, we showed that the formation of protein–PPIX complexes is much more efficient at alkaline than acidic pH and concluded that the larger efficiency is not due to the Tanford transition of BLG, which occurs at a location distant from the binding site of the porphyrin but is instead due to the fact that the PPIXs remain much more aggregated at acidic pH; thus fewer porphyrin monomers are available to bind the protein. In particular, in the case of ZnPPIX, we estimated that, for identical concentrations of protein and PS, there are twice as many ZnPPIX:BLG complexes at pH 9 than at pH 5. We have also shown that laser irradiation of the Soret band of mf-PPIX docked to BLG prompts conformational modifications of the protein in a pH-dependent manner [39,40]. The larger conformational effects at alkaline pH are the result, in part, of the larger fraction of BLG complexed with porphyrins at pH 9 compared to pH 5 and the fact that, at pH 9, BLG may be less stable [32] as a result of the Tanford transition.
In this study, we characterized the effects of laser irradiation of ZnPPIX:BLG complexes using the photosensitizing properties [28,44] of ZnPPIX. The results of our investigation reveal that laser irradiation of ZnPPIX docked to BLG modifies the Trp residues, as revealed by absorption and emission spectroscopy. This modification may prompt conformational changes of the protein that are, once again, more evident at alkaline pH. In order to establish which photophysical and photochemical mechanisms may lead to the changes in BLG, we also characterized early photophysical events using ultrafast transient absorption. We conclude that the protein changes are likely triggered by oxidation prompted by 1O2 and a photoinduced radical of ZnPPIX (ZnPPIX●+).
2. Materials and Methods
2.1. Chemicals
BLG (95% purity, Pfaltz & Bauer, Waterbury, CT, USA) was purchased as a milk lyophilized powder. ZnPPIX in powder form was purchased from Frontier Scientific (Logan, UT, USA).
2.2. Buffers and Stock Solutions
A 0.1 M citrate phosphate buffer was prepared at pH 5, and a 0.1 M Tris buffer was prepared at pH 9. The possible effects of the pH-dependent conformational states of BLG were probed using two buffers at acidic (pH 5) and alkaline (pH 9) values. Buffers were prepared using 18 MΩ deionized (DI) water and adjusted to the desired final value with H3PO4 or NaOH. Each buffer was allowed to equilibrate for at least 24 h before its use. New buffers were prepared every seven days. Given its limited solubility in aqueous solution, ZnPPIX was prepared as a millimolar stock in dimethylsulfoxide (DMSO). Stocks were stored in the dark and were stable for at least 30 days after their initial preparation.
2.3. Sample Preparation
The protocol to prepare the sample is similar to that recently reported [45]. Briefly, 100 μL of the DMSO stock of ZnPPIX was dissolved in 2 mL of buffer (pH 5 or pH 9). The solution was equilibrated at room temperature, in the dark, for 1 h. It was then transferred to 3.5-mL dialysis tubes with a molecular weight cutoff of 6 KDa (Pur-A-Lyzer Maxi dialysis kit, Sigma Aldrich, St. Louis, MO, USA). The samples were dialyzed in 1 L of the same buffer for 48 h. The dialysis step eliminates DMSO and other low molecular weight impurities from the solutions [45]. Equal amounts of the dialyzed sample were transferred into conical 1.5-mL mini-plastic tubes and centrifuged with an Eppendorf MiniSpin® (Eppendorf North America, Enfield, CT, USA) for 5 min at 14 × g. This step separates larger aggregates of ZnPPIX, which are pelleted at the bottom of the tubes [45]. Equal amounts of supernatant were recovered from the tubes and placed inside two separate amber vials. The UV-vis absorption spectrum of the supernatant was obtained, as explained below, to estimate the concentration of ZnPPIX. Once the concentration was estimated, to one of the two vials containing the ZnPPIX supernatant, an amount of BLG was added to match the concentration of the porphyrin. This mixture was left to equilibrate for at least 1 h before performing experiments. The sample in the other vial containing only ZnPPIX was used as a control.
Concentrations of ZnPPIX and BLG in the solutions were estimated from the optical density of the samples [46] utilizing a molar extinction coefficient for ZnPPIX [45] and for BLG [47]. Since the amount of ZnPPIX lost during dialysis and centrifugation was larger at acidic pH and not entirely predictable, samples had to be diluted to optimal values for each spectroscopic measurement.
2.4. Experimental Techniques
All experiments described below were carried out in triplicates on different days using fresh samples.
2.4.1. UV-Vis Absorption Spectroscopy
Absorption spectra of the samples were recorded using a Cary-100 UV-Vis spectrophotometer (Agilent Technologies Inc., Santa Clara, CA, USA). Spectra were collected between 200 and 800 nm at a speed of 600 nm/min and 1 nm spectral resolution and were corrected for the baseline. All spectra were recorded using 1-cm quartz cuvettes (FireflySci, Brooklyn, NY, USA). The range of the absorption spectra included the region of the aromatic amino acids (250–310 nm), the region of the Soret band (350–450 nm), and the region of the Q-bands (420–750 nm) of the porphyrins.
2.4.2. Steady-State Fluorescence Spectroscopy
Fluorescence emission spectra were recorded with a Perkin-Elmer L55 fluorimeter (Waltham, MA, USA). Spectra were recorded at 1.0 nm/s and 8 nm bandwidth in excitation and emission monochromators. The fluorescence emission of BLG was excited at 294 nm and recorded in the 305–450 nm range. The 294 nm excitation wavelength preferentially excites the Trp residues [48]. Monomeric BLG has two Trp residues at position 19 and 61 [33]. The proximity of Trp 61 to the Cys 60- Cys 161 disulfide bond, however, has been postulated to significantly quench the fluorescence of Trp 61. Thus, BLG can be considered a quasi-single Trp protein for fluorescence purposes [49]. The optical density of the solution at the excitation wavelength was maintained at <0.15 to ensure uniform light distribution in the sample. Corrections for the instrument response, optical density of the solution, and inner-filter effects were carried out according to published protocols [50,51].
2.4.3. Fluorescence Lifetime
Fluorescence lifetime decay was recorded using a time-correlated single photon counting (TCSPC) instrument (Fluorohub, Horiba Sci., Edison, NJ, USA). BLG and ZnPPIX were excited with pulsed LEDs at 294 nm and 388 nm, respectively (NanoLED, Horiba JobinYvon, Edison, NJ, USA). The LEDs had similar pulse durations (~1 ns) and identical repetition rates (1 MHz). The emission decay of BLG was recorded at 334 ± 4 nm (corresponding to the emission peak of the protein), while the emission decay of ZnPPIX was recorded at 630 ± 8 nm.
The 388 nm LED, albeit not matching the maximum absorption of ZnPPIX, is within the Soret band and subject to Kasha’s rule [52]. The decay data were analyzed using the deconvolution software DAS6.2 (Horiba Sci., Edison, NJ, USA), as explained elsewhere [53]. The fluorescence decay was assumed to be the sum of exponentials with amplitudes and time constants [53]:
A fitting was deemed satisfactory when, simultaneously, the values of the reduced χ2 < 1.4 and the Durbin-Watson parameter were between 1.8 and 2.0 [54]. The pulse of the NanoLEDs was recorded from a scattering solution of glycogen in DI. Data analysis was carried out for one, two, or three lifetime components. The best fit was selected based on changes in the χ2 value from one component set to the other. From the best-fitting parameters of the individual decay components (αi, τi.), the average value of the decay lifetime was calculated as follows [55]:
2.5. Laser Irradiation
Samples were irradiated using a laser diode at 405 nm (Power Technology Inc., Alexander, AR, USA). The beam was expanded and collimated to a ~1.0 cm circular cross-section. Before reaching the sample, the beam traversed a set of neutral density filters that reduced the irradiance at the sample to 22 mW/cm2. For context, this irradiance is approximately 5-fold smaller than that typically used in phototherapy [56]. Samples were irradiated at fixed irradiance for different amounts of time to produce different fluences (J/cm2). Samples were irradiated in the same cuvettes used for fluorescence and had optical densities at 405 nm of ~ 0.15 to ensure uniform energy deposition from the laser source. Spectroscopic measurements were carried out immediately following each irradiation exposure.
2.6. Femtosecond Transient Absorption Spectroscopy (FTAS)
Optical excitation of porphyrins (PP) in the region of the Soret band prompts a transition from the ground singlet state, PP, to the second excited singlet state, 1PP*(2), corresponding to a particular rearrangement of the electronic distribution within the molecule [57]. From PP*(2), porphyrins are postulated to undergo several transitions that include internal conversion (IC) to at least two nearly degenerate first-excited singlet states, 1PP*, intersystem crossing (ISC) to the excited triplet-state 3PP* as well as charge transfers that yield the formation of radical ions (e.g., PP●+). Transitions to 3PP*, as well as neutral and charged radicals, can also proceed from 1PP*. Likewise, PP●+ can be formed from 3PP* [44]. Most of these mechanisms manifest in a time range of s to s [58], and each separate transient dictates the subsequent slower steps s. For instance, 3PP* can sensitize the formation of singlet oxygen (1O2) [59], while radicals lead to other reactions [44]. The lifetime and reactivity of these species with chemical groups in proteins dictate the subsequent effects that may lead to changes in the structure of polypeptides [60,61,62].
These early photophysical mechanisms can be observed through FTAS, which is a pump–probe experimental technique created to detect optically excited transient species in the ultrafast (<10−8) time range. FTAS experiments were carried out using a HELIOS Fire (Ultrafast Systems, Sarasota, FL, USA) instrument described elsewhere [63]. The source of pump and probe beams was provided by splitting the output of a Ti:sapphire regenerative amplifier (SpitFire Ace, Spectra-Physics, Milpitas, CA, USA) at 800 nm, pulse duration of ~80 fs, and repetition rate of 1 kHz. The pump pulses were generated by sending part of the 800 nm source to an optical parametric amplifier (OPA; TOPAS-Twins, Light Conversion, Vilnius, Lithuania), where it was converted to 410 nm [64]. The pump beam radius was ~100 μm at its focal point [65] and passed through a chopper wheel to reduce the repetition rate to 500 Hz before reaching the sample. The probe beam consisted of supercontinuum (430–760 nm) pulses generated from the other fraction of the fundamental 800-nm pulse, incident upon a sapphire window. The probe beam repetition rate was 1 kHz, thus double the repetition rate of the pump. This is required so that the supercontinuum probes the sample half of the time following the pump pulse and half of the time without a prior pump pulse, thus recreating the operation of a single beam absorption spectrometer to calculate the transient absorbance of the sample. The pump and probe beams were set at the magic angle (54.7°) with respect to each other to avoid photoselection effects due to polarization. After the sample, the pump beam was discarded through a hole in a parabolic mirror that deflected the probe beam onto a second parabolic mirror and the detector [63]. From the raw values of the transmitted light under the two alternating conditions (pump-on, pump-off), the software calculated the change in absorption of the sample (ΔA) upon excitation by the pump wavelength. It thus detected the time profile of the absorption spectrum of the excited states, which we designated as the excited state absorption or ESA [63].
In our experiments, the average energy of the pump beam (410 nm) at the sample was 30 μJ, and the optical density of the samples was ~0.5. Samples were stirred with a micro-stirring magnetic bar at the bottom of the cuvette. In addition, the cuvette was mounted on a stage controlled by the instrument’s software that moved the sample in a rectangular pattern. The combination of stirring and constant motion avoided photodegradation of the sample.
2.7. 1O2 Detection
As mentioned in the introduction, one of the possible photochemical mechanisms prompted by the optical excitation of the ZnPPIX ligand is the sensitization of 1O2 [59,66]. We attempted to detect 1O2 using singlet oxygen sensor green (SOSG) [67] through established protocols [68]. Assuming an extinction coefficient of 18,000 M−1 cm−1 for SOSG at 473 nm [69], we adjusted the concentration of SOSG in the sample solution to ~3 μM. This is comparable to that of ZnPPIX:BLG and its contribution to the OD at 405 nm (laser irradiation) was extrapolated to be ~0.003, approximately 2% of the absorption of ZnPPIX:BLG. This is important to establish the contribution to the fluorescence signal due to the potential self-sensitization of 1O2 by SOSG [70,71]. In a control experiment, we also irradiated SOSG alone in buffer solution (pH 5 and pH 9) at a concentration identical to that used in the ZnPPIX:BLG sample and SOSG not exposed to irradiation.
2.8. Native Gel Electrophoresis
In order to investigate whether the photosensitized reactions cause BLG to fragment or form crosslinks, we carried out native polyacrylamide gel electrophoresis (Native-PAGE). Native-PAGE was performed as described by Schägger et al. [72]. A 15% separation gel with a 5% stacking gel was prepared with the standard concentrations of acrylamide, bis-acrylamide, TEMED, and ammonium persulfate, and an appropriate gel buffer. Samples containing 10 μg of protein were loaded in each lane. The gels were run in a Mini-Protean electrophoresis chamber (Bio-Rad, Hercules, CA, USA) at 100 volts for 90 min using a glycine Tris running buffer (pH 8.8). After electrophoresis, the gels were washed three times in DI water and stained with Bio-Safe Coomassie Brilliant Blue R-250 (Sigma Aldrich, St. Louis, MO, USA) for 1 h in a low-speed shaker. After staining, a final 1-h wash in water was performed. The gels were then digitized using an iBright CL 1500 imaging system (ThermoFisher Sci., Waltham, MA, USA). A Precision Plus Protein Dual Color Standard (BioRad) was used to estimate the molecular weights.
3. Results and Discussion
3.1. UV-Visible Absorption
The spectroscopic features displayed by the ZnPPIX:BLG show differences that depend on the pH of the solution.
3.1.1. pH 5
Prior to laser irradiation, the spectrum of ZnPPIX:BLG shows a broad peak with a maximum at 394 nm in the Soret region of the porphyrin, as well as a peak at 278 nm in the UV (Figure 1A). The peak at 394 nm is consistent with a mixture of monomeric and aggregated ZnPPIX [43] with a larger fraction of the latter, which is not eliminated by our sample preparation under acidic conditions. One also notices two Q-bands at 556 nm and 590 nm, which represent a D4h symmetry (essentially flat) for the porphyrin macrocycle [73]. Conversely, the peak at 278 nm is characteristic of the aromatic amino acid residues [74]. The saturated spectrum < 250 nm is due to the contribution of the protein amide bonds. The comparison of the absorption spectra with and without BLG shows only a very small (1 nm) blue shift of the Soret band in the presence of the protein (Figure S1A). This suggests that the ZnPPIX:BLG complex fraction is small. An exact fraction is difficult to determine optically because the extinction coefficient of ZnPPIX aggregates (likely heterogenous) cannot be determined. However, an upper limit can be estimated from the absorption peaks at 278 nm and 394 nm. If the first can be considered entirely from BLG and the second from bound ZnPPIX, using the molar extinction values listed in Section 2.3, approximately 3% of the BLG had complexed with the PS.
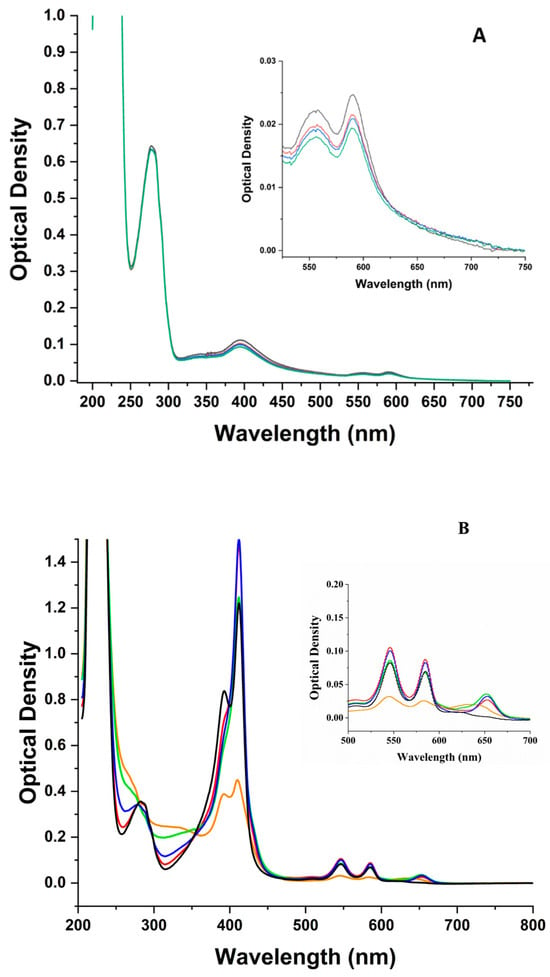
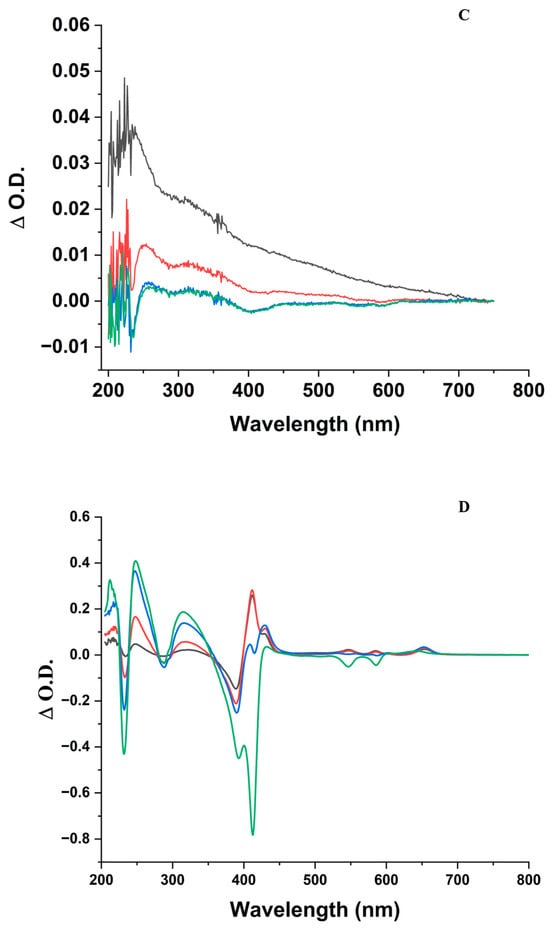
Figure 1.
Absorption spectra of the ZnPPIX:BLG complex at increasing incident fluence (J/cm2) of the 405 nm laser. (A) pH 5. (Black) = 0 J/cm2; (Red) = 48 J/cm2; (Blue) = 216 J/cm2; (Green) = 864 J/cm2. (B) pH 9. (Black) = 0 J/cm2; (Red) = 24 J/cm2; (Blue) = 72 J/cm2; (Green) = 288 J/cm2; (Orange) = 864 J/cm2. The inset of (A,B) shows the Q-band region of the spectrum on an appropriate scale. (C) Difference absorption spectra calculated by subtracting the spectrum of non-irradiated ZnPPIX:BLG from irradiated ZnPPIX:BLG at increasing fluence. pH 5. (black): after 24 J/cm2; (red): after 72 J/cm2; (blue): after 288 J/cm2; (green): after 864 J/cm. (D) Difference absorption spectra calculated by subtracting the spectrum of non-irradiated ZnPPIX:BLG from the ones irradiated at increasing fluence. pH 9. (black): after 24 J/cm2; (red): after 72 J/cm2; (blue): after 288 J/cm2; (green): after 864 J/cm2. Notice the bleaching of the Soret band (~380–450 nm) and the Q bands (~500–600 nm), the decrease in the contribution of BLG near 278 nm (aromatic amino acids) and the increase near 240 nm, 325 nm, 625 nm (black at pH 5 and green at pH 9), and 650 nm.
Laser irradiation of the solution causes an increase followed by bleaching of the Soret band by ~12% at 40 J/cm2). However, a further ~18-fold increase in fluence only causes an additional 6% photobleaching (Figure 2A). Photobleaching does not appear to be accompanied by shifts of the Soret peak. Without BLG, laser irradiation produces a very modest (~5%) photobleaching (Figure 2A), even at the largest fluence delivered in our experiments (Figure S2).
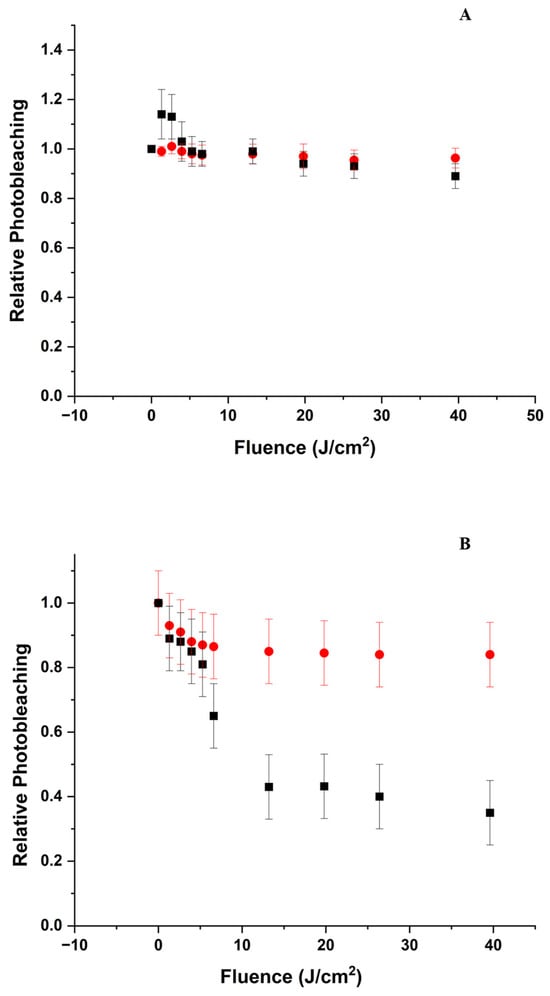
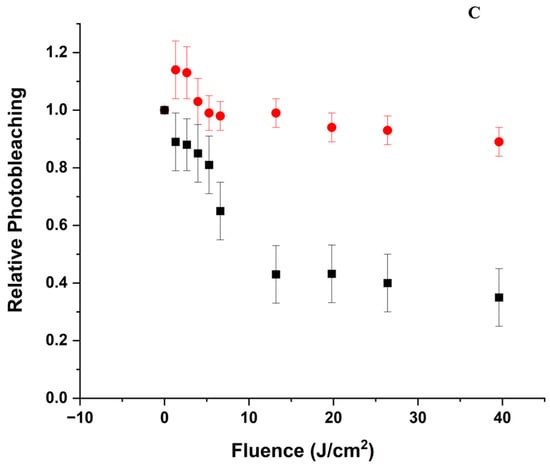
Figure 2.
Relative change in the overall intensity of the Soret band as a function of delivered fluence at 405 nm. The change is represented as the ratio between the absorption maximum in the Soret band region normalized to the absorption maximum before irradiation. (A) Relative photobleaching at pH 5 of ZnPPIX (●) and the ZnPPIX:BLG complex (■); (B) Relative photobleaching at pH 9 of ZnPPIX (●) and the ZnPPIX:BLG complex (■); (C) comparison of the relative bleaching of the ZnPPIX:BLG complex at pH 5 (■) and pH 9 (●).
By taking the difference between irradiated and non-irradiated spectra (Figure 1C), one notices that the small photobleaching in the presence of BLG does not lead to the formation of remarkable new features in the Soret region or the Q-bands. Meanwhile, in the UV region where the aromatic amino acids dominate, one notices a modest growth of two components near 250 nm and 320 nm.
3.1.2. pH 9
Prior to laser irradiation, the spectrum of ZnPPIX:BLG shows two peaks at 412 nm and 393 nm in the Soret region, as well as a peak at 278 nm (Figure 1B). Figure S1B shows the spectra of ZnPPIX in buffer and in the complex with BLG at the same pH. The spectrum of the complex is narrower and shifted by 4 nm to longer wavelengths, indicating that in the complex, ZnPPIX is mostly monomeric, while without the protein, it remains significantly aggregated [43,75]. The region > 500 nm shows two Q-bands that are significantly red-shifted and narrower compared to the pH 5 samples and indicate that even at this pH and upon forming a complex with BLG, the porphyrin maintains a flat D4h symmetry.
Irradiation with the 405 nm laser causes significant photobleaching of the Soret band accompanied by a change of the ratio between the two peaks from ~2 before laser irradiation to ~1.2 after 884 J/cm2. A remarkable change can also be seen in the region of the Q-bands with the appearance of a band near 650 nm that bleaches and shifts to 627 nm upon further irradiation (Figure 1B inset). Figure 2B shows that within the first ~40 J/cm2, the absorption of the Soret band decreases by nearly 65% (Figure 2B), and further irradiation, up to 884 J/cm2, bleaches the absorption by only an additional 5%. Conversely, the photobleaching of ZnPPIX in the absence of BLG at the same pH is limited to < 20% of the initial value (Figure 2B). Thus, the photosensitizing effects are much more significant in the complex. By estimating the fraction of complex as we described at acidic pH, we determined that ~50% of the BLG is complexed with ZnPPIX.
The comparison of the absorption data between pH 5 and pH 9 with and without BLG (Figure 1 and Figure 2) yields an initial scenario for the photoinduced effects. At acidic pH, the ZnPPIX:BLG complex fraction is very small (~3%) compared to that at pH 9 (~50). This not only means that more BLG molecules are occupied by a PS but also that since binding occurs for monomeric ZnPPIX, the fraction of monomeric PS is much larger at alkaline pH. This scenario is consistent with the data shown in Figure 2. At both pH values, the photobleaching in the presence of BLG is larger, however at pH 5, the decrease is minimal (< 10%), while at pH 9 it is > 60%, indicating that at acidic pH, ZnPPIX is mostly aggregated with or without BLG present. Aggregated porphyrins have negligible photophysical mechanisms [76] since the absorbed photon energy is dispersed mostly through heat instead of mechanisms such as the formation of triplet states or radicals, which can lead to the photobleaching of ZnPPIX. Conversely, at alkaline pH, photobleaching is much more significant with and without BLG (Figure 2B,C). At this pH, ZnPPIX (like most protoporphyrins) is more soluble [28,77]; thus, the fraction of monomeric ZnPPIX is much larger. Monomeric ZnPPIX is more likely to initiate photophysical events such as intersystem crossing to the triplet state or the formation of radicals [22,44], which may lead to the photobleaching of the porphyrin and the creation of photoproducts such as that observed at 650 nm. This band can be associated with the formation of ZnPPIX●+ [44], which, as our data suggest, requires the protein to form. The shift of this band at increasing fluence also indicates that this product may react further with the environment to create a product that absorbs at 625–627 nm, such as chlorine. As Figure 2B shows, larger solubility is not the sole factor causing the increased photochemical activity since the presence of BLG increases the photobleaching by >2-fold.
Since we want to establish the effects of PS irradiation on the protein, the data discussed raises questions on the role of BLG and whether the protein is affected by the irradiation of the PS ligand.
The difference spectra (Figure 1C,D) shed light on the mechanisms at play in the complex. At pH 9, where the photobleaching of ZnPPIX is larger (Figure 2) and a photoproduct appears at 650 nm (Figure 1B,D), a feature emerges in the UV region of the spectrum where the aromatic amino acids of BLG dominate the signal. The increase in irradiation and consequent photobleaching of the Soret band is accompanied by two peaks at 243 nm and 310 nm (Figure 1D). Comparing Figure 1C,D one notices that the same bands seem to appear at acidic pH but that the effect is at least 20-fold larger at alkaline pH. Bands in this region (i.e., ~240 nm and ~315 nm) are characteristics of N-formylkynurenine (NFK), which is a known oxidation product of Trp [78]. Therefore, the difference spectra of Figure 1D suggest that laser irradiation of the PS in the ZnPPIX:BLG complex prompts photophysical and photochemical mechanisms that lead to the modification of Trp residues to NFK. The fact that the bands can also be observed at pH 5 (Figure 1C) at a much lower intensity suggests that the same mechanisms occur at acidic pH; however, because of the much smaller ZnPPIX:BLG complex fraction (3% vs. 50%), their relative contribution in the spectra is much smaller. The fact that at pH 9, despite significant photobleaching of ZnPPIX, the same bands do not appear in the absence of BLG rules out that these bands are photoproducts of the porphyrin ligand. Also noteworthy is that the increase of the 243 nm and 310 nm bands correlates to the disappearance of the 650 nm band and its shift to shorter wavelengths. Therefore, the band at 625 nm could be a product formed by the reaction of the ZnPPIX photoproduct with amino acid residues such as Trp, leading to its modification to NFK. Unfortunately, direct attempts to confirm the presence of KYN or N-formylkynurenine (NFK) were inconclusive. Therefore, we cannot rule out that the spectral effects seen <350 nm are due to other events, including the partial unfolding of the protein [79]. However, our data are a strong initial indication that the conversion of Trp to NFK is occurring in the ZnPPIX:BLG complex. Such conversion occurs at both pH values but is much clearer at alkaline pH because the fraction of complexes is much larger than at pH 5.
If the Trp → NFK (or Kyn) conversion occurs, it should also manifest in the emission of BLG since Trp residues and, in particular, Trp19 are the leading contributors to the protein fluorescence. NFK and Kyn have photophysical properties different from their precursor (Trp) [80,81]; thus, their formation would be expected to affect the fluorescence of BLG. To address this issue, we should analyze the emission of BLG.
3.2. BLG Emission
3.2.1. pH 5
As one observes from Figure S3A, the emission spectra of BLG normalized for the optical density (OD) at = 294 nm remain virtually constant at different fluence values. This is reflected by the relative change of the integrated emission intensity of BLG, which changes marginally with increasing laser fluence (Figure 3). This behavior is consistent with the data presented in Section 3.1. At pH 5, laser irradiation causes minimal photobleaching of the Soret band and only the faint appearance of the 250 nm and 310 nm bands (Figure 1C). Since we estimated that only 3% of BLG is complexed with ZnPPIX, it is expected that the effects on BLG emission are negligible since even in the event of full conversion of Trp to NFK or Kyn, the fraction of BLG in which the conversion occurred is so small that the native unaffected BLG dominates the fluorescence spectra of the irradiated samples.
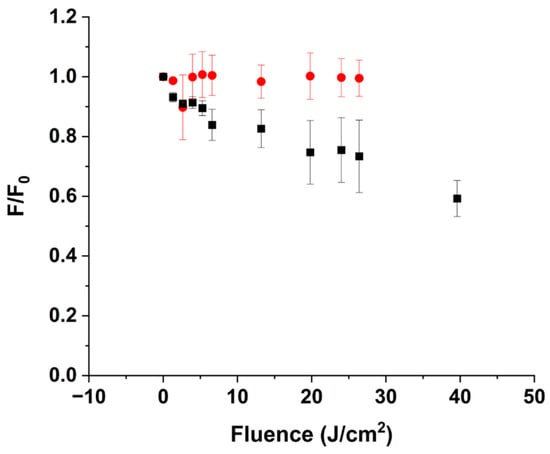
Figure 3.
Relative change in the overall intensity of the BLG emission in the BLG:ZnPPIX complex ( = 294 nm), as a function of delivered laser fluence at 405 nm. The relative change is represented as the ratio between the fluorescence intensity of BLG after irradiation () and before irradiation (). pH 5 (●); pH 9 (■).
3.2.2. pH 9
In agreement with a much larger fraction of BLG complexed with ZnPPIX and a much larger photobleaching of the PS ligand, the effects of laser irradiation on the protein host are much more significant at alkaline pH. Figure S3B shows that the BLG emission upon excitation at 294 nm decreases with increasing irradiation laser fluence. At the largest dose delivered, BLG fluorescence decreases by ~70% (Figure S3B). Since the spectra are normalized for the OD at trivial effects simply due to reduced absorption are avoided. This evidence is better analyzed within the context of the absorption changes of the band at 278 nm (Figure 1C) and the conversion of Trp residues to NFK or Kyn. The lack of blue shift of the emission maximum (Figure S3B) rules out an increased relative contribution of Tyr residues (whose fluorescence is substantially blue-shifted compared to Trp) created by the reduced contribution of Trp [82]. Therefore, the fluorescence decrease is consistent with the chemical transformation of Trp residues into NFK or Kyn. NFK and kynurenine (Kyn) have exceedingly small molar extinction coefficients at 294 nm [83] as well as an emission quantum yield in aqueous solution that is approximately 0.6% of the one of Trp in BLG [49,80] and an emission maximum > 450 nm [84]. Thus, under the conditions of our experiments, the transformation of Trp residues into NFK or Kyn would manifest as a simple decrease in protein fluorescence.
Therefore, the steady-state fluorescence emission data support the interpretation that the photophysical/photochemical mechanisms of the PS ligand lead to the conversion of Trp to NFK or Kyn.
3.3. BLG Fluorescence Decay
Additional information on the effects that the irradiation of the PS-ligand produces on the protein host can be collected from fluorescence decay. The emission decay of Trp residues in proteins is, in fact, sensitive to the chemical modification of side chains and conformational changes of the polypeptides.
3.3.1. pH 5
In agreement with previous results by our group and others [49,53], the fluorescence decay of ZnPPIX:BLG, before laser irradiation, is best fitted with two exponential components (Table 1A). A sub-nanosecond component ( in Table 1A) and a component near 1.6–1.7 ns ( in Table 1A). They combine to yield an average lifetime of ~1.40 ns, which is shorter than that of BLG alone, likely because of distance-dependent fluorescence resonance energy transfer (FRET) between the Trp residues and ZnPPIX ligand, which we reported previously [43].

Table 1.
Fluorescence decay lifetime data for BLG at acidic pH upon excitation at 294 nm at different irradiation energy densities.
As Figure 4A and Table 1A show, irradiation of ZnPPIX at 405 nm produces nearly negligible effects. The analysis of the fluorescence decay shows a 7% decrease in the average lifetime from 1.40 to 1.36 ns (Table 1A), which is within the experimental error. This evidence is consistent with our interpretation. Since the effects of laser irradiation on the steady state fluorescence of BLG in the ZnPPIX:BLG complex are nearly negligible (Figure 3A), so are the effects on the fluorescence decay of the protein.
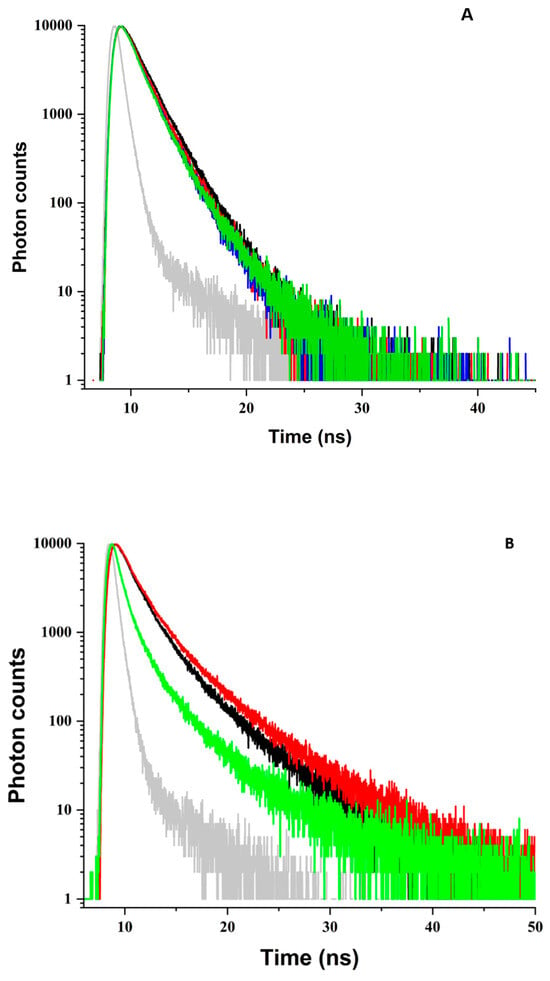
Figure 4.
Fluorescence decay, upon excitation at 294 nm, of the BLG:ZnPPIX complex as a function of laser irradiation energy density at 405 nm. (A) pH 5. (Black) = 0 J/cm2; (Red) = 48 J/cm2; (Blue) = 216 J/cm2; (Green) = 864 J/cm2. (B) pH 9. (Black) = 0 J/cm2; (Red) = 24 J/cm2; (Green) = 288 J/cm2;. In both figures the gray curve represents the instrument response or .
3.3.2. pH 9
A different scenario emerges for the complex at alkaline pH. In agreement with our previously reported results [53], at this pH, the fluorescence decay of the ZnPPIX:BLG complex before laser irradiation is better fitted by three (instead of two) exponential decays reflecting possible pH-induced conformational changes in proximity to the Trp residues [85] (Table 1B). Overall, the components yield an average fluorescence decay lifetime of 2.16 ns, which is > 800 ps longer than at pH 5. Laser irradiation of the ZnPPIX ligand causes changes in the fluorescence decay of BLG that are much more substantial than at acidic pH (Figure 4B) and in agreement with the evidence that at this pH, ~50% of BLG is complexed with PS. For irradiation up to 74 J/cm2, the average decay lifetime increases to 2.56 ns (an 18% increase from the non-irradiated sample). This is driven mostly by an increased contribution of the longer decay component (Table 1B). Upon further exposure, however, the average decay lifetime decreases sharply to 1.12 ns (a decrease of over 1 ns). The sharp decrease is driven by a 2.5-fold increase in the contribution of the shortest decay component. The sharp decrease in average lifetime is yet more evidence consistent with the photoinduced conversion of Trp to NFK or Kyn. These Trp products contribute negligibly to fluorescence upon excitation at 294 nm [84]. The decrease in overall fluorescence intensity, which contributes to the region of later channels, produces a drastic relative increase in the contribution of the shortest decay component. In fact, as we discussed in our previous study [86], the decrease in overall fluorescence intensity produces a relative increase in the contribution of scattering. Since scattering is “instantaneous” on the time scale of our instrument, it contributes significantly only in the early channels of the decays so that the net result is an apparent increase of the relative contribution of the shortest component and a decrease in the average decay lifetime (Table 1B). However, the fact that the intermediate component, τ2, decreases by ~500 ps also suggests that conformational changes may occur in BLG. The increased average lifetime at smaller irradiation values (Figure 4B) could be tied to a decreased FRET efficiency [87,88]. Photobleaching (such as that in Figure 1B) or other factors, such as conformational changes in the protein or the re-orientation of ZnPPIX, could reduce the FRET geometric factor, [55], and increase the distance between the donor and acceptor [55], which would decrease the FRET efficiency and increase the decay lifetime of the donor as we see in Figure 5B.
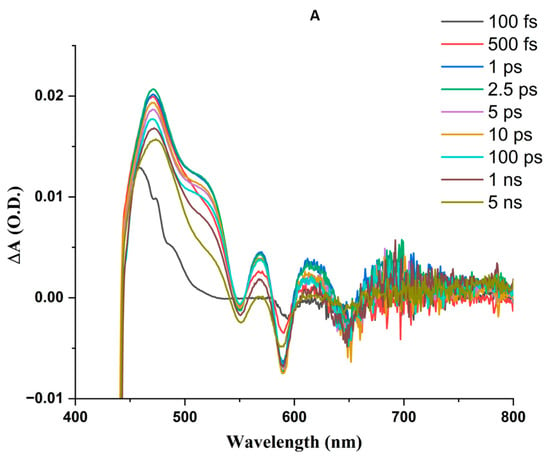
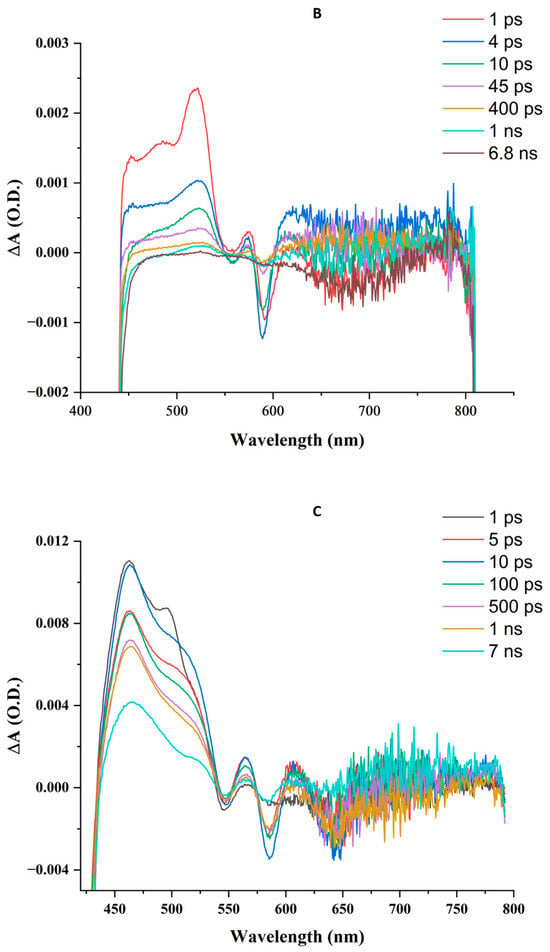
Figure 5.
Transient absorption difference spectral time courses upon femtosecond 410 nm laser excitation. Solutions are in air at similar optical densities at 410 nm, corresponding to concentrations of ZnPPIX of ~5 μM. (A) ZnPPIX in DMSO. (B) ZnPPIX in aqueous solution pH 9. (C) ZnPPIX:BLG complex at pH 9.
3.3.3. Partial Summary
It is useful at this point to summarize some of the findings and reach partial conclusions based on the optical measurements. Laser irradiation of the ZnPPIX causes effects that are much more substantial at alkaline than acidic pH (Figure 1). This is likely caused by the fact that, despite our sample preparation, smaller oligomers of the ZnPPIX are likely dominant in solution at acidic pH, whereas most PPIXs are much less soluble. These oligomers are unlikely to bind BLG [43,89,90] and prompt significant photochemical mechanisms [76]. As a result, the ZnPPIX:BLG complex is a nearly negligible fraction (<3%) of the sample. Conversely, at pH 9 the solution contains > 50% ZnPPIX:BLG complexes where the PS ligand is monomeric (Figure 1B). As a result, at alkaline pH, there is a clear correlation between laser irradiation → photobleaching of the ZnPPIX ligand (Figure 1B and Figure 2B,C) → appearance of UV absorption bands at 240 nm and 310 nm (Figure 1D) → loss of BLG fluorescence (Figure 3 and Figure S3B) → changes in fluorescence decay of BLG (Figure 4B). These correlations are consistent with ZnPPIX photosensitizing the conversion of Trp residues to NFK or Kyn. Therefore, the data presented thus far indicate that ZnPPIX induces chemical and potentially conformational modification to BLG.
These partial conclusions raise questions about which mechanisms may be responsible for the photosensitization effects.
3.4. Femtosecond Transient Absorption Spectroscopy (FTAS)
In order to better understand the nature of the photosensitizing mechanisms of the bound PS, we investigated ZnPPIX:BLG as well as ZnPPIX alone in aqueous solution and DMSO using FTAS. In all cases, the wavelength of the laser pump was 410 nm, which is the closest to the 405 nm steady-state irradiation that our FTAS system could provide.
3.4.1. ZnPPIX in DMSO
This can be considered a good model for a monomeric porphyrin [86,91]. The transient spectra (Figure 5A) display positive ΔA peaks at 471 nm, 569 nm, and 615 nm, as well as a shoulder at 515 nm and broad absorption with a peak near 685 nm. The peak at 471 nm shifts to longer wavelengths (474 nm) as it decays. The 515 nm shoulder fades at longer time points. The peaks at 569 nm and 615 nm form within 1 ps of the laser pulse and decrease at longer time points without a significant shift. Negative ΔA peaks also form near 550 nm and 650 nm, albeit in a noisy region of the supercontinuum where it is difficult to identify the exact position of the minimum. A more prominent negative peak forms within 10 ps of the laser pulse near 590 nm.
The fitting of the short kinetics (Figure 6A) reveals that all positive peaks have a sub-ps rising component of 217 ± 70 fs (470 nm) fs, 196 ± 20 fs (515 nm), 220 ± 92 fs (569 nm), and 277 ± 66 fs (615 nm). The 515 nm shoulder requires a decay of 1.47 ± 0.21 ps, which contributes <2% to the signal. Meanwhile, the 569 nm peak requires a second slower rising kinetics of 1.05 ± 0.33 ps, which contributes ~26% to the overall signal compared to the 74% contribution of the shorter component (Table 2). On a longer time scale (7 ns), the 470 nm peak has a more complex time evolution (Figure 6B) that includes two decays of time constants 253 ± 25 ps and 6.89 ± 0.88 ns, as well as a component that grows with time constant 5.24 ± 0.57 ns.
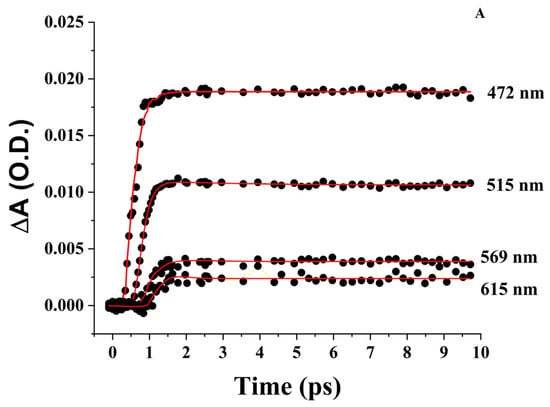
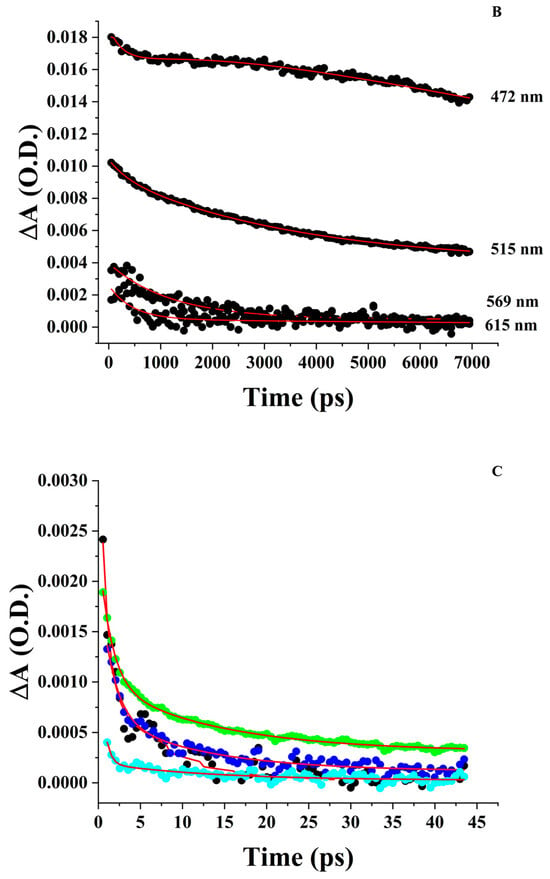
Figure 6.
FTA kinetics. (A) ZnPPIX in DMSO in the 10 ps time window. (B) ZnPPIX in DMSO in the 7 ns time window. (C) Zn ZnPPIX in aqueous solution pH 9 in the 50 ps window. (Black) = 452 nm; (Blue) = 482 nm; (Green) = 526 nm; (Cyan) = 576 nm. (D) BLG:ZnPPIX complex at pH 9 in the 8 ps window. PPIX in aqueous solution pH 9 in the 50 ps window. (Black) = 452 nm; (Blue) = 482 nm; (Green) = 526 nm; (Cyan) = 576 nm. (E) BLG:ZnPPIX complex at pH 9 in the 7 ns window. In red are the fitted curves.

Table 2.
FTAS kinetics obtained from ZnPPIX and the BLG:ZnPPIX complex. Lifetimes are in picoseconds. The arrows next to the lifetimes indicate whether the kinetics are rising or decaying. (↑ indicates kinetics of formation; ↓ indicates decay kinetics).
On the 7 ns time scale, the 515 nm peak also requires multiple decay components of 354 ± 181 ps, contributing 13% of the signal, and 3.47 ± 0.12 ns, contributing 87% of the signal (Figure 6B). Unlike the kinetics at 470 nm, those at 515 nm do not require a rising component with ps or ns duration. The same is true for the 569 nm peak that requires only decay components of 354 ± 176 ps and 1.52 ± 0.11 ns (Figure 6B). The 615 nm has decay kinetics of 226 ± 82 ps. The fitting of all kinetics requires the inclusion of an “infinitely” long-lived component, which indicates the presence of a transient with positive ΔA and a lifetime much longer than the longest time window of the FTAS experiment; this is consistent with the presence of a triplet state or a ZnPPIX radical.
The negative peaks at 550 nm and 590 nm are characterized by an initial decrease with a time constant of 411 ± 57 and fs 426 ± 72 fs, respectively. In a longer time window at 550 nm, the initial decrease is followed by a slower recovery with time constants between 0.9 and 1.8 ns. The transient absorption, however, never recovers to baseline values, indicating the permanent (or at least very long-lived) loss of absorption, which can be correlated to the transition to a long-lived triplet state or radical and is consistent with photobleaching.
The ultrafast photophysics of most porphyrins excited in the Soret region is moderately complex [92] due to a number of nearly degenerate electronic configurations. In addition, many porphyrins have considerable yields for the formation of triplet states [93,94,95] as well as intra and inter-molecular charge transfer that can proceed from the singlet and triplet excited state [96,97,98,99].
We interpreted the data based on the current understanding of the ultrafast photophysics of protoporphyrins [44,100]. The interpretation in DMSO can then be used to help discern the FTAS in an aqueous solution and the complex with BLG.
The region between 450 nm and 530 nm contains overlapping contributions from the species that peak at 470 nm and 515 nm. Considering the analysis of Benavides et al. [44], we attribute the 470 nm peak to the excited state absorption (ESA) of the triplet state (3ZnPPIX*) and the peak at 515 nm to the ESA of the singlet state 1ZnPPIX*. The differences in the peak wavelength between our sample and that of Benavides et al. [44] are likely due to the different environmental conditions (DMSO vs. bacterioferritin). The overlap between the two bands also causes the kinetics to overlap. In particular, the shorter rising femtosecond component (Table 2) is likely the overall result of the 1ZnPPIX*(2) (Soret) → 1ZnPPIX*(Q-bands) transition [100]) and direct formation of 3ZnPPIX* from 1ZnPPIX*(2) since the ESA of 1ZnPPIX* and 3ZnPPIX* overlap with each other.
The 253 ps decay time constant at 470 nm (Table 2) constitutes only a 3% contribution to the kinetics at 470 nm and could be linked to a fraction of ZnPPIX●+ formed from 3ZnPPIX*, as noted by other groups [58,101]. There are two additional components at 470 nm: a rising contribution of 5.24 ± 0.57 ns and a decay of 6.89 ± 0.88 ns. These could be interpreted, respectively, as the average time for the ESA of 3ZnPPIX* formed by the intersystem crossing from 1ZnPPIX* and ESA of 1ZnPPIX* returning to the ground state since these times are comparable to the ones observed for the fluorescence decay of ZnPPIX (Figure S4).
The kinetics at 515 nm are consistent with the ones expected for the ESA of 1ZnPPIX*. The rising time constant is likely the effect of the formation of 1ZnPPIX* from the Soret band (1ZnPPIX*(2)). The 353 ps time constant is due to the formation of ZnPPIX●+ from 1ZnPPIX*, while the 3.47 ns is consistent with the 1ZnPPIX* average lifetime measured through fluorescence lifetime decay (2.15 ns) as shown in Figure S4.
The kinetics at 569 nm are similar to those at 515 nm. The only difference is a rising component of 1.05 ps, which matches the 1.47 ps decay of the 515 nm kinetics and could be interpreted as a transition from the Q(1,0) and Q(0,0) vibrational states of 1ZnPPIX* [102]. The 515 nm represents the higher energy vibrational state that decays in ~1 ps to the lower (Q(0,0)) vibrational state at 569 nm, which is seen as rising with a similar time constant [92,102].
The two components at 615 nm can be rationalized as the formation of 3ZnPPIX* from the singlet states and ZnPPIX●+ from 3ZnPPIX*.
Finally, the peak near 685 nm, albeit in a noisier region of the spectrum, shows only increasing kinetics of 1.95 ± 0.91 ps, suggesting the formation of ZnPPIX●+ [44].
The underlying “infinitely-long” decay required by the fitting throughout the spectrum is caused by the ESA of 3ZnPPIX* and ZnPPIX●+, whose average lifetimes in organic solvents are estimated to be at least three orders of magnitude longer than the longest time window allowed by our FTAS instrumentation [103] and are, therefore, characterized as “infinite” lifetimes by the fitting software.
3.4.2. Aqueous ZnPPIX
At pH 5, too much aggregated ZnPPIX remains in solution, hindering the ability to resolve meaningful transient signals. Conversely, the transient absorption spectrum of ZnPPIX in aqueous solution at pH 9 shows positive and negative peaks. Positive ΔA peaks can be observed near 521 nm and 574 nm, as well as a broader absorption band at 630 nm (Figure 5B). Negative ΔA peaks appear near 560 nm and 590 nm. Unlike the sample in DMSO, a transient at a shorter wavelength is not clearly observed. Attributions of these transients can be made by comparison with the sample in DMSO. We attribute the peak at 526 nm to the counterpart of the peak at 515 nm in DMSO whereas that at 574 nm is the counterpart of the peak at 569 nm in DMSO. According to the previous discussion, they can be assigned to the ESA of the Q(1,0) and Q(0,0) vibrational states of 1ZnPPIX*. The broad absorption > 600 nm contains contributions from the ZnPPIX●+. Since the peak at a shorter wavelength in DMSO was attributed to 3ZnPPIX*, the absence of a similar peak in alkaline buffer suggests a much lower quantum yield of its formation or a much faster characteristic decay time.
Indeed, the decay kinetics are significantly faster than in DMSO (Figure 6C). The best fitting of the peak at 521 nm shows a complex decay behavior with no rising components and a series of decay components with time constants of 133 ± 82 fs, 1.34 ± 0.50 ps, and 13.6 ± 3.2 ps. Unique among the positive ΔA peaks, it also requires a longer 1.09 ± 0.28 ns component. We believe these are all ESA transients of the Q(1,0) vibrational state of 1ZnPPIX*. The three picosecond and sub-picosecond components are consistent with transitions to the Q(0,0) vibronic state, ISC to 3ZnPPIX*, and a transition to the radical cation (1ZnPPIX●+) [58,100]. The nanosecond decay is comparable to the 1ZnPPIX* → 1ZnPPIX transition to the ground state observed using fluorescence decay lifetime in aqueous solutions [43].
Decays with time constants ~2–3 ps and 10–20 ps are detected in the kinetics associated with other peaks (574 nm, 630 nm, and 740 nm). The 10–20 ps decay matches the recovery kinetics of the negative ΔA peak at 560 nm (13.1 ps) and 590 nm (10.5 ps), indicating that this lifetime is associated with an internal conversion back to the ground state [44]. At the longest time allowed by our instrument (7 ns), the spectrum virtually returned to zero. This is a remarkable difference compared to the case in DMSO and confirms a lower yield for the formation of long-lived transients such as 3ZnPPIX* and 1ZnPPIX●+, or their much shorter lifetime. The recovery of the spectrum back to zero also indicates the absence of other long-lived photoproducts (Figure 5B).
3.4.3. ZnPPIX:BLG at pH 9
The transient absorption spectrum of ZnPPIX complexed with BLG shows more similarities with the sample in DMSO (Figure 5C) than the aqueous sample. This indicates that our sample contains primarily the monomeric porphyrin in the form of the ZnPPIX:BLG complex. The spectrum shows positive ΔA peaks at 462 nm, 565 nm, and 607 nm. In addition, like the sample in DMSO, a prominent contribution is evident at ~510 nm. A broad contribution can also be observed at wavelengths longer than 600 nm.
The kinetics appear to be simplified compared to the sample in DMSO (Figure 6D,E). The kinetics associated with the peak at 462 nm include two decay components of 2.71 ps and 332 ps (Table 2) in addition to the long component. The kinetics associated with the peak at 510 nm require two decays with similar time constants as the 462 nm peak and the long component (Table 2). The kinetics at 565 nm require two components and the long component, but the time constants are different at 6.75 ps and 1.02 ns, respectively (Table 2). The kinetics associated with the peak at 605 nm are significantly different. This peak is the only one where a rising component can be reliably identified (118 fs, Table 2). This rise is followed by a 1.61 ps decay, a much longer but resolvable (5.79 ns) decay, and a much smaller contribution of the long-time constant.
Based on the results and the similarity with the sample in DMSO, the spectral position and kinetics of the peak at 462 nm indicate that 3ZnPPIX* is probably the major transient of interest regarding the photochemical effects on the protein [66]. The prevalent contribution of 3ZnPPIX* would suggest its subsequent reaction with diffusing O2 leads to the formation of 1O2 [104]. Singlet oxygen would then likely react with protein residues and backbone [23,61,81,105,106]. The possible formation of ZnPPIX●+ could also trigger changes in BLG by either promoting charge transfer to protein residues or by reacting with them.
We can rationalize the spectroscopic and kinetic data for ZnPPIX:BLG with the following model. Excitation of the ZnPPIX in the complex produces 3ZnPPIX* and ZnPPIX●+. The former either sensitizes 1O2 [23,107] or decays to form ZnPPIX●+ which can also cause oxidative reactions [21,23]. Either process can prompt oxidation of protein residues [21,23,108]. Since Trp is one of the major targets of photooxidation [23,59,81,109], FTAS supports our interpretation that laser irradiation of ZnPPIX bound to BLG leads to the conversion of Trp to NFK or Kyn. Oxidation of other residues (Met, Cys, Tyr, His) is also likely [23,110] but is not observable with optical spectroscopy. These oxidations may lead to conformational changes in BLG.
3.5. 1O2 Formation
To indirectly confirm FTAS evidence regarding the formation of 3ZnPPIX*, we investigated the possible photosensitization of 1O2 prompted by optical excitation at 405 nm of the ZnPPIX:BLG using SOSG [67]. In addition, because of the inability to obtain FTAS data at pH 5, the SOSG measurement would help fill the evidence gap left by the ultrafast study.
3.5.1. pH 5
Irradiation of the ZnPPIX:BLG complex at 405 nm causes a sudden increase (~30%) of the SOSG fluorescence after the first 1.56 J/cm2. Subsequent irradiation up to 46 J/cm2 produces an additional 40% increase in SOSG fluorescence and shows a tendency to plateau (Figure 7A). However, irradiation of a solution containing free ZnPPIX at concentrations similar to those in the sample containing the complex produces a similar trend in SOSG fluorescence (Figure 7A). It is well known that irradiation of SOSG in the region of the anthracene moiety (<480 nm) can cause self-sensitization of the sensor and its fluorescence increase (Figure S5) [111]. Therefore, we recorded the fluorescence of solutions containing only SOSG at the same pH and concentration as the other samples. As Figure 7A shows, direct irradiation of SOSG also causes an increase in its fluorescence, but this increase is smaller than in the other samples. The solution containing SOSG alone does not show any sudden increase in fluorescence after the first irradiation point (i.e., 1.56 J/cm2) and increases smoothly with fluence. The evidence provided in Figure 7A suggests that photosensitization of 1O2 is nearly identical in samples with and without the complex and, in both cases, is only marginally above the self-sensitization by direct excitation of SOSG (Figure S6). This is in agreement with our interpretation that at this pH, the fraction of ZnPPIX:BLG complex is relatively small, and the optical properties are likely mostly determined by small aggregates of the porphyrin which have negligible yield of triplet state formation and 1O2 photosensitization. The fraction of photosensitized 1O2 is likely a combination of direct SOSG irradiation and photosensitization from the small fraction of monomeric ZnPPIX complexed with BLG.
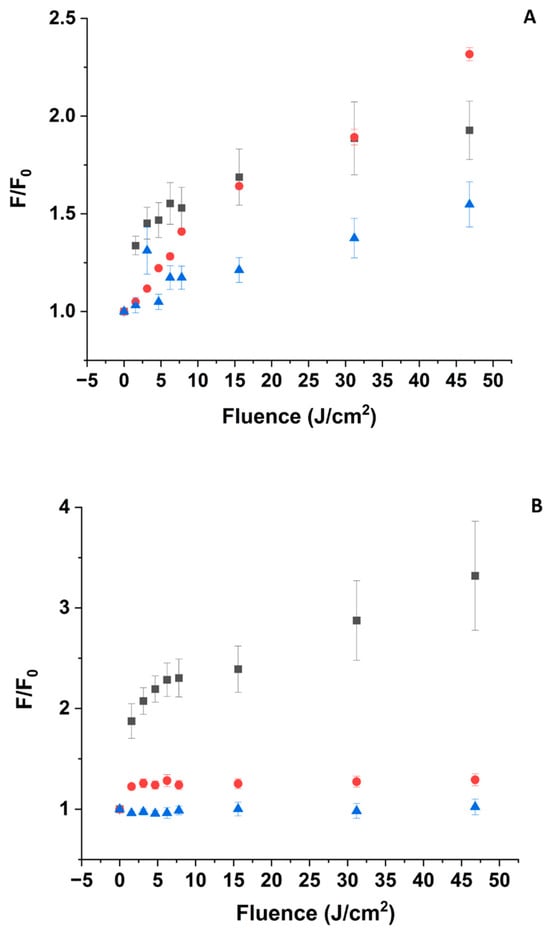
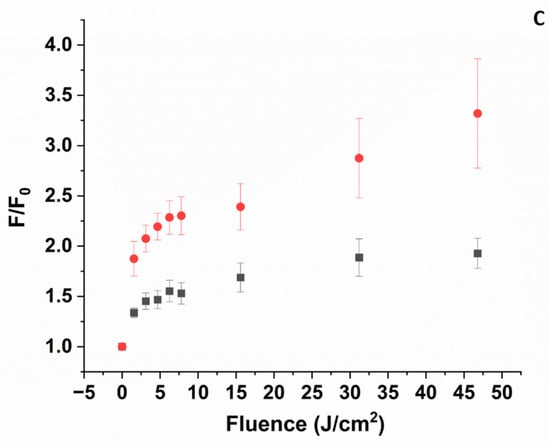
Figure 7.
SOSG fluorescence emission intensity as a function of laser irradiation at 405 nm. Fluorescence of SOSG is excited at 475 nm. [SOSG] ~3 μM. (A) pH 5. (■) in the presence of ZnPPIX:BLG (~5 μM), (●) ZnPPIX (~5 μM), (▲) SOSG alone (self-sensitization). (B) pH 9. (■) in the presence of ZnPPIX:BLG complex (~5 μM), (●) ZnPPIX (~5 μM), (▲) SOSG alone (self-sensitization). (C) Comparison of SOSG fluorescence intensity vs. fluence for ZnPPIX:BLG at pH 5 (■) and pH 9 (●).
3.5.2. pH 9
Irradiation of the complex at 405 nm causes an increase in the fluorescence of SOSG, which is much more significant than at pH 5 (Figure 7B). At 46 J/cm2, the relative increase is 4-fold or ~2.2 times larger than at acidic pH for the same complex. Another significant difference is that irradiating an equivalent concentration of SOSG alone or SOSG in the presence of only ZnPPIX increases the fluorescence of the sensor only marginally (Figure 7B). In contrast to the acidic sample, Figure 7B suggests that photosensitization of 1O2 is much larger in the presence of the ZnPPIX:BLG complex than in the presence of ZnPPIX alone or the self-sensitization of SOSG. This is also in agreement with our interpretation. At pH 9, the fraction of ZnPPIX:BLG complex is dominant. In the complex with BLG, ZnPPIX is monomeric [43]. Monomeric ZnPPIX has a larger yield of triplet state formation and, in turn, can photosensitize 1O2 more efficiently. The formation of triplet-mediated photosensitization of 1O2 is also consistent with the long-lived component of the FTAS decay (see Section 3.4.3). The more efficient photosensitization produced by the ZnPPIX:BLG complex at pH 9 is confirmed by the direct comparison of SOSG fluorescence changes upon the irradiation of the ZnPPIX:BLG complex (Figure 7C and Figure S6).
The evidence provided by FTAS and 1O2 sensing further supports the scenario suggested by absorption and fluorescence spectroscopy. At acidic pH, the fraction of ZnPPIX:BLG where the PS is monomeric is much smaller compared to its free oligomeric forms, and at the same time, most of the BLG in solution is not bound to the PS. Since oligomers do not show any significant photophysical processes in FTAS (in fact, no signal is detected), they do not substantially contribute to the photosensitization of 1O2 (Section 3.5.1). The small contribution (additional to SOSG self-sensitization and sensitization by free ZnPPIX) to the photosensitization of 1O2 (Figure 7A) is likely due to the small fraction of ZnPPIX:BLG complexes at this pH.
Conversely, at alkaline pH where the fraction of ZnPPIX:BLG is almost 20-fold larger, there is direct evidence for the formation of transient oxidizing species such as 1O2 and cation radicals from both FTAS (Figure 6 and Section 3.4.3) and SOSG fluorescence (Figure 7B,C). These species would then react with amino acid residues, leading to their conversion to oxidative products such as NFK and Kyn [81], explaining the observations from absorption and fluorescence (Figure 1, Figure 2 and Figure 3).
Having established that laser irradiation of the ZnPPIX ligand leads to photooxidation of residues in the BLG host, at least the Trp side chain, the question becomes whether the same photooxidation affects the conformation or the structural integrity of the protein. We have been unable to answer this question using circular dichroism spectroscopy because the buffers used in our experiments prevent us from collecting reliable spectra in the far UV region (<190 nm), where the information on protein secondary structures can be retrieved. Instead, we employed native gel electrophoresis (native-PAGE) to test whether photooxidation is extensive enough to cause fragmentation or aggregation of BLG. Both these effects are known to be caused by 1O2 and radicals on proteins [21,61,112,113].
3.6. Gel Electrophoresis
3.6.1. pH 5
Figure 8A shows the native-PAGE gel of the ZnPPIX:BLG complex at increasing 405 nm laser irradiation fluence. All lanes, 1 (non-irradiated BLG) to 9 (ZnPPIX:BLG, 40 J/cm2), show a similar feature: two closely spaced bands near 37 kDa. This is entirely consistent with the presence of native BLG dimers with overall molecular weight postulated to be ~36 kDa. Since we performed native PAGE, the gel in Figure 8A is evidence that BLG is indeed prevalently dimeric under the conditions of our experiments. The somewhat broad distribution and the distinct presence of a “doublet-like” pattern around 37 kDa is likely due to genetic variants of the BLG but also to the fact that native PAGE is sensitive to the orientation of the proteins during migration in the gel. Figure 8A also shows that laser irradiation of ZnPPIX:BLG does not cause significant fragmentation or aggregation as observed in other proteins upon photooxidation [61,112,114] since the same bands appear for irradiated and non-irradiated samples. The binned profiles obtained from the lanes (Figure S6) confirm the presence of two close peaks corresponding in the region of the BLG dimer.
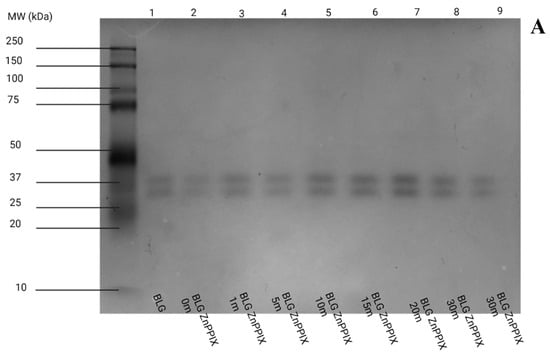
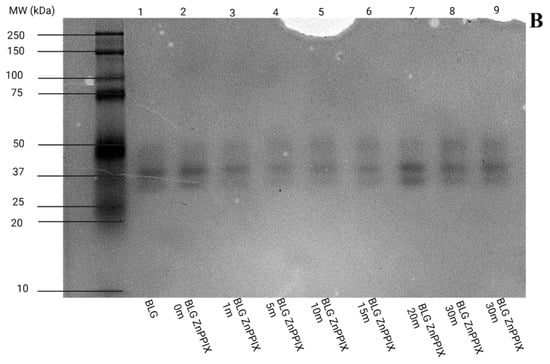
Figure 8.
Native PAGE gels as a function of laser irradiation at 405 nm. (A) pH 5, (B) pH 9. Each lane was loaded with nearly identical amounts of BLG. Lane 1 = BLG (non-irradiated); Lane 2 = ZnPPIX:BLG (non-irradiated); Lane 3 = ZnPPIX:BLG (1.32 J/cm); Lane 4 = ZnPPIX:BLG (6.60 J/cm); Lane 5 = ZnPPIX:BLG (13.2 J/cm); Lane 6 = ZnPPIX:BLG (19.8 J/cm); Lane 7 = ZnPPIX:BLG (26.4 J/cm); Lane 8 and 9 = ZnPPIX:BLG (39.6 J/cm).
3.6.2. pH 9
Compared with the gel at pH 5, one notices a significant difference, i.e., the presence of a third band (albeit fainter) around 50 kDa in addition to the two bands near 37 kDa. Such a band, however, is not the effect of protein cross-linking caused by photooxidation since the same band is present in the non-irradiated controls (Lanes 1 and 2, Figure 8B). Instead, this band is consistent with the presence of the trimeric form of native BLG (expected molecular weight ~55 kDa) in addition to the dominant dimeric form. The presence of this form may help explain some of the differences observed in the fluorescence lifetime between acidic and alkaline pH. The profiles (Figure S7) confirm the presence of a third band, although its relative contribution appears to vary considerably and does not appear to be correlated with laser irradiation.
Overall native-PAGE results suggest that laser-induced mechanisms triggered by the ZnPPIX ligand do not lead to detectable aggregation or fragmentation of BLG since the number or the position of the bands do not change significantly between irradiated and non-irradiated samples.
4. Conclusions
Our combination of experiments supports the general conclusion that the low-irradiance laser irradiation of the PS in the ZnPPIX:BLG complex prompts photochemical and photophysical events that affect the properties of the protein. Overall, we have also shown that photoinduced modifications of BLG occur more significantly at alkaline pH; the reason is a combination of three factors: the greater fraction of BLG complexed with ZnPPIX at pH 9, the consequently greater fraction of monomeric PS, and the reported greater instability of the protein at alkaline pH [32]. More specifically, our results appear to be consistent with the following sequence of events:
- Light absorption by ZnPPIX prompts the formation of transient excited state species, represented mostly by the triplet state of the PS (*ZnPPIX3) and the cation radical (ZnPPIX●+). This conclusion is supported by the evidence shown in Figure 6 and Figure 7 and the related discussion in the corresponding section of the manuscript.
- The experiments confirm the formation of 1O2 carried out with SOSG (Figure 7). The SOSG data suggest that at pH 5, the formation of 1O2 is likely negligible, while at pH 9, it is more significant.
- In the ZnPPIX:BLG complex, the photosensitized species (triplet-sensitized 1O2 or ZnPPIX●+) prompt the oxidation of various protein residues. The differential absorption spectra (Figure 1C,D), as well as the decrease in fluorescence and fluorescence lifetime (Figure 3, Figure 4, and Figure S3), suggest the oxidation of at least the Trp residues and their conversion to NFK. Oxidation is likely to occur in other susceptible residues (Met, Cys, Tyr, and His) [21], but this could not be measured directly through our experiments.
- The oxidation of the amino acids does not appear to lead to significant aggregation or fragmentation of BLG (Figure 8), which implies that the protein’s backbone is not affected by the mechanisms.
Thus, irradiation of ZnPPIX:BLG does not lead to the massive unfolding of the protein, which would be a desirable effect in FALI but not in PRPs. The changes appear to be irreversible, i.e., oxidation of amino acid residues. It remains to be investigated whether more subtle and localized conformational changes may occur in BLG due to photo-oxidation. These changes could be of interest in the development of PRPs where the structural integrity of the protein must be conserved, with the exception of localized changes.
Our conclusions reveal that ZnPPIX:BLG could be a useful model to study the effect of photooxidation on proteins and its potential biomedical applications. It could also be a useful model to create non-native PRP. For this last part, a crucial element is missing, represented by the evidence for changes in the secondary and or tertiary structure of the protein. For this purpose, investigations are ongoing using a combination of experimental tools (such as Circular Dichroism) and molecular dynamics simulations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/physchem3040027/s1. Scheme S1. 2D structure of ZnPPIX; Figure S1. Absorption spectra in the Soret band region of ZnPPIX in buffer vs. the ZnPPIX:BLG complex. (–) ZnPPIX in buffer, (–) ZnPPIX:BLG complex. (A) pH 5, (B) pH 9; Figure S2. Comparison of the relative photobleaching of the Soret band of ZnPPIX, i.e., values of the ratio of the maximum of the Soret band of irradiated samples divided by the intensity of the non-irradiated sample. (●) ZnPPIX in buffer, (●) ZnPPIX:BLG complex. (A) pH 5, (B) pH 9; Figure S3. Fluorescence spectra of the ZnPPIX:BLG complex as a function of irradiation energy density upon excitation at 294 nm. (A) pH 5. (Black) = 0 J/cm2; (Red) = 48 J/cm2; (Blue) = 216 J/cm2; (Green) = 864 J/cm2. (B) pH 9. (Black) = 0 J/cm2; (Red) = 24 J/cm2; (Blue) = 72 J/cm2; (Green) = 288 J/cm2; (Orange) = 884 J/cm2; Figure S4. Fluorescence decays of ZnPPIX in the ZnPPIX:BLG complex. The decay was obtained using λ_ex=388 nm and emission at 630 ± 16 nm. The average lifetime of the decay is 2.15 ns; Figure S5. Absorption spectra of SOSG (black) and SOSG with ZnPPIX:BLG (red) in solution at pH 9 before laser irradiation. Inset: change in OD at 507 nm (fluorescein moiety) of SOSG as a function of laser irradiation (at 405 nm) in the presence of ZnPPIX:BLG (■) and without ZnPPIX:BLG (●). The moieties of the various molecular components are indicated; Figure S6. Fluorescence intensity of SOSG in solutions as a function of laser irradiation. SOSG spectra were recorded with excitation at 470 nm. (■) pH 5, irradiation 405 nm, in the presence of ZnPPIX:BLG; (▲) pH 5, irradiation 405 nm, without ZnPPIX:BLG; (●) pH 9, irradiation 405 nm, in the presence of ZnPPIX:BLG; (▼) pH 9, irradiation 405 nm, with ZnPPIX:BLG; Figure S7. Panels of the contrast profiles obtained from the lanes of the gels of Figure 8A,B. Through ImageJ, multiple cross sections were taken for each lane and binned together to improve the overall S/N ratio. In the left panel (pH 5) one notices that the doublet (two peaks) is constant throughout all lanes. The ratio between the two peaks also appears to remain constant. The verti-cal line marks the position of the 37 kDa marker in the gels of Figure 8A. In the right panel (pH 9) one notices a third broad peak near 50 kDa (as marked by the labeled vertical line). In the left panel one also notices that the features do not change significantly except for changes in the ratio between the peaks that, however, provide no correlation with irradiation.
Author Contributions
Conceptualization, A.A. and L.B.; methodology, A.A., O.C., G.D.N. and J.M.R. and L.B.; resources, G.D.N. and J.M.R.; formal analysis, A.A., O.C. and L.B.; investigation, A.A., O.C., and L.B.; writing—original draft, L.B.; writing—review & editing, M.L.D. and L.B.; supervision, M.L.D. and L.B.; funding acquisition, L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institutes of Health grant 5SC2GM121250 (L.B.), the United States Air Force Research Laboratory (AFRL) and the United States Air Force Office of Scientific Research (AFOSR) Summer Faculty Fellowship Program (to L.B.) and the Saudi Arabian Cultural Mission Fellowship (to A.A.). Additional support was provided by the AFRL Airman System Directorate contract FA8650-19_C-6024 (G.D.N. and J.M.R.) and the AFOSR grant 19RHCOR067 (M.L.D.)
Data Availability Statement
Data are contained within the article and supplementary materials.
Acknowledgments
The opinions expressed in this document, electronic or otherwise, are solely those of the author(s). They do not represent an endorsement by or the views of the United States Air Force, the Department of Defense, or the United States Government. This manuscript is approved for public release (PA Case Number: AFRL-2023-4928). We would also like to thank G. Zhang and R. Palanisamy for the use of the gel imager.
Conflicts of Interest
Authors John Michael Rickman and Gary D. Noojin were employed by the company SAIC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Bozovic, O.; Jankovic, B.; Hamm, P. Using azobenzene photocontrol to set proteins in motion. Nat. Rev. Chem. 2022, 6, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Azagarsamy, M.A.; Alge, D.L.; Radhakrishnan, S.J.; Tibbitt, M.W.; Anseth, K.S. Photocontrolled Nanoparticles for On-Demand Release of Proteins. Biomacromolecules 2012, 13, 2219–2224. [Google Scholar] [CrossRef] [PubMed]
- Abbruzzetti, S.; Sottini, S.; Viappiani, C.; Corrie, J.E. Acid-induced unfolding of myoglobin triggered by a laser pH jump method. Photochem. Photobiol. Sci. 2006, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Brieke, C.; Rohrbach, F.; Gottschalk, A.; Mayer, G.; Heckel, A. Light-controlled tools. Angew. Chem. Int. Ed. 2012, 51, 8446–8476. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.; Folini, M.; Prasmickaite, L.; Selbo, P.K.; Bonsted, A.; Engesaeter, B.Ø.; Zaffaroni, N.; Weyergang, A.; Dietze, A.; Maelandsmo, G.M.; et al. Photochemical internalization: A new tool for drug delivery. Curr. Pharm. Biotechnol. 2007, 8, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Calzaferri, G. Artificial photosynthesis. Top. Catal. 2010, 53, 130–140. [Google Scholar] [CrossRef]
- Chen, J.; Zhu, Y.; Kaskel, S. Porphyrin-Based Metal–Organic Frameworks for Biomedical Applications. Angew. Chem. Int. Ed. 2021, 60, 5010–5035. [Google Scholar] [CrossRef]
- Lee, C.T., Jr.; Smith, K.A.; Hatton, T.A. Photocontrol of protein folding: The interaction of photosensitive surfactants with bovine serum albumin. Biochemistry 2005, 44, 524–536. [Google Scholar] [CrossRef]
- Möglich, A.; Moffat, K. Engineered photoreceptors as novel optogenetic tools. Photochem. Photobiol. Sci. 2010, 9, 1286–1300. [Google Scholar] [CrossRef]
- Kraiselburd, I.; Moyano, L.; Carrau, A.; Tano, J.; Orellano, E.G. Bacterial Photosensory Proteins and Their Role in Plant–pathogen Interactions. Photochem. Photobiol. 2017, 93, 666–674. [Google Scholar] [CrossRef]
- Ernst, O.P.; Lodowski, D.T.; Elstner, M.; Hegemann, P.; Brown, L.S.; Kandori, H. Microbial and Animal Rhodopsins: Structures, Functions, and Molecular Mechanisms. Chem. Rev. 2014, 114, 126–163. [Google Scholar] [CrossRef]
- Losi, A. Flavin-based blue-light photosensors: A photobiophysics update. Photochem. Photobiol. 2007, 83, 1283–1300. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, Z.; Ma, C.; Zhu, Z. Photoswitchable Enzymatic Biofuel Cell Based on Fusion Protein with Natural Photoreceptor Vivid. ACS Appl. Bio Mater. 2022, 5, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.P. Photosynthesis. Essays Biochem. 2016, 60, 255–273. [Google Scholar] [CrossRef] [PubMed]
- Palczewski, K. Chemistry and biology of vision. J. Biol. Chem. 2012, 287, 1612–1619. [Google Scholar] [CrossRef] [PubMed]
- Bonnett, R.; Martinez, G. Photobleaching of the compounds of the 5,10,15,20-tetrakis(m-hydroxyphenyl)-porphyrin series (m-THPP, m-THPC and m-THPBC). Org. Lett. 2002, 4, 2013–2016. [Google Scholar] [CrossRef]
- Ericson, M.B.; Grapengiesser, S.; Gudmundson, F.; Wennberg, A.-M.; Larko, O.; Moan, J.; Rosen, A. A spectroscopic study of the photobleaching of protoporphyrin IX in solution. Lasers Surg. Med. 2003, 18, 56–62. [Google Scholar] [CrossRef]
- Finlay, J.C.; Conover, D.L.; Hull, E.L.; Foster, T.H. Porphyrin bleaching and PDT-induced spectral changes are irradiance dependent in ALA-sensitized normal rat skin in vivo. Photochem. Photobiol. 2001, 73, 54–63. [Google Scholar] [CrossRef]
- Jacobson, K.; Rajfur, Z.; Vitriol, E.; Hahn, K. Chromophore-assisted laser inactivation in cell biology. Trends Cell Biol. 2008, 18, 443–450. [Google Scholar] [CrossRef]
- Yan, P.; Xiong, Y.; Chen, B.; Negash, S.; Squier, T.C.; Mayer, M.U. Fluorophore-Assisted Light Inactivation of Calmodulin Involves Singlet-Oxygen Mediated Cross-Linking and Methionine Oxidation. Biochemistry 2006, 45, 4736–4748. [Google Scholar] [CrossRef]
- Davies, M.J. Protein oxidation and peroxidation. Biochem. J. 2016, 473, 805–825. [Google Scholar] [CrossRef] [PubMed]
- Bensasson, R.V.; Land, E.J.; Truscott, T.G. Excited States and Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 1993. [Google Scholar]
- Davies, M.J. Singlet oxygen-mediated damage to proteins and its consequences. Biochem. Biophys. Res. Commun. 2003, 305, 761–770. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.J. Reactive species formed on proteins exposed to singlet oxygen. Photochem. Photobiol. Sci. 2004, 3, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.-F.; Chen, Y.; Yi, L.; Brantley, T.; Stanley, B.; Sosic, Z.; Zang, L. Discovery and Characterization of Histidine Oxidation Initiated Cross-links in an IgG1 Monoclonal Antibody. Anal. Chem. 2017, 89, 7915–7923. [Google Scholar] [CrossRef]
- Majiya, H.; Adeyemi, O.O.; Stonehouse, N.J.; Millner, P. Photodynamic inactivation of bacteriophage MS2: The A-protein is the target of virus inactivation. J. Photochem. Photobiol. B 2018, 178, 404–411. [Google Scholar] [CrossRef]
- Lee, A.J.; Ensign, A.A.; Krauss, T.D.; Bren, K.L. Zinc Porphyrin as a Donor for FRET in Zn(II)cytochrome c. J. Am. Chem. Soc. 2010, 132, 1752–1753. [Google Scholar] [CrossRef] [PubMed]
- Ndong Ntoutoume, G.M.A.; Granet, R.; Mbakidi, J.-P.; Constantin, E.; Bretin, L.; Léger, D.Y.; Liagre, B.; Chaleix, V.; Brégier, F.; Sol, V. Design and synthesis of zinc protoporphyrin IX-adamantane/cyclodextrin/cellulose nanocrystals complexes for anticancer photodynamic therapy. Bioorgan. Med. Chem. Lett. 2021, 41, 128024. [Google Scholar] [CrossRef]
- Tangar, A.; Derrien, V.; Lei, R.; Santiago Estevez, M.J.; Sebban, P.; Bernad, S.; Miksovska, J. Utility of fluorescent heme analogue ZnPPIX to monitor conformational heterogeneity in vertebrate hexa-coordinated globins. Metallomics 2019, 11, 906–913. [Google Scholar] [CrossRef]
- Shiraki, K.; Nishikawa, K.; Goto, Y. Trifluoroethanol-induced stabilization of the a-helical structure of b-lactoglobulin: Implication for non-hierirchical protein folding. J. Mol. Biol. 1995, 245, 180–194. [Google Scholar] [CrossRef]
- Barbiroli, A.; Iametti, S.; Bonomi, F. Beta-Lactoglobulin as a Model Food Protein: How to Promote, Prevent, and Exploit Its Unfolding Processes. Molecules 2022, 27, 1131. [Google Scholar] [CrossRef]
- Uhrinova, S.; Smith, M.H.; Jameson, G.B.; Uhrin, D.; Sawyer, L.; Barlow, P.N. Structural changes accompanying pH-induced dissociation of the β-lactoglobulin dimer. Biochemistry 2000, 39, 3565–3574. [Google Scholar] [CrossRef]
- Qin, B.Y.; Bewley, M.C.; Creamer, L.K.; Baker, H.M.; Baker, E.N.; Jameson, G.B. Structural basis of the tanford tyransition of bovine β-lactoglobulin. Biochemistry 1998, 37, 14014–14023. [Google Scholar] [CrossRef] [PubMed]
- Fenner, K.; Redgate, A.; Brancaleon, L. A 200 Nanoseconds All-Atom Simulation of the pH-dependent EF Loop Transition in Bovine β-Lactoglobulin. The Role of the Orientation of the E89 Side Chain. J. Biomol. Struct. Dyn. 2020, 40, 549–564. [Google Scholar] [CrossRef]
- Shafaei, Z.; Ghalandari, B.; Vaseghi, A.; Divsalar, A.; Haertlé, T.; Saboury, A.A.; Sawyer, L. β-Lactoglobulin: An efficient nanocarrier for advanced delivery systems. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1685–1692. [Google Scholar] [CrossRef] [PubMed]
- Lohcharoenkal, W.; Wang, L.; Chen, Y.C.; Rojanasakul, Y. Protein nanoparticles as drug delivery carriers for cancer therapy. BioMed Res. Int. 2014, 2014, 180549. [Google Scholar] [CrossRef] [PubMed]
- Teng, Z.; Xu, R.; Wang, Q. Beta-lactoglobulin-based encapsulating systems as emerging bioavailability enhancers for nutraceuticals: A review. RSC Adv. 2015, 5, 35138–35154. [Google Scholar] [CrossRef]
- Rodríguez-Amigo, B.; Delcanale, P.; Rotger, G.; Juárez-Jiménez, J.; Abbruzzetti, S.; Summer, A.; Agut, M.; Luque, F.J.; Nonell, S.; Viappiani, C. The complex of hypericin with β-lactoglobulin has antimicrobial activity with perspective applications in dairy industry. J. Dairy Sci. 2015, 98, 89–94. [Google Scholar] [CrossRef]
- Belcher, J.; Sansone, S.; Fernandez, N.F.; Haskins, W.E.; Brancaleon, L. Photoinduced Unfolding of Beta-Lactoglobulin Mediated by a Water-Soluble Porphyrin. J. Phys. Chem. B 2009, 113, 6020–6030. [Google Scholar] [CrossRef]
- Fernandez, N.F.; Sansone, S.; Mazzini, A.; Brancaleon, L. Irradiation of the porphyrin causes unfolding of the protein in the Protoporphyrin IX/β-lactoglobulin non covalent complex. J. Phys. Chem. B 2008, 112, 7592–7600. [Google Scholar] [CrossRef]
- McMicken, B.; Thomas, R.J.; Brancaleon, L. Photoinduced partial unfolding of tubulin bound to meso-tetrakis(sulfonatophenyl) porphyrin leads to inhibition of microtubule formation in vitro. J. Biophotonics 2014, 7, 874–888. [Google Scholar] [CrossRef]
- Tian, F.; Johnson, K.; Lesar, A.E.; Moseley, H.; Ferguson, J.; Samuel, I.D.W.; Mazzini, A.; Brancaleon, L. The pH-Dependent Conformational Transition of β-Lactoglobulin Modulates the Binding of Protoporphyrin IX. Biochim. Biophys. Acta 2005, 1760, 38–46. [Google Scholar] [CrossRef]
- Castillo, O.; Mancillas, J.; Hughes, W.; Brancaleon, L. Characterization of the Interaction of Metal-Protoporphyrins Photosensitizers with β-Lactoglobulin. Biophys. Chem. 2023, 292, 106918. [Google Scholar] [CrossRef]
- Benavides, B.S.; Acharya, R.; Clark, E.R.; Basak, P.; Maroney, M.J.; Nocek, J.M.; Schanze, K.S.; Kurtz, D.M., Jr. Structural, Photophysical, and Photochemical Characterization of Zinc Protoporphyrin IX in a Dimeric Variant of an Iron Storage Protein: Insights into the Mechanism of Photosensitized H(2) Generation. J. Phys. Chem. B 2019, 123, 6740–6749. [Google Scholar] [CrossRef]
- Johnson, C.N.; Gorbet, G.E.; Ramsower, H.; Urquidi, J.; Brancaleon, L.; Demeler, B. Multi-wavelength analytical ultracentrifugation of human serum albumin complexed with porphyrin. Eur. Biophys. J. 2018, 47, 789–797. [Google Scholar] [CrossRef]
- Swinehart, D.F. The Beer-Lambert Law. J. Chem. Educ. 1962, 39, 333. [Google Scholar] [CrossRef]
- Collini, M.; D’Alfonso, L.; Baldini, G. New insight on β-lactoglobulin binding sites by 1-anilinonaphtalene-8-sulfonate fluorescence decay. Prot. Sci. 2000, 9, 1968–1974. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. On spectral relaxation in proteins. Photochem. Photobiol. 2000, 72, 421–437. [Google Scholar] [CrossRef]
- Albani, J.R.; Vogelaer, J.; Bretesche, L.; Kmiecik, D. Tryptophan 19 residue is the origin of bovine β-lactoglobulin fluorescence. J. Pharm. Biomed. Anal. 2014, 91, 144–150. [Google Scholar] [CrossRef]
- Hu, J.; Allen, R.; Rozinek, S.; Brancaleon, L. Experimental and computational characterization of photosensitized conformational effects mediated by protoporphyrin ligands on human serum albumin. Photochem. Photobiol. Sci. 2017, 16, 694–710. [Google Scholar] [CrossRef]
- van de Weert, M.; Stella, L. Fluorescence quenching and ligand binding: A critical discussion of a popular methodology. J. Mol. Struct. 2011, 998, 144–150. [Google Scholar] [CrossRef]
- Kasha, M. Characterization of Electronic Transitions in Complex Molecules. Discuss. Faraday Soc. 1950, 9, 14–19. [Google Scholar] [CrossRef]
- Harvey, B.J.; Bell, E.; Brancaleon, L. A Tryptophan Rotamer Located in a Polar Environment Probes pH-Dependent Conformational Changes in Bovine beta-Lactoglobulin A. J. Phys. Chem. B 2007, 111, 2610–2620. [Google Scholar] [CrossRef] [PubMed]
- Durbin, J.; Watson, G.S. Testing for serial correlation in least squares regression. II. Biometrika 1951, 38, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Brancaleon, L.; Moseley, H. Laser and Non-Laser Light Sources for Photodynamic Therapy. Lasers Med. Sci. 2002, 17, 173–186. [Google Scholar] [CrossRef]
- Wamser, C.C.; Ghosh, A. The Hyperporphyrin Concept: A Contemporary Perspective. JACS Au 2022, 2, 1543–1560. [Google Scholar] [CrossRef]
- Chang, C.-W.; Luo, L.; Chou, C.-K.; Lo, C.-F.; Lin, C.-Y.; Hung, C.-S.; Lee, Y.-P.; Diau, E.W.-G. Femtosecond Transient Absorption of Zinc Porphyrins with Oligo(phenylethylnyl) Linkers in Solution and on TiO2 Films. J. Phys. Chem. C 2009, 113, 11524–11531. [Google Scholar] [CrossRef]
- Schmidt, R. Photosensitized Generation of Singlet Oxygen. Photochem. Photobiol. 2006, 82, 1161–1177. [Google Scholar] [CrossRef]
- Jones, M.; Talfournier, F.; Bobrov, A.; Grossmann, J.G.; Vekshin, N.; Sutcliffe, M.J.; Scrutton, N.S. Electron transfer and conformational change in complexes of trimethylamine dehydrogenase and electron transferring flavoprotein. J. Biol. Chem. 2002, 277, 8457–8465. [Google Scholar] [CrossRef]
- Jiang, S.; Carroll, L.; Mariotti, M.; Hägglund, P.; Davies, M.J. Formation of protein cross-links by singlet oxygen-mediated disulfide oxidation. Redox Biol. 2021, 41, 101874. [Google Scholar] [CrossRef]
- Prinsze, C.; Dubbelman, T.M.A.R.; Van Steveninck, J. Protein damage induced by small amounts of photodynamically generated singlet oxygen or hydroxyl radicals. Biochim. Biophys. Acta 1990, 1038, 152–157. [Google Scholar] [CrossRef]
- O’Connor, S.P.; Powell, S.M.; Rickman, J.M.; Pope, N.J.; Noojin, G.D.; Scully, M.O.; Denton, M.L.; Yakovlev, V.V. Transient absorption spectroscopy to explore cellular pathways to photobiomodulation. J. Photochem. Photobiol. B Biol. 2021, 222, 112271. [Google Scholar] [CrossRef]
- Yakovlev, V.V.; Kohler, B.; Wilson, K.R. Broadly tunable 30-fs pulses produced by optical parametric amplification. Opt. Lett. 1994, 19, 2000–2002. [Google Scholar] [CrossRef]
- Johnston, T.F. Beam propagation (M2) measurement made as easy as it gets: The four-cuts method. Appl. Opt. 1998, 37, 4840–4850. [Google Scholar] [CrossRef]
- Mazur, L.M.; Roland, T.; Leroy-Lhez, S.; Sol, V.; Samoc, M.; Samuel, I.; Matczyszyn, K. Efficient Singlet Oxygen Photogeneration by Zinc Porphyrin Dimers upon One- and Two-Photon Excitation. J. Phys. Chem. B 2019, 123, 4271–4277. [Google Scholar] [CrossRef]
- Kim, S.; Fujitsuka, M.; Majima, T. Photochemistry of Singlet Oxygen Sensor Green. J. Phys. Chem. B 2013, 117, 13985–13992. [Google Scholar] [CrossRef]
- Gollmer, A.; Arnbjerg, J.; Blaikie, F.H.; Pedersen, B.W.; Breitenbach, T.; Daasbjerg, K.; Glasius, M.; Ogilby, P.R. Singlet oxygen sensor green: Photochemical behaviour in solution and in mammalian cell. Photochem. Photobiol. 2011, 87, 671–679. [Google Scholar] [CrossRef]
- Knoblauch, R.; Moskowitz, J.; Hawkins, E.; Geddes, C.D. Fluorophore-Induced Plasmonic Current: Generation-Based Detection of Singlet Oxygen. ACS Sens. 2020, 5, 1223–1229. [Google Scholar] [CrossRef]
- Prasad, A.; Sedlářová, M.; Pospíšil, P. Singlet oxygen imaging using fluorescent probe Singlet Oxygen Sensor Green in photosynthetic organisms. Sci. Rep. 2018, 8, 13685. [Google Scholar] [CrossRef]
- Liu, H.; Carter, P.J.H.; Laan, A.C.; Eelkema, R.; Denkova, A.G. Singlet Oxygen Sensor Green is not a Suitable Probe for 1O2 in the Presence of Ionizing Radiation. Sci. Rep. 2019, 9, 8393. [Google Scholar] [CrossRef]
- Schägger, H.; Cramer, W.A.; von Jagow, G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 1994, 217, 220–230. [Google Scholar] [CrossRef]
- Jarzecki, A.A.; Spiro, T.G. Porphyrin distorsion from resonance raman intensities of out-of-plane modes: Computation and modeling of N-methylmesoporphyrin, a ferrochelatase transition state analogue. J. Phys. Chem. A 2005, 109, 421–430. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Prot. Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Hu, J.; Hernandez Soraiz, E.; Johnson, C.N.; Demeler, B.; Brancaleon, L. Novel combinations of experimental and computational analysis tested on the binding of metalloprotoporphyrins to albumin. Int. J. Biol. Macromol. 2019, 134, 445–457. [Google Scholar] [CrossRef]
- Monsù Scolaro, L.; Castriciano, M.; Romeo, A.; Patanè, S.; Cefali, E.; Allegrini, M. Aggregation behavior of protoporphyrin IX in aqueous solutions: Clear evidence of vescicle formation. J. Phys. Chem. B 2002, 106, 2453–2459. [Google Scholar] [CrossRef]
- Sandberg, S.; Romslo, I. Porphyrin-induced photodamage at the cellular and the subcellular level as related to the solubility of the porphyrin. Clin. Chim. Acta 1981, 109, 193–201. [Google Scholar] [CrossRef]
- Dreaden, T.M.; Chen, J.; Rexroth, S.; Barry, B.A. N-formylkynurenine as a marker of high light stress in photosynthesis. J. Biol. Chem. 2011, 286, 22632–22641. [Google Scholar] [CrossRef]
- Afaghi, P.; Lapolla, M.A.; Ghandi, K. Determining the degree of denaturation of bovine serum albumin using a new UV analysis technique. Chem. Rep. 2021, 3, 173–176. [Google Scholar] [CrossRef]
- Fukunaga, Y.; Katsuragi, Y.; Izumi, T.; Sakiyama, F. Fluorescence characteristics of kynurenine and N′-formylkynurenine. Their use as reporters of the environment of tryptophan 62 in hen egg-white lysozyme. J. Biochem. 1982, 92, 129–141. [Google Scholar] [CrossRef]
- Gracanin, M.; Hawkins, C.L.; Pattison, D.I.; Davies, M.J. Singlet-oxygen-mediated amino acid and protein oxidation: Formation of tryptophan peroxides and decomposition products. Free Radic. Biol. Med. 2009, 47, 92–102. [Google Scholar] [CrossRef]
- Zhdanova, N.G.; Shirshin, E.A.; Maksimov, E.G.; Panchishin, I.M.; Saletsky, A.M.; Fadeev, V.V. Tyrosine fluorescence probing of the surfactant-induced conformational changes of albumin. Photochem. Photobiol. Sci. 2015, 14, 897–908. [Google Scholar] [CrossRef]
- Han, Q.; Robinson, H.; Li, J. Biochemical identification and crystal structure of kynurenine formamidase from Drosophila melanogaster. Biochem. J. 2012, 446, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Sherin, P.S.; Grilj, J.; Kopylova, L.V.; Yanshole, V.V.; Tsentalovich, Y.P.; Vauthey, E. Photophysics and photochemistry of the UV filter kynurenine covalently attached to amino acids and to a model protein. J. Phys. Chem. B 2010, 114, 11909–11919. [Google Scholar] [CrossRef] [PubMed]
- Taulier, N.; Chalikian, T.V. Characterization of pH-induced transitions of beta-lactoglobulin: Ultrasonic, densimetric, and spectroscopic studies. J. Mol. Biol. 2001, 314, 873–889. [Google Scholar] [CrossRef] [PubMed]
- Rozinek, S.E.; Thomas, R.J.; Brancaleon, L. Biophysical Characterization of the Interaction of Human Albumin with a Cationic Porphyrin. Biochem. Biophys. Rep. 2016, 7, 295–302. [Google Scholar] [CrossRef]
- Khrenova, M.; Topol, I.; Collins, J.; Nemukhin, A. Estimating Orientation Factors in the FRET Theory of Fluorescent Proteins: The TagRFP-KFP Pair and Beyond. Biophys. J. 2015, 108, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Piston, D.W.; Kremers, G.J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem. Sci. 2007, 32, 407–414. [Google Scholar] [CrossRef]
- Grabolle, M.; Pauli, J.; Brehm, R.; Resch-Genger, U. Structural control of dye–protein binding, aggregation and hydrophilicity in a series of asymmetric cyanines. Dye. Pigment. 2014, 103, 118–126. [Google Scholar] [CrossRef]
- Maitra, D.; Pinsky, B.M.; Soherawardy, A.; Zheng, H.; Banerjee, R.; Omary, M.B. Protoporphyrin-IX nanostructures modulate their protein aggregation ability via differential oxidation and protein binding. bioRxiv 2021. [Google Scholar] [CrossRef]
- Iyer, J.K.; Shi, L.; Shankar, A.H.; Sullivan, D.J., Jr. Zinc protoporphyrin IX binds heme crystals to inhibit the process of crystallization in Plasmodium falciparum. Mol. Med. 2003, 9, 175–182. [Google Scholar] [CrossRef]
- Gouterman, M. Spectra of porphyrins. J. Mol. Spectrosc. 1961, 6, 138–163. [Google Scholar] [CrossRef]
- Andrés, G.O.; Cabrerizo, F.M.; Martínez-Junza, V.; Braslavsky, S.E. A Large Entropic Term Due to Water Rearrangement is Concomitant with the Photoproduction of Anionic Free-Base Porphyrin Triplet States in Aqueous Solutions. Photochem. Photobiol. 2007, 83, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Finikova, O.S.; Chen, P.; Ou, Z.; Kadish, K.M.; Vinogradov, S.A. Dynamic quenching of porphyrin triplet-states by two-photon absorpting dyes: Towards two-photon-enhanced oxygen nanosensors. J. Photochem. Photobiol. A 2008, 198, 75–84. [Google Scholar] [CrossRef]
- Togashi, D.M.; Costa, S.M.B. Absorption, fluorescence and transient triplet-triplet absorption spectra of zinc tetramethylpyridylporphyrin in reverse micellesand microemulsions of aerosol OT-(AOT). Phys. Chem. Chem. Phys. 2000, 2, 5437–5444. [Google Scholar] [CrossRef]
- Ballard, S.G.; Mauzerall, D. Photo-initiated ion formation from octaethyl-porphyrin and its zinc chelate as a model for electron transfer in reaction centers. Biophys. J. 1978, 24, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Mukherjee, D.; De, S.; Kathiravan, A. Probing the Highly Efficient Electron Transfer Dynamics between Zinc Protoporphyrin IX and Sodium Titanate Nanosheets. J. Phys. Chem. A 2016, 120, 7121–7129. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, J.L.; Kuciauskas, D. Charge transfer enhances two-photon absorption in transition metal porphyrins. J. Am. Chem. Soc. 2006, 128, 3902–3903. [Google Scholar] [CrossRef]
- Laia, C.A.T.; Costa, S.M.B.; Ferreira, L.F.V. Electron-transfer mechanism of the triplet state quenching of aluminum tetrasulfonated phthalocyanine by cytochrome C. Biophys. Chem. 2006, 122, 143–155. [Google Scholar] [CrossRef]
- Kumar, P.H.; Venkatesh, Y.; Siva, D.; Ramakrishna, B.; Bangal, P.R. Ultrafast Relaxation Dynamics of 5,10,15,20-meso-Tetrakis Pentafluorophenyl Porphyrin Studied by Fluorescence Up-Conversion and Transient Absorption Spectroscopy. J. Phys. Chem. A 2015, 119, 1267–1278. [Google Scholar] [CrossRef]
- Shimizu, D.; Osuka, A. Porphyrinoids as a platform of stable radicals. Chem. Sci. 2018, 9, 1408–1423. [Google Scholar] [CrossRef]
- Hashimoto, T.; Choe, Y.-K.; Nakano, H.; Hirao, K. Theoretical Study of the Q and B Bands of Free-Base, Magnesium, and Zinc Porphyrins, and Their Derivatives. J. Phys. Chem. A 1999, 103, 1894–1904. [Google Scholar] [CrossRef]
- Feitelson, J.; Barboy, N. Triplet-state reactions of zinc protoporphyrins. J. Phys. Chem. 1986, 90, 271–274. [Google Scholar] [CrossRef]
- Nonell, S.B.; Braslavsky, S.E. Time-resolved singlet oxygen detection. Methods Enzymol. 2000, 319, 37–49. [Google Scholar] [PubMed]
- Montfort, C.V.; Sharov, V.S.; Metzger, S.; Schöneich, C.; Sies, H.; Klotz, L.-O. Singlet oxygen inactivates protein tyrosine phosphatase-1B by oxidation of the active site cysteine. Biol. Chem. 2006, 387, 1399–1404. [Google Scholar] [CrossRef]
- Ronsein, G.E.; Oliveira, M.C.B.; Miyamoto, S.; Medeiros, M.H.G.; Di Mascio, P. Tryptophan Oxidation by Singlet Molecular Oxygen [O2 (1Δg)]: Mechanistic Studies Using 18O-Labeled Hydroperoxides, Mass Spectrometry, and Light Emission Measurements. Chem. Res. Toxicol. 2008, 21, 1271–1283. [Google Scholar] [CrossRef]
- Boreen, A.L.; Edhlund, B.L.; Cotner, J.B.; McNeill, K. Indirect photodegradation of dissolved free amino acids: The contribution of singlet oxygen and differential reactivity of DOM from various sources. Environ. Sci. Technol. 2008, 42, 5492–5498. [Google Scholar] [CrossRef]
- Ding, Y.; Zhao, Z.; Matysik, J.; Gärtner, W.; Losi, A. Mapping the role of aromatic amino acids within a blue-light sensing LOV domain. Phys. Chem. Chem. Phys. 2021, 23, 16767–16775. [Google Scholar] [CrossRef]
- Jensen, R.L.; Arnbjerg, J.; Ogilby, P.R. Reaction of Singlet Oxygen with Tryptophan in Proteins: A Pronounced Effect of the Local Environment on the Reaction Rate. J. Am. Chem. Soc. 2012, 134, 9820–9826. [Google Scholar] [CrossRef]
- Gokhale, N.H.; Bradford, S.; Cowan, J.A. Catalytic inactivation of human carbonic anhydrase I by a metallopeptide-sulfonamide conjugate is mediated by oxydation of active site residues. J. Am. Chem. Soc. 2008, 130, 2388–2389. [Google Scholar] [CrossRef]
- Ragàs, X.; Jiménez-Banzo, A.; Sánchez-García, D.; Batllori, X.; Nonell, S. Singlet oxygen photosensitisation by the fluorescent probe Singlet Oxygen Sensor Green®. Chem. Commun. 2009, 2920–2922. [Google Scholar] [CrossRef]
- Pattison, D.I.; Rahmanto, A.S.; Davies, M.J. Photo-oxidation of proteins. Photochem. Photobiol. Sci. 2012, 11, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.; Bubb, W.A.; Hawkins, C.L.; Davies, M.J. Singlet oxygen-mediated protein oxidation: Evidence for the formation of reactive side chain peroxides on tyrosine residues. Photochem. Photobiol. 2002, 76, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Leinisch, F.; Mariotti, M.; Rykaer, M.; Lopez-Alarcon, C.; Hägglund, P.; Davies, M.J. Peroxyl radical- and photo-oxidation of glucose 6-phosphate dehydrogenase generates cross-links and functional changes via oxidation of tyrosine and tryptophan residues. Free Radic. Biol. Med. 2017, 112, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Kochevar, I.E.; Redmond, R.W. Photosensitized production of singlet oxygen. Methods Enzymol. 2000, 319, 20–28. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).