Abstract
While genetic mechanisms in neurodevelopmental disorders are well studied, the mechanisms of environmental factors such as prenatal stress are less understood. Our lab previously characterized miRNA changes associated with stress during pregnancy in mouse brains and in maternal blood from mothers of children with ASD and indicated that prenatal stress can be linked to epigenetic markers. These miRNAs could be used as discovery biomarkers for stress exposure, as well as predictors of neurodevelopmental outcomes. In this pilot study, we gathered saliva samples and stress survey questionnaires from 83 pregnant African American women (ages 18–40) at the time of their ultrasound performed at 20 weeks. miRNA analysis was performed on the 10 highest- and 10 lowest-stress subjects. Out of 6631 miRNAs examined, 34 had significant differential expression, with 5 being upregulated and 29 downregulated in the high-stress group. Predicted targets of differentially expressed miRNAs revealed significant enrichment in neurodevelopmental pathways, including forebrain development, sensory system development, and neuronal growth regulation. This may suggest the potential developmental salience of these miRNA profiles. Future research will examine the neurodevelopmental outcomes of these pregnancies to determine the predictive potential of these miRNAs. This may help identify individuals at greatest risk after stress exposure during pregnancy.
1. Introduction
Autism Spectrum Disorder (ASD) is a neurodevelopmental disorder characterized by social communication deficits (e.g., perspective-taking, synchrony, rigid language) and repetitive, stereotyped behavior [1]. The prevalence of ASD, particularly amongst minority populations, has been rising in recent years. In 2014, data collected across 11 sites in the United States by the Autism and Developmental Disease Monitoring Network (ADDM) reported 1 in 59 8-year-old children living within ADDM sites were diagnosed with ASD, and White children were 20% more likely to be identified with ASD than Hispanic and 10% more likely than non-Hispanic Black 8 year olds [2]. Then, in 2018, sites reported a rise to 1 in 44, with researchers finding no differences in prevalence between Hispanic, non-Hispanic Black, Asian/Pacific Islander, nor non-Hispanic White children [3]. In its most recent report, released in 2023, the ADDM Network identified significantly higher prevalence per 1000 children amongst Hispanic (31.6), non-Hispanic Black (29.3), and Asian/Pacific Islander (33.4) than in non-Hispanic white (24.3) 8 year olds for the first time. Furthermore, two of these sites, Wisconsin and Missouri, showed the highest percent change from 2018 to 2020 (49.5% and 48.5%, respectively), attributed to education data access and no changes in geographic area. Wisconsin and Missouri also had the lowest percentage of diagnoses, 1.9% and 1.7% of 8 year olds, respectively, fitting ASD criteria in 2018 (compared to 2.3% across all communities tracked that year). However, in 2020, Wisconsin’s percentage of diagnoses (2.8%) was similar to the 2.8% average across ADDM sites, but Missouri still fell short (2.5%). As diagnostic tools and accessibility improve across racial and ethnic backgrounds, rates amongst minorities increase in the diagnosis of ASD, yet non-Hispanic Black children show much higher rates of comorbid intellectual disability (ID). In fact, the same ADDM report from 2023 found 50.8% of Black 8-year-old children diagnosed with ASD were also diagnosed with ID compared to 31.8% of white and 34.9% of Hispanic 8 year olds, similarly echoed in previous ADDM reports as well as in the 2020 data collected by the MO-ADDM. This has been suggested to be due to low identification and diagnosis of ASD without ID in Black populations, necessitating further work towards the identification of biomarkers relevant to this community [4].
The developmental origins of health and disease hypothesis propose that exposures to the environment during fetal development in utero determine postnatal health [5]. Emerging evidence indicates that environmental adversity, including psychosocial stressors, can have a negative impact on neurodevelopment, including ASD [6]. While genetic influences exert considerable influence on the causation of ASD, as observed in the 60–92% monozygotic concordance, environmental influences are increasingly recognized to have a possible role in ASD [7]. Epigenetic modifications—heritable changes in gene expressions that do not include a change in DNA sequence—are increasingly thought to be involved in stress-exposure-associated neurodevelopmental disorders [8]. Dysregulation of microRNAs (miRNAs) may contribute to epigenetic alterations, such as changes in DNA methylation, that affect the expression of autism-related genes [9]. Identifying epigenetic markers could provide a way to assess the biological impact of maternal stress during pregnancy in real time, with potential implications for fetal neurodevelopment.
However, not all mothers exposed to stress during pregnancy have children who develop ASD or other neurodevelopmental disorders. Our lab has previously examined the potential role of maternal genetics interacting with stress exposure for the risk of developing clinical ASD, or ASD-associated behaviors in animal models [10,11,12,13,14,15].
Further research analyzed epigenetic markers in maternal stress-exposed mice born from genetically at-risk dams, revealing increased expression level of specific miRNAs in the brains of the offspring. A similar pattern was detected in blood samples from mothers of children with ASD, where a specific set of miRNAs were differentially expressed in prenatal stress-associated ASD with stress susceptibility in contrast to ASD without such exposure [16,17]. Since blood is a primary method through which maternal stress effects can be relayed to the fetus and given that miRNAs can cross the placental barrier [18], miRNAs present in maternal blood may mediate mechanisms underlying ASD in some instances. These findings led to our investigation of maternal miRNA as a biomarker for exposure to stress in a clinical population. This may serve as an indicator of the subsequent emergence of neurodevelopmental effects.
Historically, research on prenatal stress and neurodevelopment has been scant in minority and marginalized populations. More recent research, though, is starting to examine the effects of stress due to economic and social disparities, tracking mothers along the course of their pregnancy. To fill this gap, our research targets an underserved population where exposure to stress is carefully tracked, enabling us to more clearly examine these relationships. The findings of this first investigation of the relationship between miRNA and stress exposure in pregnancy will lay the groundwork for subsequent studies to investigate their possible impacts on neurodevelopment. Additionally, we utilized salivary collection to obtain miRNA samples, a reliable method of assessment for miRNA [19,20], highly correlated with the miRNA profiles of other biofluids such as adult blood plasma and cord blood plasma [21], and which has significant advantages for scalability and minimizing invasiveness of procedures during pregnancy.
2. Results
Of the 83 participants, the top 10 highest and lowest scores on the Holmes–Rahe Life Stress Inventory had their miRNA profiles developed then comparatively analyzed. Scores in the low-stress group (n = 10) ranged from 0 to 89 (M = 25.30 ± 9.90) while the scores in the high-stress group (n = 10) ranged from 231 to 555 (M = 342.50 ± 27.84). The most frequent stressors reported were changes in sleep habits, changes in social relationships, and changes in financial status (Table 1). Scores from the SRRS were negatively correlated with the scores from the LSS (r = −0.79, p < 0.001, Pearson correlation), with the HS (M = 3.57 ± 0.10) self-reports reporting significantly lower LSS scores, indicating higher stress, than the LS group (M = 4.55 ± 0.32).

Table 1.
Responses on Holmes–Rahe’s Life Stress Scale.
All samples passed quality control metrics for hybridization, spike-in controls, and other QC thresholds. Of 6631 miRNAs analyzed, 34 (0.51%) met the applied filtering criteria for differential expression between the HS and LS groups. Of these, 5 (14.71%) were upregulated, and 29 (85.29%) were downregulated in the HS condition compared to LS. After removing one precursor-miRNA (pre-miRNA) and two reference miRNA signatures, 4 (12.9%) upregulated miRNAs and 27 (87.1%) downregulated miRNAs remained in the high stress group compared to the low-stress groups (Figure 1). Table 2 provides a list of all differentially expressed miRNAs, including average expression (log2–transformed sample signal), fold change relative to the comparison group (LS), and associated p-values. In the HS group, hsa-miR-1184 (2.82-fold) and hsa-miR-668–5p (2.13-fold) were among the most upregulated miRNAs, while hsa-miR-1229–5p (2.54-fold) and hsa-miR-8063 (2.41-fold) were among the most significantly downregulated.
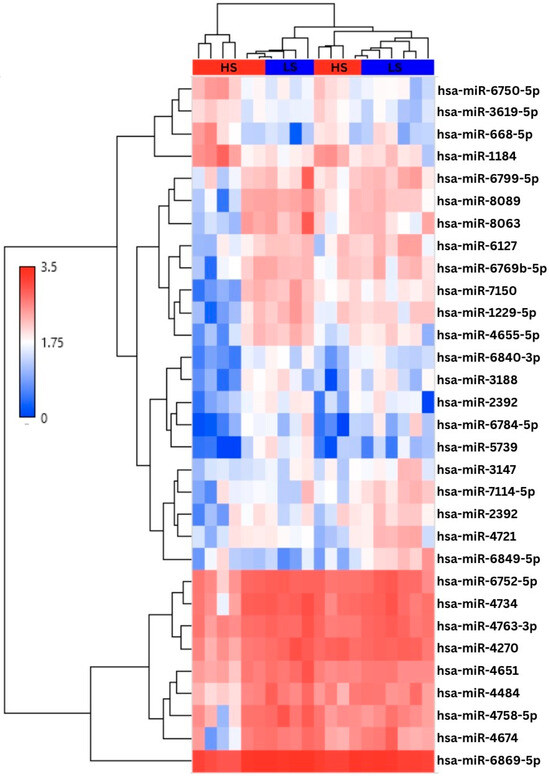
Figure 1.
Hierarchical clustering analysis of the 31 differentially expressed miRNAs. Groups are segmented at the top of the chart where high stress (HS) is represented in red and low stress (LS) is in blue. The first 4 miRNAs are upregulated in the high-stress conditions, followed by 27 downregulated miRNAs. Values are expressed via log transformation (log2Signal).

Table 2.
Differentially expressed miRNA profiles and fold change values.
Target prediction analysis identified 2116 and 6839 predicted genes for the upregulated and downregulated miRNAs, respectively, including 1210 overlapping targets. Both group’s lists of predicted targets were also assessed for GO and using KEGG pathways that were then used to compare enriched clusters for biological processes (BP), molecular function (MF), and cellular component (CC). Both up- and downregulated predicted targets showed particularly high enrichment in biological processes such as the development of the forebrain, telencephalon, and sensory systems as well as dendritic growth and neuronal projection regulation, as shown in Figure 2 below.
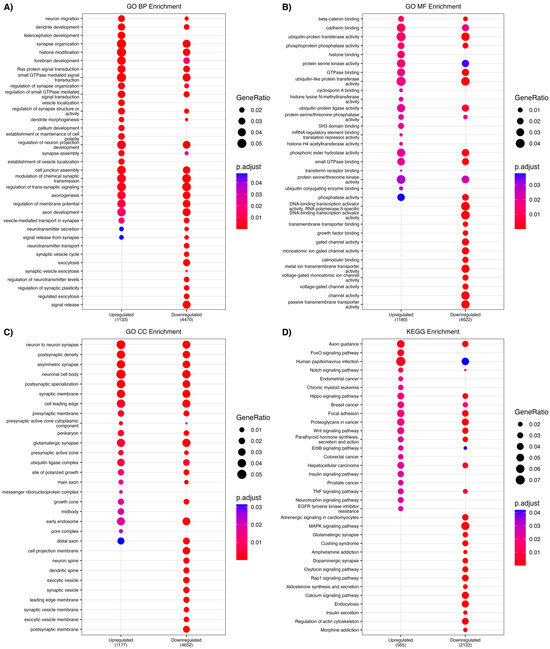
Figure 2.
Gene ontology (GO) analysis was conducted separately on predicted target genes of both the 4 upregulated miRNAs and the 27 downregulated miRNAs. Each plot using compareClusters identifies enriched GO clusters from biological processes, molecular functions, cellular components and in both the targets of the upregulated and the downregulated miRNAs. (A) Biological processes enriched in both the up and down gene projections implicate various neurodevelopmental processes such as dendritic development, neuronal migration and telencephalon development, amongst others. (B) Molecular functions enriched include cadherin binding, histone binding and histone methyltransferase binding. (C) Cellular components implicated include neuronal cell body, glutamatergic synapses and both pre-and post-synaptic membranes. (D) KEGG analysis identified enriched clusters related to axonal guidance, TNF, FoxO, MAPK, and Ras signaling pathways.
3. Discussion
These findings identify a pilot cohort of African American expecting mothers with differing levels of prenatal stress measured by two separate prenatal stress surveys, The primary survey, the SSRS, has been used previously to illuminate relationships between prenatal stressors and subsequent ASD diagnoses [13,15,16], and was therefore utilized to stratify the population for stress exposure. The LSS, originally developed to assess socioecological environmental stressors experienced by historically marginalized communities, was strongly correlated with the SRRS. This suggests that commonplace stressful life events and perceived environmental stressors may be related in this cohort of pregnant African American individuals, supporting the convergent validity of these measures.
Our work identified several miRNAs that may be clinically relevant to stress-related pathways expressed during pregnancy. In placentas complicated by intrauterine growth restriction, miR-3619–5 p, miR-6127, miR-3147, miR-4734, and miR-4484 are consistently dysregulated—miR-3619–5p targets β-catenin while miR-6127 and miR-3147 are upregulated and miR-4734 and miR-4484 are, respectively, up- and downregulated—highlighting roles in trophoblast function and fetal growth pathways [22,23]. Pregnancy-specific stressors further modulate this signature; intrahepatic cholestasis elevates exosomal miR-6127 but reduces miR-7150 and miR-1229–5p in maternal serum, while preeclampsia is marked by decreased circulating miR-2392 and miR-4758–5p reflecting vascular and immune dysregulation characteristic of hypertensive disorders of pregnancy [24,25]. Assisted-reproduction contexts reveal upregulation of miR-7150 and miR-4721 in frozen-thawed embryo transfer placentae, suggesting epigenetic or adaptive placental responses, whereas prenatal smoking induces methylation-mediated silencing of fetal miR-4721, linking environmental toxicants to impaired placental vascular programming [26,27]. Prenatal alcohol exposure downregulates miR-1184 in maternal serum, while adult stress paradigms show miR-668–5p upregulated in anterior cingulate cortex in depressed patients and miR-8063 downregulated by particulate matter equal to a work week worth of exposure [28,29,30].
Targets of these miRNA that are up- and downregulated include biological processes implicating various neurodevelopmental processes such as dendritic development, neuronal migration and telencephalon development. Additionally, targets also include molecular functions including cadherin binding, histone binding and histone methyltransferase binding, as well as targeting cellular components including neuronal cell bodies, glutamatergic synapses and both pre-and post-synaptic membranes (Figure 2). Dendritic and pre-and post-synaptic adhesion as well as glutamate as part of the excitatory/inhibitory balance are prominent features of the neurobiology of autism [31], and epigenetic changes as well as neuronal migration are readily implicated as well [6,17,19,20]. There was high overlap with the targets in the SFARI database, related to intellectual disability in ASD, with MECP2 and DAGLA genes targeted 5 times, and genes related to DNA methylation/epigenetic regulatory/chromatin remodeling and synaptic regulation frequently targeted (Table 3).

Table 3.
microRNA predicted targets by miRDB identified in the SFARI database of ASD–related genes. Predicted targets used for GO Analysis were then overlapped with the 1230 genes currently listed in the SFARI database. Listed are only those found on both lists. Bolded genes indicate the gene was moderately to highly predicted by both an upregulated and a downregulated microRNA listed. Italicized genes notate predicted genes found within the same direction of microRNA. Bold and italicized indicates the gene meets both of the above criteria.
Exposure to both acute and chronic stress has been investigated for its capacity to induce miRNA expression changes as biological manifestations of psychological experiences. In murine models, acute restraint stress triggers frontal cortex-specific miRNA alterations in CD1 mice, though repeated exposure demonstrates transient effects, with expression levels normalizing within five days [32]. Chronic stress instead may present variably based on previous and continuing stressors; a recent analysis of four miRNAs in a cohort of African American women from a historically Black college (HBCU) stratified participants into low chronic stress (LCS), high chronic stress (HCS), and high chronic stress with high adverse childhood experiences (HCS + HA) using the Social Readjustment Rating Scale (SRRS) and Adverse Childhood Experiences (ACES) survey. Three miRNAs showed reduced expression in both high-stress groups compared to LCS controls, while miR-135 exhibited elevated expression exclusively in the HCS group–a finding the authors suggested to be consistent with prior observations of blunted cortisol responses in adults exposed to early-life adversity [33]. Furthermore, pregnancy itself introduces unique miRNA dynamics, with temporal expression shifts across gestation that may illuminate complications like pre-eclampsia and preterm birth [34]. Cohort studies such as the Maternal and Developmental Risks from Environmental and Social Stressors (MADRES) and the PRogramming of Intergenerational Stress Mechanisms (PRISM) are working to expand the common subject pool and investigate perceived stress or maternal life stressors as they relate to miRNA expression in these underrepresented populations [35,36,37]. Notably, African American women experience disproportionate rates of pregnancy-related morbidity [38], yet miRNA research in this demographic remains sparse, growing the importance of early biomarkers informed by diverse groups that may be at a higher risk for neurodevelopmental disorders.
The growing prevalence of neurodevelopmental disorders, such as ASD, has been accompanied by a demographic shift. Prior to 2010, non-Hispanic white children were frequently identified as the predominantly affected demographic [39]. However, in recent years, Hispanic, African American, and Asian/Pacific Islander (API) children were found to match in prevalence and have now surpassed white children in the most recent report released by the CDC. According to this 2023 report, across 10 of the 11 ADDM reported sites, prevalence in non-Hispanic African American 8-year-old children surpassed non-Hispanic white child prevalence in 7 states (Arkansas, California, Georgia, Maryland, Missouri, New Jersey and Tennessee). Similarly, the Hispanic and non-Hispanic API children prevalence ratio compared to non-Hispanic white children was 1.3 and 1.4, respectively [3,40]. This difference also persists in the perception of life stressors, as minority groups are more likely to identify events related to financial and housing instability as more stressful than their white counterparts [40,41].
However, the extent to which these differences in stressful events and their perception may influence stress-related miRNA expression during pregnancy and subsequent neurodevelopmental disorder diagnoses has not been explored. With historically underrepresented groups, such as non-Hispanic Black individuals, now representing a growing proportion of recent ASD diagnoses, it is increasingly important to understand how stress-related factors may contribute to these disparities [42]. Given the impact of miRNAs on the downregulation of target genes, our miRNA differential expression analysis is of particular interest as it revealed predominantly downregulated signatures, suggesting these fine-turning post-transcriptional regulators of key regulatory pathways can be altered by varying levels of prenatal stress.
4. Limitations
This study’s findings should be interpreted considering several limitations. First, the sample size was relatively small and drawn exclusively from African American women attending two clinics in a single geographic region, which limits the generalizability of the results to other populations and settings. However, this population was targeted in this study due to the lack of representation in the literature for this population, in addition to their elevated risk of stress exposure. In addition, we implemented narrow inclusion criteria—restricting participation to pregnant individuals aged 18–40 and excluding those with prior children diagnosed with neurodevelopmental conditions—which further limits the extent to which findings may generalize to higher-risk or more diverse populations, including adolescent and geriatric pregnancies. However, these restrictions were selected to limit potential confounds that might arise from the extreme ends of the age range and due to potential confounds from genetic factors that might lead to neurodevelopmental conditions. These exclusions reflect the pilot nature of the study and funding limitations at the time of data collection.
Additionally, data was collected at a single gestational time point, precluding analysis of how stress exposure and miRNA expression may fluctuate throughout pregnancy. Second, stress exposure was assessed using self-report measures, which are inherently subject to recall and reporting biases. Although the strong correlation between the two stress surveys supports their convergent validity, self-reported data may not fully capture the complexity of stress experiences during pregnancy. Technical variability also arose from differences in RNA yield and the use of microarray-based miRNA profiling, which may not detect all relevant or low-abundance miRNAs. Furthermore, logistical issues necessitated our focus on the 10 most stress exposed and the 10 least stress-exposed individuals for the contrast examined. This may have limited the richness of data that could be gleaned by not examining the entire sample. However, we consider this pilot a proof of principle for the ability of miRNA to detect the effects of stress exposure in this setting. Future studies will need to examine this across the entire sample population. Finally, while several differentially expressed miRNAs were identified in relation to prenatal stress, their functional roles and biological targets were not experimentally validated in this study. Future research should address these limitations by employing larger, more diverse cohorts, longitudinal designs, and integrated approaches that combine genetic, epigenetic, and clinical outcome data.
5. Materials and Methods
5.1. Participants
Eighty-three pregnant African American women were recruited from the University of Missouri’s Thompson Center for Autism & Neurodevelopment and Maternal-Fetal Medicine clinics. The subjects were women aged 18 to 40 years (high stress: M = 26 ± 5.9 years; low stress: M = 26.5 ± 6.3 years), who were approximately 20 weeks pregnant at the time of sampling, as they were all coming in for their 20-week ultrasound. Exclusion factors were a prior child with ASD or being outside of the predetermined age group, as the age group was established to avoid potential confounding variables related to adolescent or advanced maternal age during pregnancy. All procedures were conducted in accordance with the approval of the Institutional Review Board at the University of Missouri (IRB #2090123) on 14 April 2022. All participants provided informed consent under this protocol to participate.
5.2. Procedures and Materials
When participants first arrived for their ultrasound appointments, they filled out two self-report measures: the Life Stress Survey (LSS) [43,44] and the Holmes–Rahe Life Stress Inventory (also known as the Social Readjustment Rating Scale–SRRS) [45]. The SRRS asked participants to identify stressful events that had occurred throughout their pregnancy, the month and year of each event, or check them as “ongoing” if not yet resolved. The Life Stress Inventory assessed the overall stress levels endured in the last three months using a reverse-coded Likert scale, where 1 indicated extreme stress and 5 represented no stress.
Once participants finished the questionnaires, researchers ensured that they had not eaten or consumed any beverage for at least 30 min to minimize the possibility of sample contamination. Saliva samples were collected from all the participants. The saliva swab was swirled ten times in the gum pockets on both sides of the mouth. The samples were marked with subject identification numbers and collection dates, then stored in cryovials and transferred directly to a −80 °C freezer. Saliva swabs were transported on dry ice to the University of Kansas Medical Center–Genomics Core for analysis. After vigorously shaking the sample in the ORAcollect●RNA Kit collection tube (DNA Genotek (Stittsville, ON K2V 1C2, Canada) ORE-100), a 250 μL aliquot of stabilized RNA from the cheek swab samples was used to perform the Extraction of RNA from an ORAcollect●RNA device using the Qiagen miRNeasy Mini Kit protocol (PD-PR-01416) according to the manufacturer’s instructions with the addition of an on-column DNase treatment using the RNase-free DNase Set (Qiagen 79254 Hilden, Germany). The purified RNA was eluted from the miRNeasy purification column in 30 μL nuclease-free water. Concentration and RNA integrity assessment was determined using a Bioanalyzer 2100 running the RNA Pico Chip assay (Agilent Technologies 5067–1513 Santa Clara, CA, USA). The total RNA isolates were normalized to 65 ng for the higher-yielding samples. For RNA isolates with total RNA yields below 65 ng, all available RNA ranging from 6.8 ng–34.4 ng (6 of 20 samples) was used for biotin labeling. Biotin labeling was performed using the FlashTag Biotin HSR RNA Labeling Kit (Life Technologies #901910 Carlsbad, CA, USA). The miRNA fraction of the biotin labeled RNAs was interrogated with GeneChip miRNA 4.0 expression cartridge arrays (Life Technologies 902412) using the GeneChip System composed of 645 hybridization oven, GeneChip Fluidics Station 450 and GeneChip Scanner 3000 7 G with autoloader following manufacturer’s instructions. We used Affymetrix® Transcriptome Analysis Console (TAC) software (Version 4.0.2, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturers instructions to identify differentially expressed (DE) miRNAs. Normalization and summarization of the microarray data were performed using the Robust Multi-array Average and Detected Above Background (RMA + DABG) method native to TAC. A total of 6631 miRNAs were analyzed using TAC, and significantly differentially expressed genes were identified using a p-value threshold of 0.05 and fold change cutoffs of ≥1.5 or ≤−1.5.
5.3. Analytical Plan
Subsequent analysis further characterized differentially expressed miRNAs using an integrative bioinformatics approach whereby known mRNA and miRNA interactions were identified by miRDB [46]. Predicted targets were identified for both upregulated and downregulated miRNAs with a predicted target score >60 [47]. Using the PANTHER classification system, we examined the functional annotations of these projections using Gene Ontology (GO) analysis of biological processes, molecular functioning and cellular components and their corresponding pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG) databases [48,49]. Finally, enriched clusters of these annotations were compared with clusterProfiler [50] to identify relevant overlapping target functions.
6. Conclusions
This serves as a proof of concept, demonstrating that miRNA profiles have the potential to distinguish between pregnancies characterized by high- vs. low-stress exposure during pregnancy. In future work, we aim to follow up with the mothers in this study to assess the neurodevelopmental outcomes of their children, thereby enabling a better understanding of the predictive value of the identified miRNAs. Subsequent larger studies will be needed to determine these relationships across a demographic spectrum to enhance our understanding of these relationships, with the ultimate goal of identifying readily accessible biomarkers that can help detect those at greatest developmental risk after exposure to major life stressors during pregnancy.
Author Contributions
Conceptualization, D.Q.B., Z.T.; methodology, D.Q.B., C.B., R.R., Z.T., J.G. (Jean Goodman); software, C.B., R.R.; validation, C.B.; formal analysis, B.V.B., N.I.A., C.K., J.G. (Jahnavi Godavarthi); investigation, R.B., K.L., K.W., M.C., S.H., N.T.; resources, C.B., N.T., D.Q.B.; data curation, B.V.B., N.I.A., C.K., C.B.; writing—original draft preparation, B.V.B., N.I.A., C.K., D.Q.B.; writing—review and editing, ALL AUTHORS; visualization, B.V.B., N.I.A., C.K.; supervision, R.R., N.T., D.Q.B.; project administration, R.R., N.T., D.Q.B.; funding acquisition, Z.T., D.Q.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by BioNexus KC Collaborate2Cure Research Grant.
Institutional Review Board Statement
All procedures were conducted in accordance with and approval of the Institutional Review Board at the University of Missouri (IRB #2090123) approved on 14 April 2022.
Informed Consent Statement
All participants provided informed consent under this protocol to participate.
Data Availability Statement
Data will be made available by the corresponding author to all valid requests. Data will not be posted due to confidentiality issues.
Conflicts of Interest
The author declares no conflicts of interest.
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®); American Psychiatric Pub: Washington, DC, USA, 2013. [Google Scholar]
- Baio, J.; Wiggins, L.; Christensen, D.L.; Maenner, M.J.; Daniels, J.; Warren, Z.; Kurzius-Spencer, M.; Zahorodny, W.; Robinson, C.; Rosenberg, N.; et al. Prevalence of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2014. MMWR Surveill. Summ. 2018, 67, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Maenner, M.J.; Shaw, K.A.; Bakian, A.V.; Bilder, D.A.; Durkin, M.S.; Esler, A.; Furnier, S.M.; Hallas, L.; Hall-Lande, J.; Hudson, A.; et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2018. MMWR Surveill. Summ. 2021, 70, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Maenner, M.J.; Warren, Z.; Williams, A.R.; Amoakohene, E.; Bakian, A.V.; Bilder, D.A.; Durkin, M.S.; Fitzgerald, R.T.; Furnier, S.M.; Hughes, M.M.; et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—Autism and Developmental Disabilities Monitoring Network, 11 sites, United States, 2020. MMWR Surveill. Summ. 2023, 72, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Gage, S.H.; Munafò, M.R.; Smith, G.D. Causal Inference in Developmental Origins of Health and Disease (DOHaD) Research. Annu. Rev. Psychol. 2016, 67, 567–585. [Google Scholar] [CrossRef]
- Beversdorf, D.Q.; Stevens, H.E.; Jones, K.L. Prenatal Stress, Maternal Immune Dysregulation, and Their Association with Autism Spectrum Disorders. Curr. Psychiatry Rep. 2018, 20, 76. [Google Scholar] [CrossRef]
- Pardo, C.A.; Eberhart, C.G. The neurobiology of autism. Brain Pathol. 2007, 17, 434–447. [Google Scholar] [CrossRef]
- Howerton, C.L.; Morgan, C.P.; Fischer, D.B.; Bale, T.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. USA 2013, 110, 5169–5174. [Google Scholar] [CrossRef]
- Mor, M.; Nardone, S.; Sams, D.S.; Elliott, E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol. Autism 2015, 6, 46. [Google Scholar] [CrossRef]
- Heils, A.; Teufel, A.; Petri, S.; Stöber, G.; Riederer, P.; Bengel, D.; Lesch, K.P. Allelic variation of human serotonin transporter gene expression. J. Neurochem. 1996, 66, 2621–2624. [Google Scholar] [CrossRef]
- Nakamura, M.; Ueno, S.; Tanabe, H.; Sano, A. The human serotonin transporter gene linked polymorphism (5-HTTLPR) shows ten novel allelic variants. Mol. Psychiatry 2000, 5, 32–38. [Google Scholar] [CrossRef]
- Alexander, N.; Wankerl, M.; Hennig, J.; Miller, R.; Zänkert, S.; Steudte-Schmiedgen, S.; Stalder, T.; Kirschbaum, C. DNA methylation profiles within the serotonin transporter gene moderate the association of 5-HTTLPR and cortisol stress reactivity. Transl. Psychiatry 2014, 4, e443. [Google Scholar] [CrossRef] [PubMed]
- Duman, E.A.; Canli, T. Influence of life stress, 5-HTTLPR genotype, and SLC6A4methylation on gene expression and stress response in healthy Caucasian males. Biol. Mood Anxiety Disord. 2015, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Beversdorf, D.Q.; Manning, S.E.; Hillier, A.; Anderson, S.L.; Nordgren, R.E.; Walters, S.E.; Nagaraja, H.N.; Cooley, W.C.; Gaelic, S.E.; Bauman, M.L. Timing of prenatal stressors and autism. J. Autism Dev. Disord. 2005, 35, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.L.; Smith, R.M.; Edwards, K.S.; Givens, B.; Tilley, M.R.; Beversdorf, D.Q. Combined effect of maternal serotonin transporter genotype and prenatal stress in modulating offspring social interaction in mice. Int. J. Dev. Neurosci. 2010, 28, 529–536. [Google Scholar] [CrossRef]
- Hecht, P.M.; Hudson, M.; Connors, S.L.; Tilley, M.R.; Liu, X.; Beversdorf, D.Q. Maternal serotonin transporter genotype affects risk for ASD with exposure to prenatal stress. Autism Res. 2016, 9, 1151–1160. [Google Scholar] [CrossRef]
- Beversdorf, D.Q.; Shah, A.; Jhin, A.; Noel-MacDonnell, J.; Hecht, P.; Ferguson, B.J.; Bruce, D.; Tilley, M.; Talebizadeh, Z. microRNAs and Gene–Environment Interactions in Autism: Effects of Prenatal Maternal Stress and the SERT Gene on Maternal microRNA Expression. Front. Psychiatry 2021, 12, 668577. [Google Scholar] [CrossRef]
- Sjaarda, C.P.; Hecht, P.; McNaughton, A.J.M.; Zhou, A.; Hudson, M.L.; Will, M.J.; Smith, G.; Ayub, M.; Liang, P.; Chen, N.; et al. Interplay between maternal Slc6a4 mutation and prenatal stress: A possible mechanism for autistic behavior development. Sci. Rep. 2017, 7, 8735. [Google Scholar] [CrossRef]
- Hicks, S.D.; Carpenter, R.L.; Wagner, K.E.; Pauley, R.; Barros, M.; Tierney-Aves, C.; Barns, S.; Greene, C.D.; Middleton, F.A. Saliva MicroRNA differentiates children with autism from peers with typical and atypical development. J. Am. Acad. Child Adolesc. Psychiatry 2019, 59, 296–308. [Google Scholar] [CrossRef]
- Hicks, S.D.; Middleton, F.A. A Comparative review of microRNA expression patterns in autism spectrum Disorder. Front. Psychiatry 2016, 7, 176. [Google Scholar] [CrossRef]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large differences in small RNA composition between human biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.F.; Dang, Y.J.; Shi, X.M.; Chen, L.; Wang, N.; Cai, Y.; Zhao, Y.Y. Differentially expressed circular RNAs in maternal and neonatal umbilical cord plasma from SGA compared with AGA. J. Cell. Biochem. 2020, 121, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Chen, L.; He, J.; Lin, J. MicroRNA expression profiles and networks in placentas complicated with selective intrauterine growth restriction. Mol. Med. Rep. 2017, 16, 6650–6673. [Google Scholar] [CrossRef] [PubMed]
- Dong, R.; Ye, N.; Wang, J.; Zhao, S.; Wang, T.; Wang, G.; Shi, X.; Cheng, J.; Zhang, Y.; Yao, T.; et al. Serum Exosomes MicroRNAs Are Novel Non-Invasive Biomarkers of Intrahepatic Cholestasis of Pregnancy. Front. Endocrinol. 2022, 13, 832577. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhu, F.; Ding, Y. Differential microRNA expression profile in the plasma of preeclampsia and normal pregnancies. Exp. Ther. Med. 2019, 18, 826–832. [Google Scholar] [CrossRef]
- Hiura, H.; Hattori, H.; Kobayashi, N.; Okae, H.; Chiba, H.; Miyauchi, N.; Kitamura, A.; Kikuchi, H.; Yoshida, H.; Arima, T. Genome-wide microRNA expression profiling in placentae from frozen-thawed blastocyst transfer. Clin. Epigenetics 2017, 9, 79. [Google Scholar] [CrossRef]
- Barrio, E.; Quirós, A.; Lerma-Puertas, D.; Labarta, J.I.; Gascón-Catalán, A. Identification of miRNAs Involved in Foetal Growth Restriction Due to Maternal Smoking during Pregnancy. J. Clin. Med. 2022, 11, 5808. [Google Scholar] [CrossRef]
- Kaurani, L. Clinical Insights into MicroRNAs in Depression: Bridging Molecular Discoveries and Therapeutic Potential. Int. J. Mol. Sci. 2024, 25, 2866. [Google Scholar] [CrossRef]
- Gardiner, A.S.; Gutierrez, H.L.; Luo, L.; Davies, S.; Savage, D.D.; Bakhireva, L.N.; Perrone-Bizzozero, N.I. Alcohol Use During Pregnancy is Associated with Specific Alterations in MicroRNA Levels in Maternal Serum. Alcohol. Clin. Exp. Res. 2016, 40, 826–837. [Google Scholar] [CrossRef]
- Finicelli, M.; Squillaro, T.; Galderisi, U.; Peluso, G. Micro-RNAs: Crossroads between the Exposure to Environmental Particulate Pollution and the Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2020, 21, 7221. [Google Scholar] [CrossRef]
- Beversdorf, D.Q.; Wang, P.; Barnes, G.; Weisskopf, M.; Hardan, A.; Hu, V.; Mazurek, M.O.; Talebedizah, Z.; Goldberg, W.; Jones, K.L.; et al. Phenotyping, etiological factors, and biomarkers: Toward precision medicine in autism spectrum disorder. J. Dev. Behav. Pediatr. 2016, 37, 659–673. [Google Scholar] [CrossRef]
- Rinaldi, A.; Vincenti, S.; De Vito, F.; Bozzoni, I.; Oliverio, A.; Presutti, C.; Fragapane, P.; Mele, A. Stress induces region specific alterations in microRNAs expression in mice. Behav. Brain Res. 2010, 208, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Holliday, E.; Bagasra, A.; Bagasra, O.; Pandey, P. Assessing the feasibility of using salivary microRNAs as biomarkers to distinguish between chronic stress and childhood trauma in African American young women in an exploratory pilot study. Front. Psychiatry 2025, 16, 1507064. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Weiss, D.; Nyhan, K.; Dewan, A.; Jukic, A.M.Z. Circulating miRNAs in the first trimester and pregnancy complications: A systematic review. Epigenetics 2023, 18, 2152615. [Google Scholar] [CrossRef] [PubMed]
- Bozack, A.K.; Colicino, E.; Rodosthenous, R.; Bloomquist, T.R.; Baccarelli, A.A.; Wright, R.O.; Wright, R.J.; Lee, A.G. Associations between maternal lifetime stressors and negative events in pregnancy and breast milk-derived extracellular vesicle microRNAs in the programming of intergenerational stress mechanisms (PRISM) pregnancy cohort. Epigenetics 2020, 16, 389–404. [Google Scholar] [CrossRef]
- Illarionov, R.A.; Pachuliia, O.V.; Vashukova, E.S.; Tkachenko, A.A.; Maltseva, A.R.; Postnikova, T.B.; Nasykhova, Y.A.; Bespalova, O.N.; Glotov, A.S. Plasma miRNA Profile in High Risk of Preterm Birth during Early and Mid-Pregnancy. Genes 2022, 13, 2018. [Google Scholar] [CrossRef]
- Foley, H.B.; Howe, C.G.; Eckel, S.P.; Chavez, T.; Gevorkian, L.; Reyes, E.G.; Kapanke, B.; Martinez, D.; Xue, S.; Suglia, S.F.; et al. Depression, perceived stress, and distress during pregnancy and EV-associated miRNA profiles in MADRES. J. Affect. Disord. 2023, 323, 799–808. [Google Scholar] [CrossRef]
- Njoku, A.; Evans, M.; Nimo-Sefah, L.; Bailey, J. Listen to the Whispers before They Become Screams: Addressing Black Maternal Morbidity and Mortality in the United States. Healthcare 2023, 11, 438. [Google Scholar] [CrossRef]
- Gallin, Z.; Kolevzon, A.M.; Reichenberg, A.; Hankerson, S.H.; Kolevzon, A. Racial Differences in the Prevalence of Autism Spectrum Disorder: A Systematic review. J. Autism Dev. Disord. 2024, 1–14, (epub ahead of print). [Google Scholar] [CrossRef]
- Grosvenor, L.P.; Croen, L.A.; Lynch, F.L.; Marafino, B.J.; Maye, M.; Penfold, R.B.; Simon, G.E.; Ames, J.L. Autism diagnosis among US children and adults, 2011–2022. JAMA Netw. Open 2024, 7, e2442218. [Google Scholar] [CrossRef]
- Brown, L.L.; Mitchell, U.A.; Ailshire, J.A. Disentangling the stress process: Race/Ethnic differences in the exposure and appraisal of chronic stressors among older adults. J. Gerontol. Ser. B 2018, 75, 650–660. [Google Scholar] [CrossRef]
- Frost, D.M.; Meyer, I.H. Minority stress theory: Application, critique, and continued relevance. Curr. Opin. Psychol. 2023, 51, 101579. [Google Scholar] [CrossRef] [PubMed]
- Ashing-Giwa, K.; Ganz, P.A.; Petersen, L. Quality of life of African-American and white long term breast carcinoma survivors. Cancer 1999, 85, 418–426. [Google Scholar] [CrossRef]
- Ashing-Giwa, K.T.; Padilla, G.V.; Tejero, J.S.; Kim, J. Breast cancer survivorship in a multiethnic sample: Challenges in recruitment and measurement. Cancer 2004, 101, 450–465. [Google Scholar] [CrossRef] [PubMed]
- Holmes, T.H.; Rahe, R.H. The social readjustment rating scale. J. Psychosom. Res. 1967, 11, 213–218. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).