Effective Applications of Bacillus subtilis and B. amyloliquefaciens as Biocontrol Agents of Damping-Off Disease and Biostimulation of Tomato Plants
Abstract
1. Introduction
2. Results
2.1. Identification of Pathogenic Fungi
2.2. Identification of Bacillus Isolates
2.3. In Vitro Antagonistic Test
2.4. Pot Trial
2.4.1. Antagonistic Effect
2.4.2. Photosynthetic Pigments
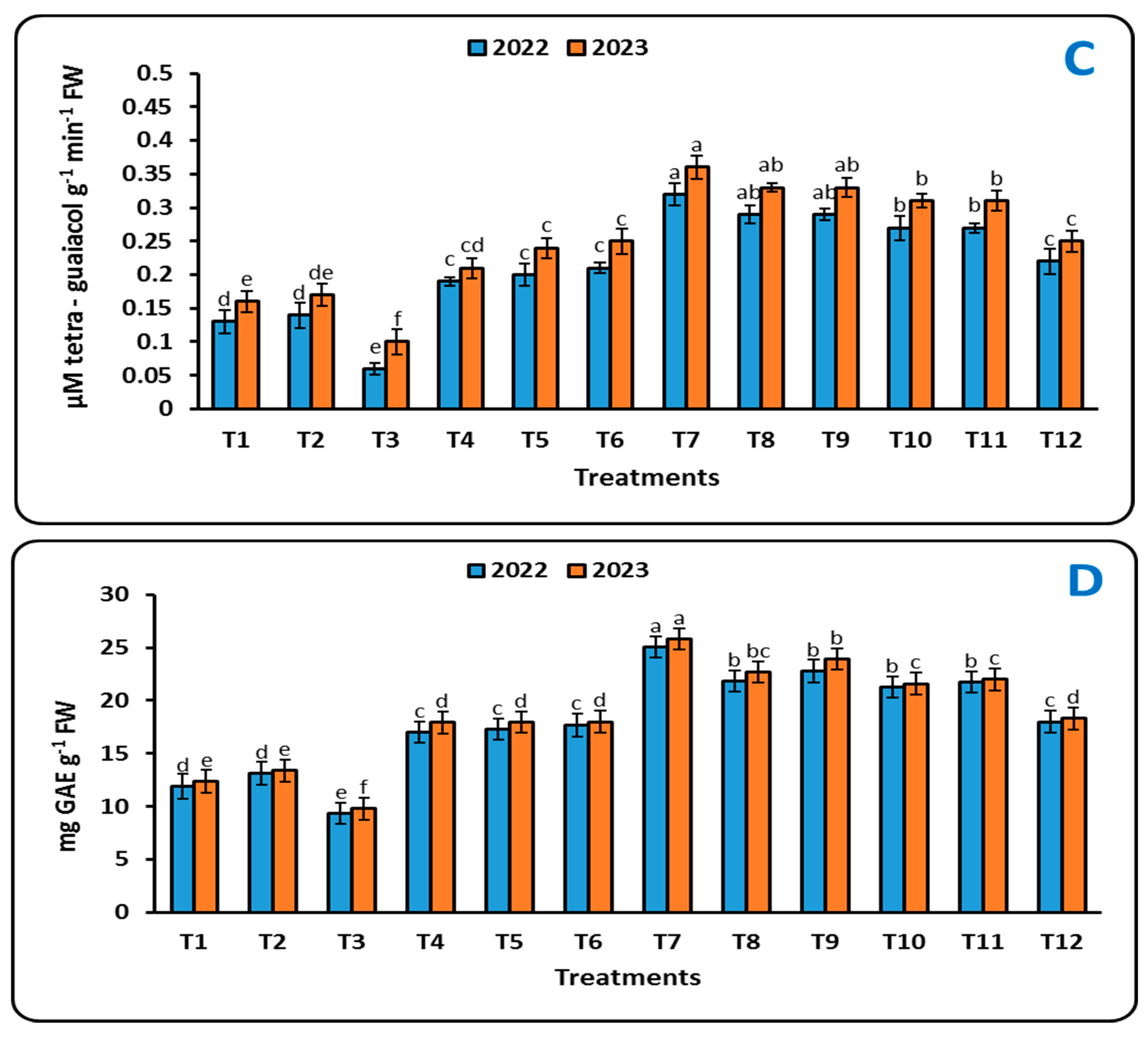
2.4.3. Antioxidant Enzymes
2.4.4. N, P, and K (%) of Leaves
2.4.5. Growth Characteristics and Yield
3. Discussion
4. Materials and Methods
4.1. Sources of Samples
4.2. Isolation and Identification of Pathogenic Fungi
4.3. Isolation and Identification of Bacillus spp.
4.4. In Vitro Antagonistic Assay
4.5. Inoculum Preparation
4.6. Greenhouse Trial
4.7. Measurements
4.7.1. Disease Assessment
4.7.2. Physiological Features
Photosynthetic Pigments
TSS
4.7.3. Antioxidant Enzymes
Peroxidase Activity (PO)
Polyphenol Oxidase Assay (PPO)
Phenylalanine Ammonia Lyase Assay (PAL)
Total Phenolic Content (TPC)
Chemical Contents of Leaves
4.8. Plant Growth and Yield
4.9. Statistical Analyses
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hashem, A.H.; Abdelaziz, A.M.; Askar, A.A.; Fouda, H.M.; Khalil, A.M.; Abd-Elsalam, K.A.; Khaleil, M.M. Bacillus megaterium-mediated synthesis of selenium nanoparticles and their antifungal activity against Rhizoctonia solani in Faba Bean Plants. J. Fungi 2021, 7, 195. [Google Scholar] [CrossRef] [PubMed]
- Abdelaziz, A.M.; Kalaba, M.H.; Hashem, A.H.; Sharaf, M.H.; Attia, M.S. Biostimulation of tomato growth and biocontrol of Fusarium wilt disease using certain endophytic fungi. Bot. Stud. 2022, 63, 34. [Google Scholar] [CrossRef]
- FAOSTAT Database. Food and Agriculture Organization Statistics. Available online: https://www.fao.org/faostat/en/ (accessed on 15 November 2023).
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Matić, S. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the Mediterranean Basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Montenegro, I.; Madrid, A.; Cuellar, M.; Seeger, M.; Alfaro, J.F.; Besoain, X.; Martínez, J.P.; Ramirez, I.; Olguín, Y.; Valenzuela, M. Biopesticide activity from drimanic compounds to control tomato pathogens. Molecules 2018, 23, 2053. [Google Scholar] [CrossRef]
- Attia, M.S.; El-Wakil, D.A.; Hashem, A.H.; Abdelaziz, A. Antagonistic effect of plant growth-promoting fungi against fusarium wilt disease in tomato: In vitro and in vivo study. Appl. Biochem. Biotechnol. 2022, 194, 5100–5118. [Google Scholar] [CrossRef] [PubMed]
- Renu, J. A review of Fusarium oxysporum on its plant interaction and industrial use. J. Med. Plants Stud. 2018, 6, 112–115. [Google Scholar]
- Salman, M.; Abuamsha, R. Potential for integrated biological and chemical control of damping-off disease caused by Pythium ultimum in tomato. BioControl 2012, 57, 711–718. [Google Scholar] [CrossRef]
- Balghouthi, A.; Jonathan, R.; Gognies, S.; Mliki, A.; Belarbi, A. A new species, Pythium echinogynum, causing severe damping-off of tomato seedlings, isolated from Tunisia, France, and India: Morphology, pathology, and biological control. Ann. Microbiol. 2013, 63, 253–258. [Google Scholar] [CrossRef]
- Sarhan, E.A.D.; El-Far, E.M.M.; Ebrahiem, A.M.Y. Systemic resistance in snap bean (Phaseolus vulgaris L.) elicited by some chemicals and biotic inducers against white mold disease caused by Sclerotinia sclerotiorum. Egypt. J. Phytopathol. 2018, 46, 61–84. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Macarisin, D.; Wilson, C. Twenty years of postharvest biocontrol research: Is it time for a new paradigm? Postharvest Biol. Technol. 2009, 52, 137–145. [Google Scholar] [CrossRef]
- Singh, A.; Jain, A.; Sarma, B.K.; Upadhyay, R.S.; Singh, H.B. Rhizosphere microbes facilitate redox homeostasis in Cicer arietinum against biotic stress. Ann. Appl. Biol. 2013, 163, 33–46. [Google Scholar] [CrossRef]
- Gupta, R.; Singh, A.; Srivastava, M.; Singh, V.; Gupta, M.M.; Pandey, R. Microbial modulation of bacoside a biosynthetic pathway and systemic defense mechanism in Bacopa monnieri under Meloidogyne incognita stress. Sci. Rep. 2017, 7, 41867. [Google Scholar] [CrossRef] [PubMed]
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gao, J.; Zhao, F.; Zhao, Y.; Li, Z. Characterization of a salt-tolerant bacterium Bacillus sp. from a membrane bioreactor for saline wastewater treatment. J. Environ. Sci. 2014, 26, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Zhang, Z.; Li, Y.; Zhang, X.; Duan, Y.; Wang, Q. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactinmediated antibacterial activity and colonization. Front. Microbiol. 2017, 8, 1973. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Zhang, Y.; He, L.L.; Ji, Z.L.; Tong, Y.H. Identification of a small antimycotic peptide produced by Bacillus amyloliquefaciens 6256. Pesticide Biochem. Physiol. 2018, 150, 78–82. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, R.; Zhang, R.; Jiang, W.; Chen, X.; Yin, C.; Mao, Z. Isolation, identification, and antibacterial mechanisms of Bacillus amyloliquefaciens QSB-6 and its effect on plant roots. Front. Microbiol. 2021, 12, 746799. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fraile, P.; Menendez, E.; Rivas, R. Role of bacterial biofertilizers in agriculture and forestry. AIMS Bioeng. 2015, 2, 183–205. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; Lee, I.J. Bacillus amyloliquefaciens subsp. plantarum GR53, a potent biocontrol agent resists Rhizoctonia disease on Chinese cabbage through hormonal and antioxidants regulation. World J. Microbiol. Biotechnol. 2015, 31, 1517–1527. [Google Scholar] [PubMed]
- Ahmed, W.; Zhou, G.; Yang, J. Bacillus amyloliquefaciens WS-10 as a potential plant growth-promoter and biocontrol agent for bacterial wilt disease of flue-cured tobacco. Egypt. J. Biol. Pest. Control 2022, 32, 25. [Google Scholar] [CrossRef]
- Soliman, S.A.; Khaleil, M.M.; Metwally, R.A. Evaluation of the antifungal activity of Bacillus amyloliquefaciens and B. velezensis and characterization of the bioactive secondary metabolites produced against plant pathogenic fungi. Biology 2022, 11, 1390. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Singh, R.K.; Srivastava, S.; Kumar, S.; Kashyup, P.L.; Srivastava, A.K. Characterization of antagonistic-potential of two Bacillus strains and their biocontrol activity against Rhizoctonia solani in tomato. J. Basic. Microbiol. 2013, 53, 82–90. [Google Scholar] [CrossRef]
- Zouari, I.; Jlaiel, L.; Tounsi, S.; Trigui, M. Biocontrol activity of the endophytic Bacillus amyloliquefaciens strain CEIZ-11 against Pythium aphanidermatum and purification of its bioactive compounds. Biol. Control 2016, 100, 54–62. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Shulga, E.U.; Validov, S.Z.; Afordoanyi, D.M. Genetic characteristics and enzymatic activities of Bacillus velezensis KS04AU as a stable biocontrol agent against phytopathogens. Int. J. Plant Biol. 2022, 13, 201–222. [Google Scholar] [CrossRef]
- Rashad, Y.M.; Abdalla, S.A.; Sleem, M.M. Endophytic Bacillus subtilis SR22 triggers defense responses in tomato against rhizoctonia root rot. Plants 2022, 11, 2051. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Bouchard-Rochette, M.; Machrafi, Y.; Cossus, L.; Nguyen, T.T.A.; Antoun, H.; Droit, A.; Tweddell, R.J. Bacillus pumilus PTB180 and Bacillus subtilis PTB185: Production of lipopeptides, antifungal activity, and biocontrol ability against Botrytis cinerea. Biol. Control 2022, 170, 104925. [Google Scholar] [CrossRef]
- Ahmad, A.G.; Attia, A.Z.; Mohamed, M.S.; Elsayed, H.E. Fermentation, formulation and evaluation of PGPR Bacillus subtilis isolate as a bioagent for reducing occurrence of peanut soil-borne diseases. J. Integr. Agric. 2019, 18, 2080–2092. [Google Scholar] [CrossRef]
- Khalifa, N.A.; Saleh, R.A.; Mahmoud, N.A. Efficiency of some bio-control agents and plant extracts against beans (Phaseolus vulgaris L.) damping-off and root rot diseases under greenhouse and field conditions. Egypt. J. Phytopathol. 2021, 49, 152–167. [Google Scholar] [CrossRef]
- Samaras, A.; Roumeliotis, E.; Ntasiou, P.; Karaoglanidis, G. Bacillus subtilis MBI600 promotes growth of tomato plants and induces systemic resistance contributing to the control of soilborne pathogens. Plants 2021, 10, 1113. [Google Scholar] [CrossRef]
- Gaya Karunasinghe, T.; Hashil Al-Mahmooli, I.; Al-Sadi, A.M.; Velazhahan, R. The effect of salt-tolerant antagonistic bacteria from tomato rhizosphere on plant growth promotion and damping-off disease suppression under salt-stress conditions. Acta Agric. Scand. Sect. B Soil. Plant Sci. 2020, 70, 69–75. [Google Scholar] [CrossRef]
- Al-Daghari, D.S.; Al-Sadi, A.M.; Al-Mahmooli, I.H.; Janke, R.; Velazhahan, R. Biological control efficacy of indigenous antagonistic bacteria isolated from the rhizosphere of cabbage grown in biofumigated soil against Pythium aphanidermatum damping-off of cucumber. Agriculture 2023, 13, 626. [Google Scholar] [CrossRef]
- Abdelhameed, R.E.; Metwally, R.A.; Soliman, S.A. Prospects of Bacillus amyloliquefaciens (MZ945930) mediated enhancement of Capsicum annuum L. plants under stress of Alternaria alternata in terms of physiological traits, thiol content, antioxidant defense, and phytohormones. J. Plant Growth Regul. 2024, 43, 3265–3281. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, I.; Pati, P. Post-infectional changes associated with the progression of leaf spot disease in Withania somnifera (L.) Dunal. J. Plant Pathol. 2011, 93, 397–405. [Google Scholar]
- Kazerooni, E.A.; Maharachchikumbura, S.S.N.; Al-Sadi, A.M.; Kang, S.M.; Yun, B.W.; Lee, I.J. Biocontrol potential of Bacillus amyloliquefaciens against Botrytis pelargonii and Alternaria alternata on Capsicum annuum. J. Fungi 2021, 7, 472. [Google Scholar] [CrossRef] [PubMed]
- Alwadi, H.M.; Baka, Z.A.M. Microorganisms associated with Withania somnifera leaves. Microbiol. Res. 2001, 156, 303–309. [Google Scholar] [CrossRef]
- Yildirim, E.; Turan, M.; Donmez, M.F. Mitigation of salt stress in radish (Raphanus sativus L.) by plant growth promoting rhizobacteria.—Roman. Biotechnol. Lett. 2008, 13, 3933–3943. [Google Scholar]
- Srivastava, S.; Bist, V.; Srivastava, S.; Singh, P.C.; Trivedi, P.K.; Asif, M.H.; Chauhan, P.S.; Nautiyal, C.S. Unraveling aspects of Bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of Rhizoctonia solani. Front. Plant Sci. 2016, 6, 587. [Google Scholar] [CrossRef] [PubMed]
- Metwally, R.A.; Soliman, S.A. Alleviation of the adverse effects of NaCl stress on tomato seedlings (Solanum lycopersicum L.) by Trichoderma viride through the antioxidative defense system. Bot. Stud. 2023, 64, 4. [Google Scholar] [CrossRef]
- Omara, A.E.; El-maghraby, F.M. Novel Bioformulations with Trichoderma lixii to Improve the Growth Dynamics and Biocontrol of the Cowpea Damping-Off Disease. Microbiol. Res. 2023, 14, 2041–2066. [Google Scholar] [CrossRef]
- Adhilakshmi, M.; Paranidharan, V.; Balachandar, D.; Ganesamurthy, K.; Velazhahan, R. Suppression of root rot of mung bean (Vigna radiata L.) by Streptomyces sp. is associated with induction of peroxidase and polyphenol oxidase. Arch. Phytopathol. Plant Prot. 2014, 47, 71–583. [Google Scholar] [CrossRef]
- Kavitha, R.; Umesha, S. Regulation of defense-related enzymes associated with bacterial spot resistance in tomato. Phytoparasitica 2008, 36, 144. [Google Scholar] [CrossRef]
- Perincherry, L.; Lalak-Kańczugowska, J.; Stępień, Ł. Fusarium-produced mycotoxins in plant-pathogen interactions. Toxins 2019, 11, 664. [Google Scholar] [CrossRef]
- Inayati, A.; Sulistyowati, L.; Aini, L.Q.; Yusnawan, E. Trichoderma virens-Tv4 enhances growth promoter and plant defenserelated enzymes of mungbean (Vigna radiata) against soil-borne pathogen Rhizoctonia solani. Biodiversitas J. Biol. Divers. 2020, 21, 2410–2419. [Google Scholar] [CrossRef]
- Constabel, C.P.; Barbehenn, R. Defensive roles of polyphenol oxidase in plants. In Induced Plant Resistance to Herbivory; Schaller, A., Ed.; Springer: Dordrecht, Germany, 2008. [Google Scholar]
- Mousa, E.S.; Elbagory, M.; Mahdy, M.E.; Abo-Koura, H.A.; Omara, A.E. Microencapsulation of Bacillus megaterium in Humic Acid-Supplied Alginate Beads Enhances Tomato Growth and Suppresses the Root-Knot Nematode Meloidogyne javanica Under Greenhouse Conditions. Horticulturae 2024, 10, 1284. [Google Scholar] [CrossRef]
- Kubalt, K. The role of phenolic compounds in plant resistance. Biotechnol. Food Sci. 2016, 80, 97–108. [Google Scholar]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Sood, P.; Citovsky, V. The roles of plant phenolics in defense and communication during Agrobacterium and Rhizobium infection. J. Mol. Plant Pathol. 2010, 11, 705–719. [Google Scholar] [CrossRef]
- Hesham, A.E.-L.; Upadhyay, R.S.; Sharma, G.D.; Manoharachary, C.; Gupta, V.K. Fungal Biotechnology and Bioengineering; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Radhakrishnan, R.; Lee, I.J. Gibberellins producing Bacillus methylotrophicus KE2 supports plant growth and enhances nutritional metabolites and food values of lettuce. Plant Physiol. Biochem. 2016, 109, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Egamberdieva, D.; Wirth, S.J.; Shurigin, V.V.; Hashem, A.; Abd-Allah, E.F. Endophytic bacteria improve plant growth, symbiotic performance of chickpea (Cicer arietinum L.) and induce suppression of root rot caused by Fusarium solani under salt stress. Front. Microbiol. 2017, 8, 1887. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montano, F.; Alias-Villegas, C.; Bellogin, R.A.; del Cerro, P.; Espuny, M.R.; Jimenez-Guerrero, I.; Lopez-Baena, F.J.; Ollero, F.J.; Cubo, T. Plant growth promotion in cereal and leguminous agricultural important plants: From microorganism capacities to crop production. Microbiol. Res. 2014, 169, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Almaghrabi, O.A.; Massoud, S.I.; Abdelmoneim, T.S. Influence of inoculation with plant growth promoting rhizobacteria (PGPR) on tomato plant growth and nematode reproduction under greenhouse conditions. Saudi J. Biol. Sci. 2013, 20, 57–61. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Chun, S.C.; Oh, J.W.; Paramasivan, M.; Saini, R.K.; Sahayarayan, J.J. Bacillus subtilis CBR05 for tomato (Solanum lycopersicum) fruits in South Korea as a novel plant probiotic bacterium (PPB): Implications from total phenolics, flavonoids, and carotenoids content for fruit quality. Agronomy 2019, 9, 838. [Google Scholar] [CrossRef]
- Rahi, A.A.; Anjum, M.A.; Iqbal Mirza, J.; Ahmad Ali, S.; Marfo, T.D.; Fahad, S.; Datta, R. Yield enhancement and better micronutrients uptake in tomato fruit through potassium humate combined with micronutrients mixture. Agriculture 2021, 11, 357. [Google Scholar] [CrossRef]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud. Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Ait Rahou, Y.; Douira, A.; Tahiri, A.; Cherkaoui, E.M.; Benkirane, R.; Meddich, A. Application of plant growth-promoting rhizobacteria combined with compost as a management strategy against Verticillium dahliae in tomato. Can. J. Plant Pathol. 2022, 44, 806–827. [Google Scholar] [CrossRef]
- Adinarayana, M.; Narasegowda, A.N.; Devappa, V. Effect of Bacillus subtilis var. amyloliquefaciens on Growth, Yield and Control of Early Blight in Tomato (Solanum lycopersicum L.). Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 656–664. [Google Scholar]
- Sinclair, J.B.; Dhingra, O.D. Basic Plant Pathology Methods; CRC Press: Boca Raton, FL, USA, 2017; 22p. [Google Scholar]
- Thi Minh Le, T.; Thi Hong Hoang, A.; Thi Bich Le, T.; Thi Bich Vo, T.; Van Quyen, D.; Hoang Chu, H. Isolation of endophytic fungi and screening of Huperzine A–producing fungus from Huperzia serrata in Vietnam. Sci. Rep. 2019, 9, 16152. [Google Scholar] [CrossRef] [PubMed]
- Burgess, N.; Recce, M.; O’Keefe, J. A model of hippocampal function. Neural Netw. 1994, 7, 1065–1081. [Google Scholar] [CrossRef]
- Margosch, D.; Gänzle, M.G.; Ehrmann, M.A.; Vogel, R.F. Pressure inactivation of Bacillus endospores. Appl. Environ. Microbiol. 2004, 70, 7321–7328. [Google Scholar] [CrossRef]
- Lu, Z.; Guo, W.; Liu, C. Isolation, identification and characterization of novel Bacillus subtilis. J. Vet. Med. Sci. 2018, 80, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Sadfi, N.; Cherif, M.; Fliss, I.; Boudabous, A.; Antoun, H. Evaluation of bacterial isolates from salty soils and Bacillus thuringiensis strains for the biocontrol of Fusarium dry rot of potato tubers. J. Plant Pathol. 2001, 83, 101–118. [Google Scholar]
- Aghighi, S.; Bonjar, G.S.; Rawashdeh, R.; Batayneh, S.; Saadoun, I. First report of antifungal spectra of activity of Iranian actinomycetes strains against Alternaria solani, Alternaria alternate, Fusarium solani, Phytophthora megasperma, Verticillium dahliae and Saccharomyces cerevisiae. Asian J. Plant Sci. 2004, 3, 463–471. [Google Scholar] [CrossRef]
- El-Shabrawy, E.; Heba, S. Controlling maize late-wilt and enhancing plant salinity tolerance by some rhizobacterial strains. Egypt. J. Phytopathol. 2018, 46, 235–255. [Google Scholar] [CrossRef]
- El-Nahrawy, S.; Elhawat, N.; Alshaal, T. Biochemical traits of Bacillus subtilis MF497446: Its implications on the development of cowpea under cadmium stress and ensuring food safety. Ecotoxicol. Environ. Saf. 2019, 180, 384–395. [Google Scholar] [CrossRef]
- Arafa, M.K. Studies on Fusarium Wilt of Cumin. Master’s Thesis, Assiut University, Asyut, Egypt, 1985. [Google Scholar]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology; Academic Press: San Diego, CA, USA, 1987; Volume 148, pp. 350–382. [Google Scholar]
- Hendrix, D.L. Rapid extraction and analysis of nonstructural carbohydrates in plant tissues. Crop. Sci. 1993, 33, 1306–1311. [Google Scholar] [CrossRef]
- Srivastava, S.K. Peroxidase and polyphenol-oxidase in Brassica juncea plants infected with Macrophomina phaseolina (Tassi): Goid of and their implication in disease resistance. Phytopathology 1987, 120, 249–254. [Google Scholar] [CrossRef]
- Matta, A.I.; Dimond, A.F. Symptoms of Fusarium wilt in relation to quantity of fungus and enzyme activity in tomato stems. Phytopathology 1963, 53, 574–578. [Google Scholar]
- Zucker, M. Sequential induction of phenylalanine ammonia layse and lyase in inactivating system in potato tuber disks. Plant Physiol. 1968, 43, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Zieslin, N.; Ben-Zaken, R. Peroxidase activity and presence of phenolic substances in peduncles of rose flowers. Plant Physiol. Biochem. 1993, 31, 333–339. [Google Scholar]
- Page, A.L.; Miller, R.H.; Keeny, D.R. Methods of Soil Analysis, Part II, Agronomy Monographs ASA and SSSA, 2nd ed.; Madison Book Company: Madison, WI, USA, 1982. [Google Scholar]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- USDA. Soil Survey Laboratory Methods Manual Soil Survey Investigation Report, No. 42, Version 4; USDA-NRCS: Lincoln, NE, USA, 2004. [Google Scholar]
- Duncan, D.B. Multiple range and multiple F tests. Biometrics 1955, 11, 1–42. [Google Scholar] [CrossRef]
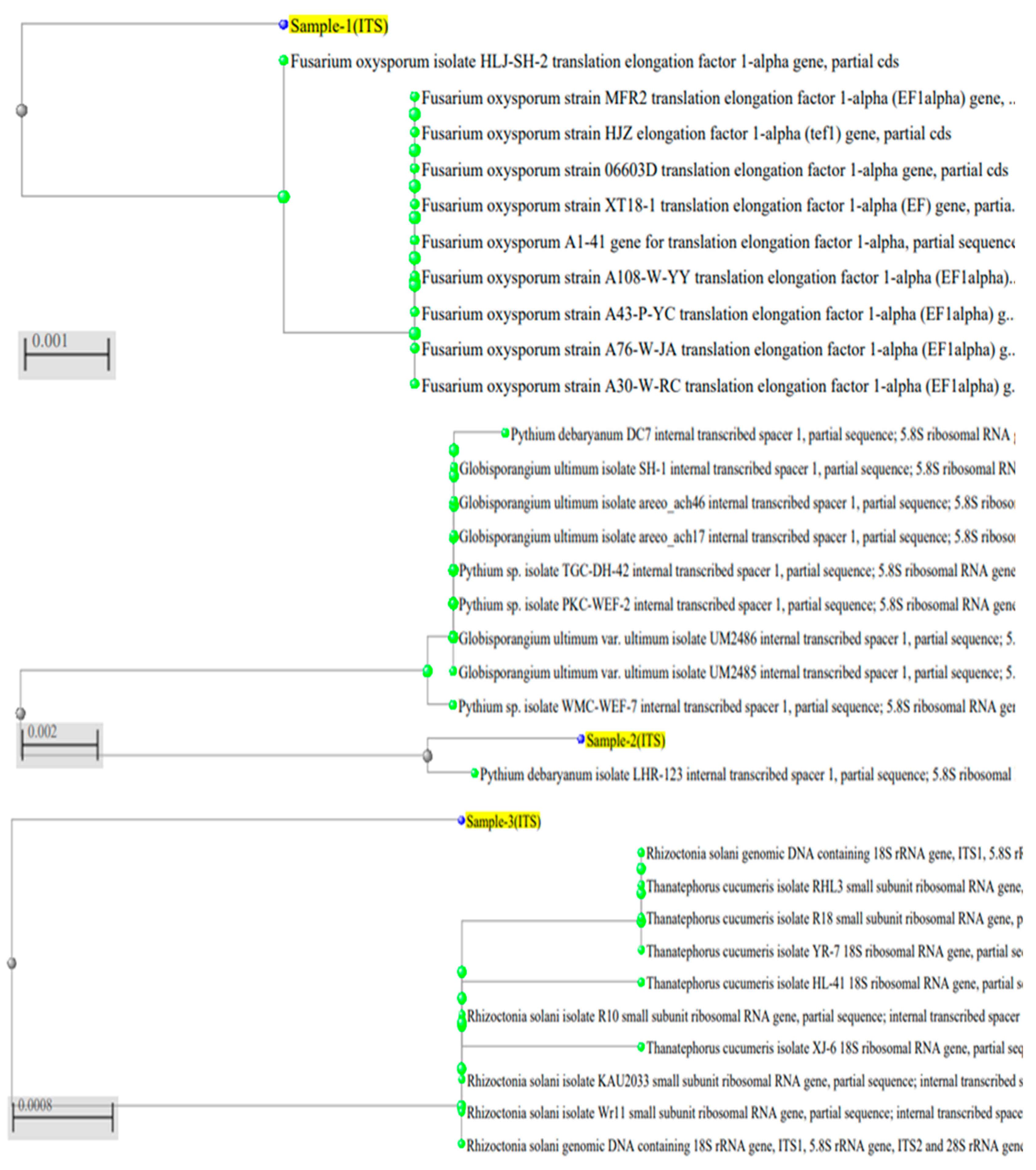
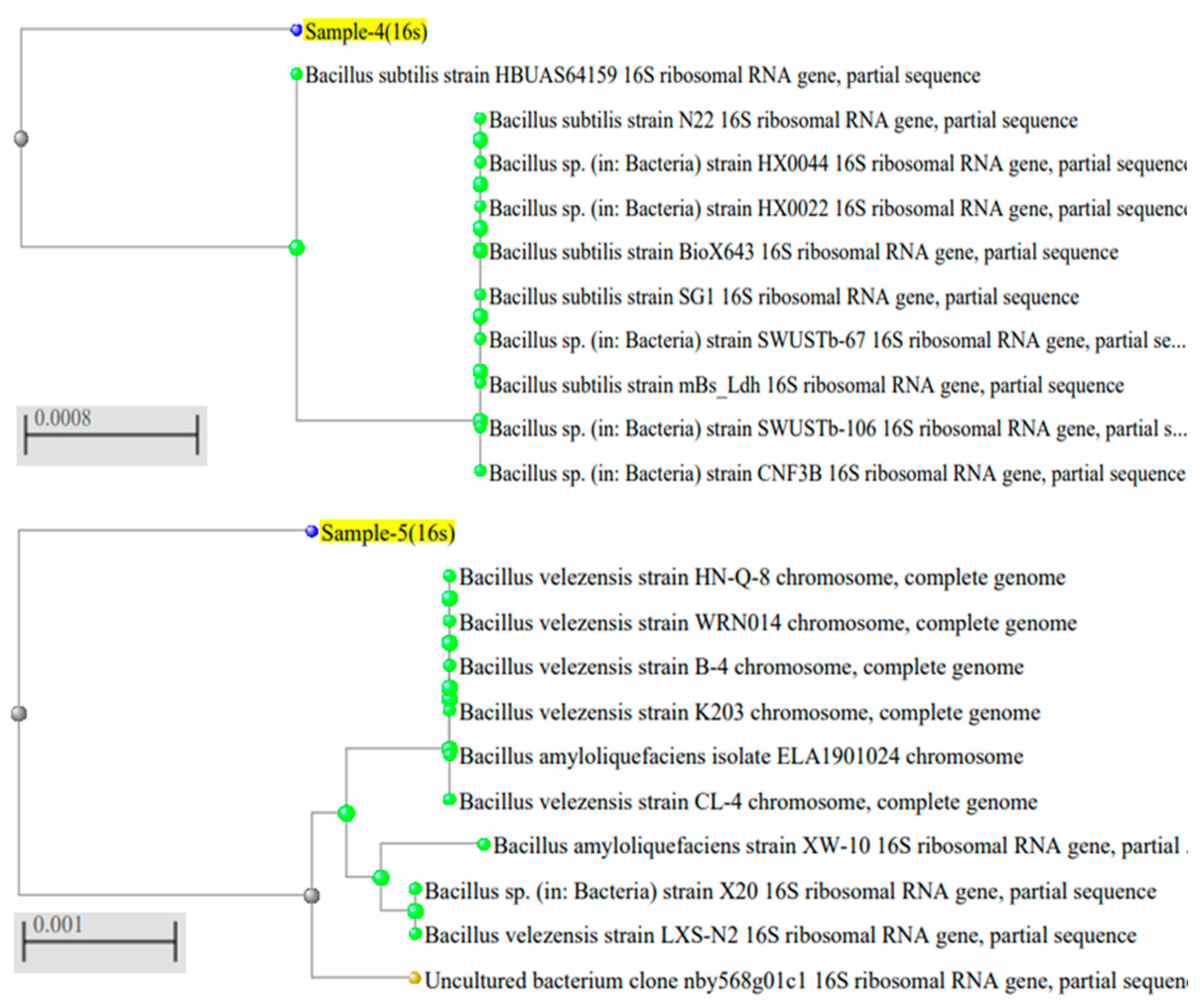



| Pathogenic Fungi | Antagonistic Impacts (Zone, mm) | |
|---|---|---|
| Bacillus subtilis | Bacillus amyloliquefaciens | |
| Rhizoctonia solani | 28.33 ± 2.08 a | 23.00 ± 2.00 b |
| Pythium debaryanum | 33.33 ± 1.15 b | 39.00 ± 1.00 a |
| Fusarium oxysporum | 33.00 ± 2.00 b | 15.33 ± 1.52 c |
| Treatments | Total Chlorophyll (mg g−1 FW) | Carotenoids (µg g−1 FW) | TSS (µg g−1 FW) | |||
|---|---|---|---|---|---|---|
| 2022 | 2023 | 2022 | 2023 | 2022 | 2023 | |
| T1 | 3.11 ± 0.85 e | 2.89 ± 0.49 e | 0.55 ± 0.09 g | 0.62 ± 0.10 e | 1.80 ± 0.32 e | 1.90 ± 0.85 e |
| T2 | 3.46 ± 0.37 e | 3.30 ± 0.39 e | 0.65 ± 0.11 fg | 0.70 ± 0.17 de | 1.98 ± 0.39 e | 2.05 ± 0.88 e |
| T3 | 1.39 ± 0.29 f | 1.22 ± 0.21 f | 0.29 ± 0.05 gh | 0.31 ± 0.07 f | 1.01 ± 0.28 f | 1.09 ± 0.27 f |
| T4 | 4.31 ± 0.89 d | 4.43 ± 0.38 d | 0.77 ± 0.15 ef | 0.83 ± 0.19 cde | 3.13 ± 0.57 d | 3.20 ± 0.68 d |
| T5 | 4.47 ± 0.97 d | 4.60 ± 0.49 d | 0.85 ± 0.19 ef | 0.93 ± 0.15 cd | 3.19 ± 0.65 d | 3.27 ± 0.26 d |
| T6 | 4.80 ± 0.87 d | 5.01 ± 0.51 d | 0.91 ± 0.12 e | 0.94 ± 0.14 cd | 3.32 ± 0.59 d | 3.40 ± 0.46 d |
| T7 | 8.03 ± 1.09 a | 8.49 ± 0.94 a | 1.57 ± 0.25 a | 1.77 ± 0.28 a | 5.25 ± 0.69 a | 5.58 ± 0.94 a |
| T8 | 6.49 ± 0.94 bc | 6.52 ± 0.78 c | 1.27 ± 0.31 bc | 1.36 ± 0.36 b | 4.52 ± 0.77 bc | 4.55 ± 0.91 bc |
| T9 | 7.07 ± 0.94 b | 7.35 ± 0.91 b | 1.38 ± 0.29 ab | 1.50 ± 0.41 b | 4.79 ± 0.81 b | 4.83 ± 0.83 b |
| T10 | 6.02 ± 0.91 c | 6.13 ± 0.76 c | 1.12 ± 0.22 cd | 1.28 ± 0.32 b | 4.27 ± 0.85 c | 4.34 ± 0.76 c |
| T11 | 6.33 ± 0.83 c | 6.46 ± 0.79 c | 1.21 ± 0.32 bc | 1.32 ± 0.27 b | 4.35 ± 0.80 bc | 4.41 ± 0.59 bc |
| T12 | 5.01 ± 0.59 d | 5.13 ± 0.94 d | 0.95 ± 0.19 de | 0.95 ± 0.21 c | 3.39 ± 0.46 d | 3.55 ± 0.48 d |
| LSD 0.05 | 0.72 | 0.74 | 0.19 | 0.24 | 0.43 | 0.41 |
| Treatments | N (%) | P (%) | K (%) | |||
|---|---|---|---|---|---|---|
| 2022 | 2023 | 2022 | 2023 | 2022 | 2023 | |
| T1 | 1.21 ± 0.25 e | 1.34 ± 0.23 e | 0.090 ± 0.01 d | 0.106 ± 0.02 f | 2.15 ± 0.25 e | 2.42 ± 1.15 b |
| T2 | 1.30 ± 0.19 de | 1.44 ± 0.35 de | 0.094 ± 0.03 d | 0.126 ± 0.01 ef | 2.24 ± 0.19 de | 2.66 ± 1.15 ab |
| T3 | 1.17 ± 0.09 e | 1.25 ± 0.05 e | 0.060 ± 0.01 e | 0.066 ± 0.02 g | 2.11 ± 0.09 e | 2.15 ± 0.15 b |
| T4 | 1.39 ± 0.22 cde | 1.48 ± 0.21 cde | 0.124 ± 0.02 c | 0.140 ± 0.03 de | 2.33 ± 0.22 cde | 2.66 ± 0.13 ab |
| T5 | 1.61 ± 0.27 bcd | 1.69 ± 0.19 bcd | 0.127 ± 0.01 c | 0.143 ± 0.02 cde | 2.55 ± 0.27 bcd | 2.66 ± 0.31 ab |
| T6 | 1.68 ± 0.22 bc | 1.75 ± 0.21 bcd | 0.130 ± 0.02 c | 0.160 ± 0.02 bcd | 2.62 ± 0.22 bc | 2.70 ± 0.36 ab |
| T7 | 2.30 ± 0.11 a | 2.48 ± 0.10 a | 0.187 ± 0.01 a | 0.216 ± 0.02 a | 3.24 ± 0.11 a | 3.29 ± 0.06 a |
| T8 | 1.82 ± 0.24 b | 1.92 ± 0.18 b | 0.171 ± 0.02 ab | 0.18 ± 0.02 ab | 2.76 ± 0.24 b | 2.93 ± 0.29 ab |
| T9 | 1.91 ± 0.15 b | 1.95 ± 0.11 b | 0.171 ± 0.01 ab | 0.206 ± 0.02 a | 2.85 ± 0.15 b | 2.95 ± 0.07 ab |
| T10 | 1.72 ± 0.13 b | 1.82 ± 0.16 b | 0.160 ± 0.02 b | 0.170 ± 0.01 bcd | 2.66 ± 0.13 b | 2.73 ± 0.05 ab |
| T11 | 1.78 ± 0.18 b | 1.90 ± 0.13 b | 0.161 ± 0.03 b | 0.173 ± 0.02 bc | 2.72 ± 0.18 b | 2.85 ± 0.15 ab |
| T12 | 1.71 ± 0.10 bc | 1.79 ± 0.08 bc | 0.135 ± 0.01 c | 0.160 ± 0.02 bcd | 2.65 ± 0.10 bc | 2.70 ± 0.12 ab |
| LSD 0.05 | 0.31 | 0.29 | 0.019 | 0.031 | 0.31 | 0.85 |
| Treatments | SL (cm Plant−1) | RL (cm Plant−1) | SDW (g Plant−1) | RDW (g Plant−1) | Yield (g Plant−1) |
|---|---|---|---|---|---|
| 2022 Season | |||||
| T1 | 65.06 ± 7.12 de | 22.44 ± 2.54 de | 188.67 ± 20.64 de | 88.98 ± 9.96 de | 780.70 ± 85.40 de |
| T2 | 61.28 ± 5.55 e | 21.08 ± 1.98 e | 177.70 ± 16.08 e | 83.69 ± 7.76 e | 735.33 ± 66.54 e |
| T3 | 62.43 ± 2.65 e | 21.50 ± 0.95 e | 181.05 ± 7.68 e | 85.30 ± 3.71 e | 749.17 ± 31.77 e |
| T4 | 78.88 ± 6.31 b | 27.37 ± 2.25 b | 228.75 ± 18.31 b | 108.33 ± 8.84 b | 946.56 ± 75.76 b |
| T5 | 74.09 ± 7.69 bcd | 23.78 ± 2.75 cde | 214.85 ± 22.29 bcd | 101.62 ± 10.76 bcd | 889.05 ± 92.24 bcd |
| T6 | 75.97 ± 6.27 bc | 27.20 ± 2.24 bc | 220.32 ± 18.19 bc | 104.26 ± 8.78 bc | 911.67 ± 75.27 bc |
| T7 | 76.89 ± 3.09 bc | 26.40 ± 1.10 bc | 222.99 ± 8.97 bc | 105.55 ± 4.33 bc | 922.72 ± 37.11 bc |
| T8 | 67.74 ± 7.02 cde | 23.85 ± 2.51 cde | 196.44 ± 20.35 cde | 92.60 ± 9.82 cde | 812.84 ± 84.19 cde |
| T9 | 77.23 ± 4.39 b | 25.74 ± 1.57 bcd | 223.98 ± 12.73 b | 105.63 ± 6.15 bc | 926.80 ± 52.67 b |
| T10 | 82.57 ± 3.84 b | 27.59 ± 1.37 ab | 239.46 ± 11.15 b | 113.10 ± 5.38 b | 990.88 ± 46.12 b |
| T11 | 80.14 ± 5.25 b | 28.03 ± 1.87 ab | 232.40 ± 15.22 b | 109.69 ± 7.35 b | 961.64 ± 62.98 b |
| T12 | 94.13 ± 2.76 a | 31.08 ± 0.99 a | 272.97 ± 8.00 a | 129.28 ± 3.86 a | 1129.52 ± 33.12 a |
| LSD 0.05 | 9.33 | 3.52 | 27.07 | 13.06 | 112.01 |
| 2023 Season | |||||
| T1 | 69.55 ± 7.61 de | 25.02 ± 2.74 de | 201.68 ± 22.06 de | 97.59 ± 10.67 de | 793.71 ± 86.82 de |
| T2 | 65.50 ± 5.93 e | 23.57 ± 2.13 e | 189.96 ± 17.19 e | 91.92 ± 8.32 e | 747.58 ± 67.65 e |
| T3 | 66.74 ± 2.83 e | 24.01 ± 1.02 e | 193.54 ± 8.21 e | 93.65 ± 3.97 e | 761.66 ± 32.30 e |
| T4 | 84.32 ± 6.75 b | 30.34 ± 2.43 b | 244.53 ± 19.57 b | 118.32 ± 9.47 b | 962.33 ± 77.03 b |
| T5 | 79.20 ± 8.22 bcd | 28.50 ± 2.96 bcd | 229.67 ± 23.83 bcd | 111.13 ± 11.53 bcd | 903.87 ± 93.78 bcd |
| T6 | 81.21 ± 6.70 bc | 29.22 ± 2.41 bc | 235.52 ± 19.44 bc | 113.96 ± 9.41 bc | 926.87 ± 76.52 bc |
| T7 | 82.20 ± 3.31 bc | 29.57 ± 1.19 bc | 238.37 ± 9.59 bc | 115.34 ± 4.64 bc | 938.10 ± 37.73 bc |
| T8 | 72.41 ± 7.50 cde | 26.05 ± 2.70 cde | 209.98 ± 21.75 cde | 101.61 ± 10.52 cde | 826.39 ± 85.59 cde |
| T9 | 82.56 ± 4.69 b | 29.71 ± 1.69 b | 239.42 ± 13.61 b | 115.85 ± 6.58 b | 942.24 ± 53.55 b |
| T10 | 88.27 ± 4.11 b | 31.76 ± 1.48 b | 255.98 ± 11.91 b | 123.86 ± 5.77 b | 1007.40 ± 46.89 b |
| T11 | 85.66 ± 5.61 b | 30.82 ± 2.02 b | 248.42 ± 16.27 b | 120.20 ± 7.87 b | 977.67 ± 64.03 b |
| T12 | 100.62 ± 2.95 a | 36.20 ± 1.06 a | 291.79 ± 8.56 a | 141.19 ± 4.14 a | 1148.34 ± 33.67 a |
| LSD 0.05 | 9.97 | 3.59 | 28.93 | 14.00 | 113.87 |
| Season | Mechanical Analysis (%) | Texture | pH (1:2.5) | EC (dSm−1) | OM (g Kg−1) | Available Elements (mg Kg−1) | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sand | Silt | Clay | N | P | K | |||||
| 2022 | 21.14 | 25.69 | 53.17 | Clayey | 7.71 | 2.61 | 16.95 | 8.96 | 8.22 | 391.31 |
| 2023 | 21.33 | 25.02 | 53.65 | Clayey | 7.62 | 2.89 | 17.82 | 9.32 | 8.76 | 372.27 |
| Symbol | Description |
|---|---|
| T1 | Seedlings grown in soil infested with F. oxysporum (3%) |
| T2 | Seedlings grown in soil infested with P. debaryanum (3%) |
| T3 | Seedlings grown in soil infested with R. solani (3%) |
| T4 | Seedlings dipped with Topsin-M70 (fungicide, 2 g L−1, 90 min) + Soil infested with F. oxysporum (3%) |
| T5 | Seedlings dipped with Topsin-M70 (fungicide, 2 g L−1, 90 min) + Soil infested with P. debaryanum (3%) |
| T6 | Seedlings dipped with Topsin-M70 (fungicide, 2 g L−1, 90 min) + Soil infested with R. solani (3%) |
| T7 | Seedlings dipped with B. subtilis (90 min) + Soil infested with F. oxysporum (3%) |
| T8 | Seedlings dipped with B. subtilis (90 min) + Soil infested with P. debaryanum (3%) |
| T9 | Seedlings dipped with B. subtilis (90 min) + Soil infested with R. solani (3%) |
| T10 | Seedlings dipped with B. amyloliquefaciens (90 min) + Soil infested with F. oxysporum (3%) |
| T11 | Seedlings dipped with B. amyloliquefaciens (90 min) + Soil infested with P. debaryanum (3%) |
| T12 | Seedlings dipped with B. amyloliquefaciens (90 min) + Soil infested with R. solani (3%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, D.F.I.; El-Nahrawy, S.; EL-Zawawy, H.A.H.; Omara, A.E.-D. Effective Applications of Bacillus subtilis and B. amyloliquefaciens as Biocontrol Agents of Damping-Off Disease and Biostimulation of Tomato Plants. Stresses 2025, 5, 9. https://doi.org/10.3390/stresses5010009
Ali DFI, El-Nahrawy S, EL-Zawawy HAH, Omara AE-D. Effective Applications of Bacillus subtilis and B. amyloliquefaciens as Biocontrol Agents of Damping-Off Disease and Biostimulation of Tomato Plants. Stresses. 2025; 5(1):9. https://doi.org/10.3390/stresses5010009
Chicago/Turabian StyleAli, Dina Fathi Ismail, Sahar El-Nahrawy, Hassan A. H. EL-Zawawy, and Alaa El-Dein Omara. 2025. "Effective Applications of Bacillus subtilis and B. amyloliquefaciens as Biocontrol Agents of Damping-Off Disease and Biostimulation of Tomato Plants" Stresses 5, no. 1: 9. https://doi.org/10.3390/stresses5010009
APA StyleAli, D. F. I., El-Nahrawy, S., EL-Zawawy, H. A. H., & Omara, A. E.-D. (2025). Effective Applications of Bacillus subtilis and B. amyloliquefaciens as Biocontrol Agents of Damping-Off Disease and Biostimulation of Tomato Plants. Stresses, 5(1), 9. https://doi.org/10.3390/stresses5010009






