Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens
Abstract
1. Introduction
2. The Genus Bacillus as a Lipopeptide Producer
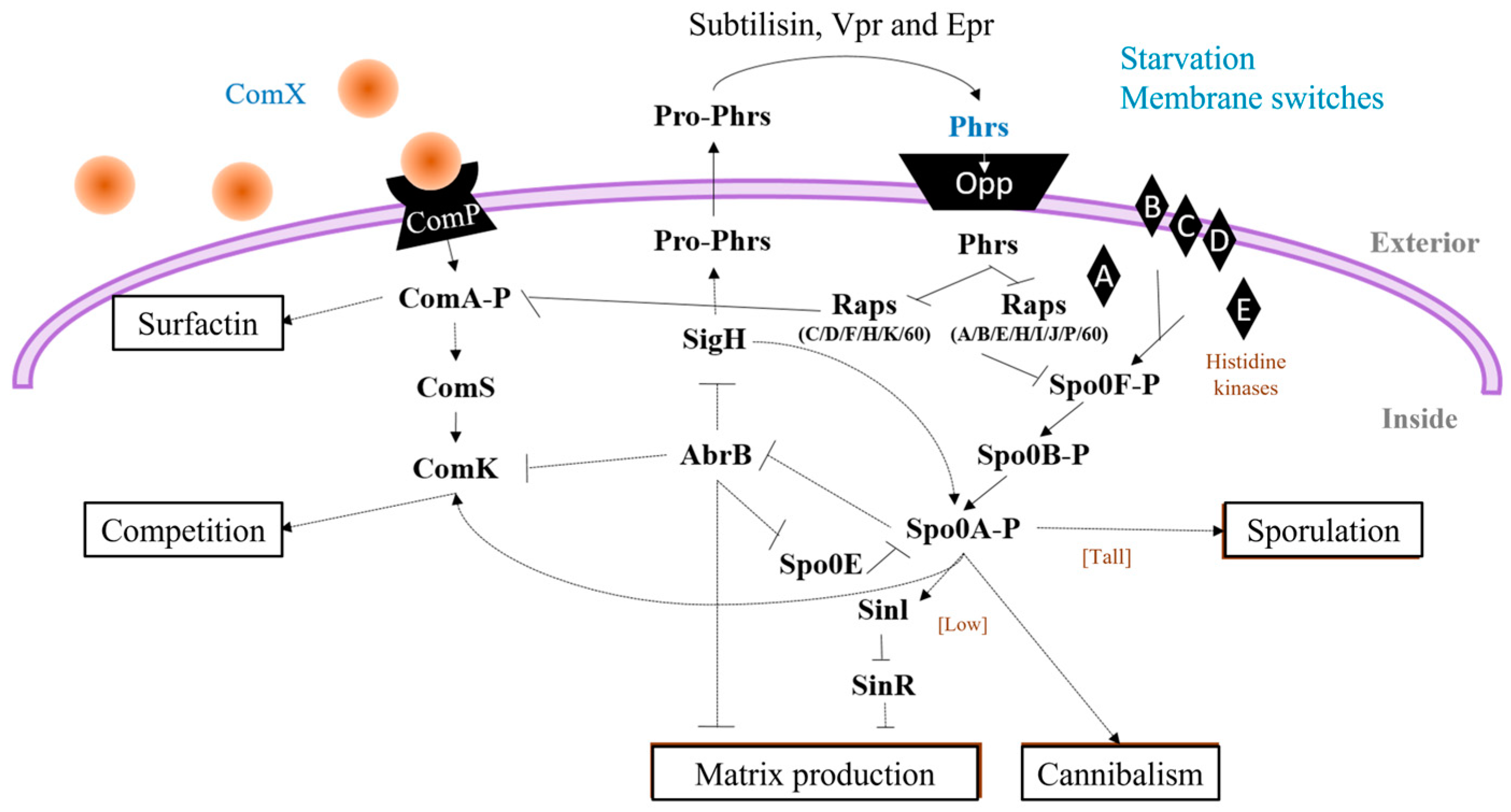
2.1. Surfactin Biosynthesis
- ComA (ComA-P): ComX is a pheromone and ComP, a membrane kinase that phosphorylates ComA, induces the pathway for the production of surfactin [50] (Figure 3). In addition, a fraction of the surfactin producers undergo a secondary process of cell differentiation to convert a subpopulations of cells that are competent and capable of incorporating exogenous DNA, to cells with the ability to acquire characteristics that benefit them under stress conditions [51].
- DegU (DegU-P): DegQ activates cytoplasmic kinase DegS with the addition of ComA-P Spacapan et al., [52] DegS is the one which phosphorylates DegU. Activation of DegU-P leads to the expression of the machinery responsible for the production and secretion of proteases, constituting the subpopulation of miners, and is also responsible for providing more assimilable peptides to the community through the hydrolysis of the most complex molecules [53].
- Spo0A (Spo0A-P): five kinases (Kin AE) are responsible for sensing the signals that activate the Spo0A-P protein to differentiate subpopulations towards the cell matrix producers and cannibalism when there are low levels of phosphorylated protein in the cell, and when there are levels high Spo0A-P, sporulation genes are induced [54].
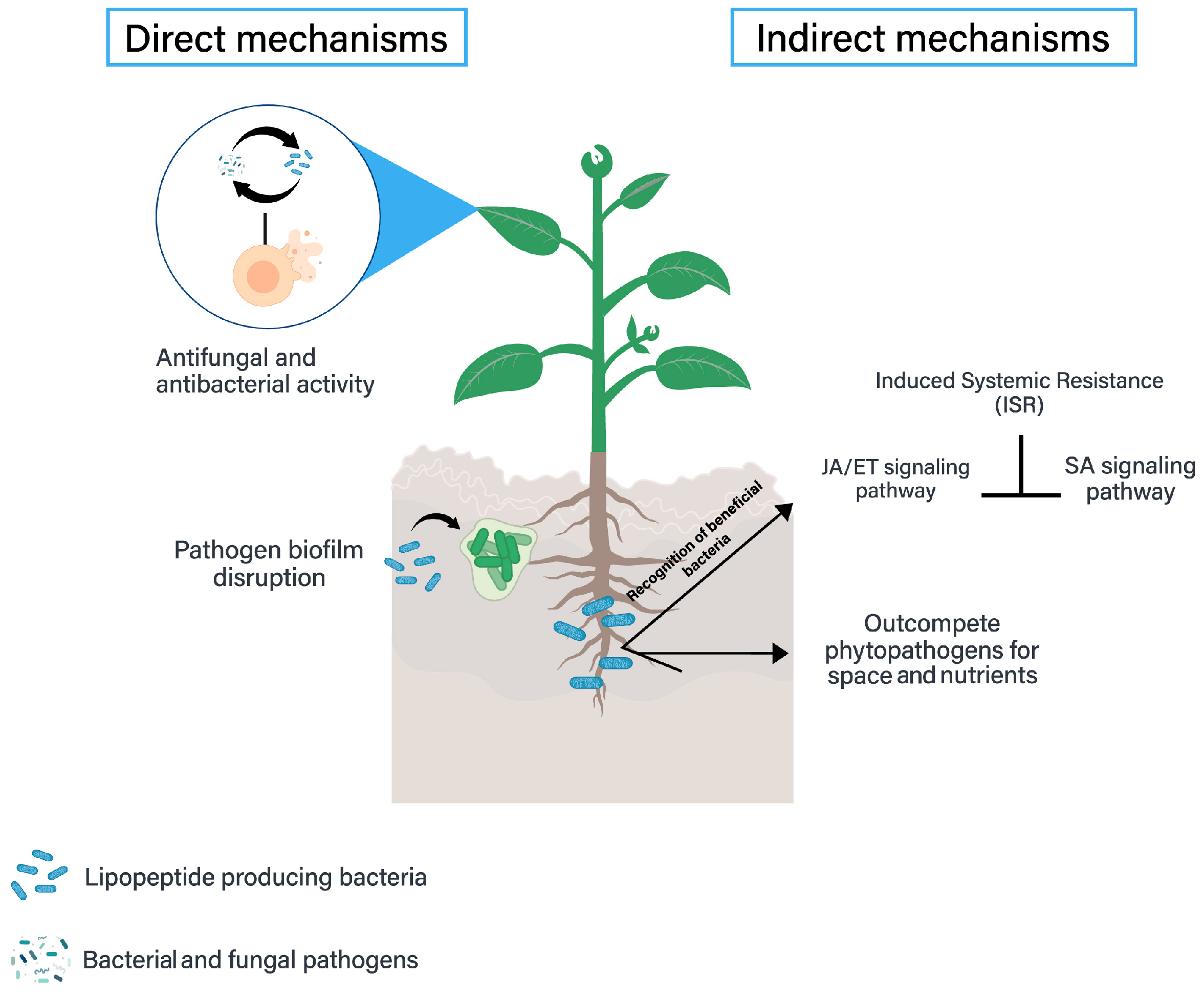
Surfactin Biocontrol Activity
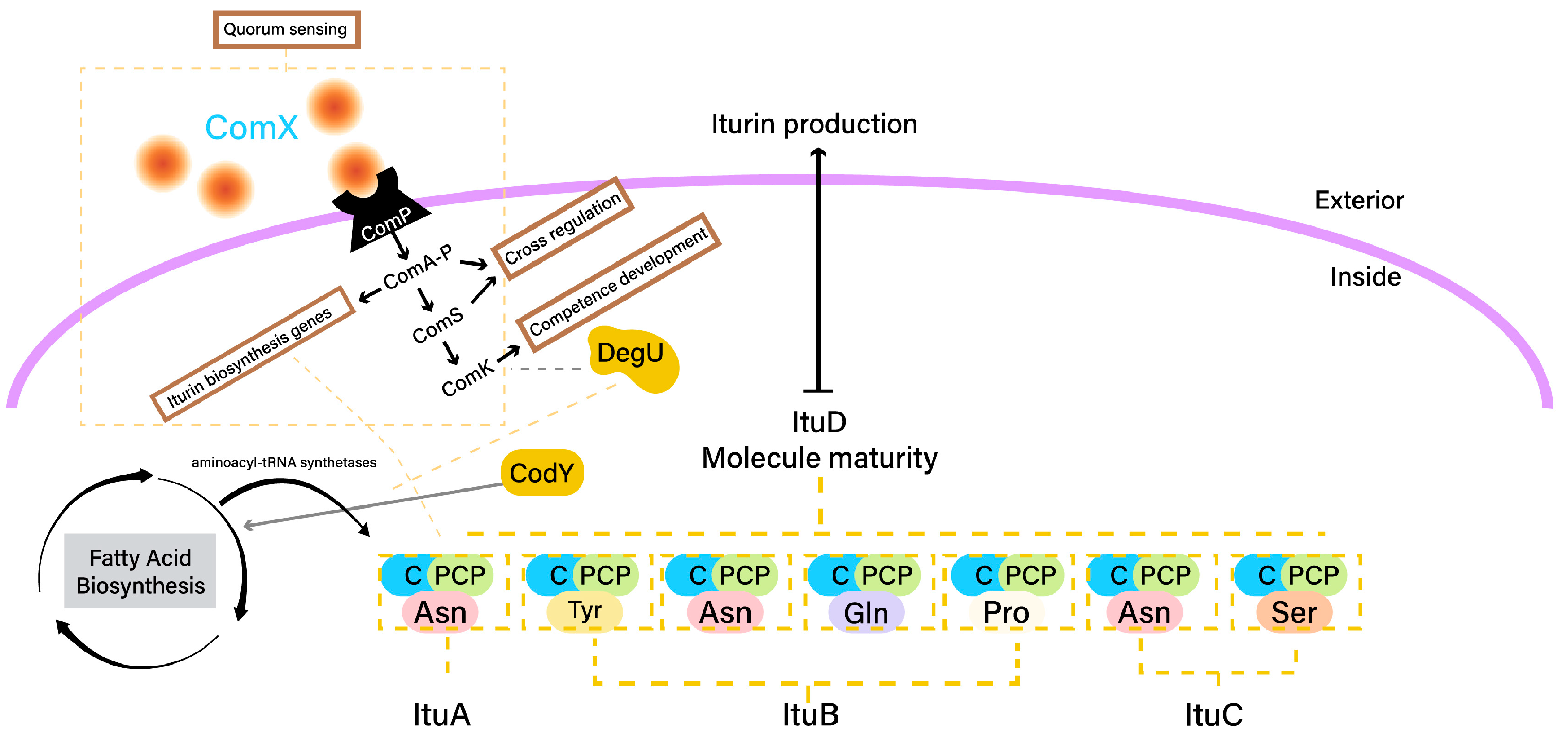
2.2. Iturin Biosynthesis
- ComA: The response regulator ComA is a key player in quorum sensing, a mechanism that coordinates gene expression based on cell density. When quorum-sensing signaling peptides (i.e., ComX) reach a certain concentration, ComA becomes phosphorylated and binds to the promoters of target genes, including those involved in iturin biosynthesis [78]. This activates the transcription of iturin biosynthetic genes.
- DegU: The response regulator DegU is part of a two-component regulatory system. Phosphorylated DegU activates the transcription of genes involved in iturin biosynthesis, enhancing their expression [79].
- CodY: The transcriptional regulator CodY senses nutrient availability. In nutrient-rich conditions, CodY binds to the promoters of genes related to secondary metabolism, including iturin biosynthetic genes, repressing their expression [80].
Iturin Biocontrol Activity
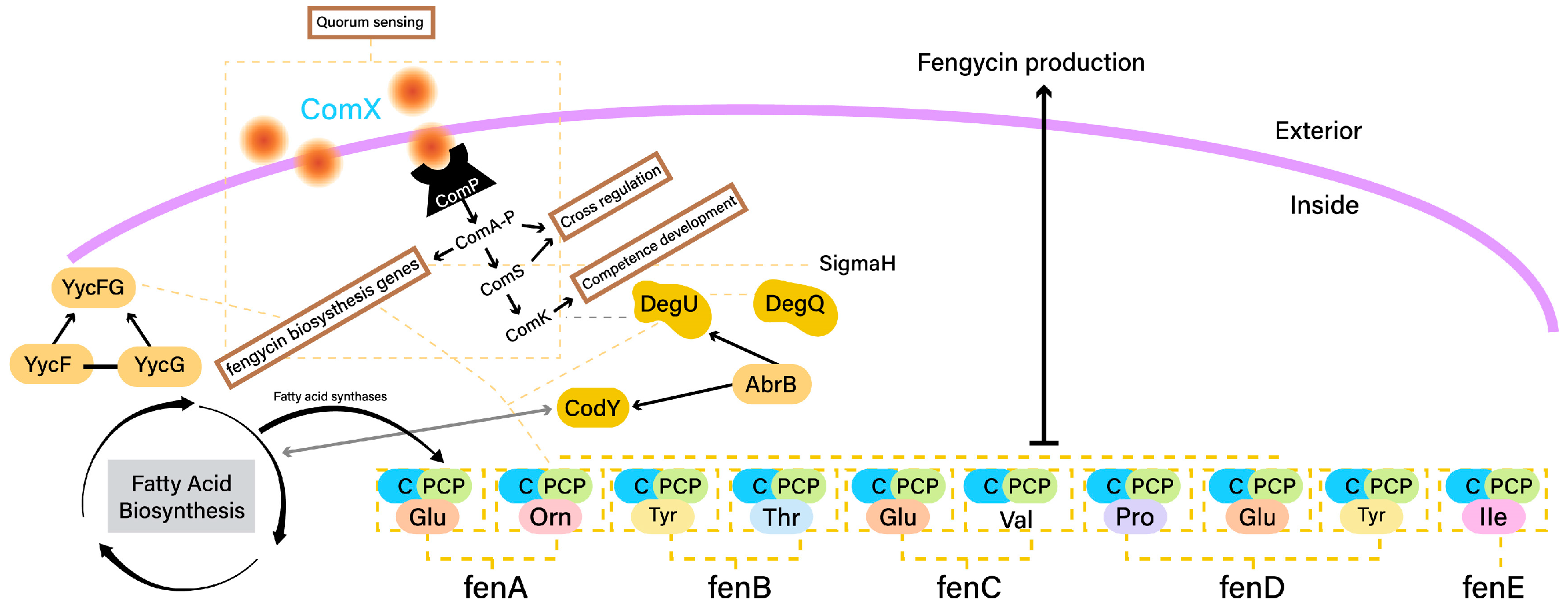
2.3. Fengycin Biosynthesis
- CodY is a global regulator that plays a pivotal role in coordinating fengycin production with nutrient availability. CodY responds to changes in intracellular amino acid levels and acts as a sensor for nutrient sufficiency. In response to limiting nutrients, CodY negatively regulates the expression of fengycin biosynthetic genes, thus adjusting fengycin production to match cellular metabolic status [71].
- The ComA-ComP quorum sensing system is central to fengycin production regulation in response to cell population density. The ComP histidine kinase senses external signaling peptides, and upon reaching a certain threshold, activates ComA, a response regulator. Activated ComA influences the expression of genes, including those involved in fengycin biosynthesis, in a density-dependent manner. This system ensures coordinated fengycin production within a microbial community [85].
- DegU is a response regulator involved in fengycin regulation and environmental adaptation. The DegU phosphorylation status determines its activity as a transcription factor. In response to specific environmental cues, such as cell wall stress, DegU influences fengycin biosynthesis by directly affecting the expression of fengycin biosynthetic genes and other regulators [86].
- AbrB is a pleiotropic transcriptional regulator that modulates the activity of CodY and DegU. It indirectly impacts fengycin production by influencing the regulatory cascades controlled by CodY and DegU. AbrB’s role in coordinating various regulatory pathways adds complexity to the control of fengycin biosynthesis [83].
Fengycin Biocontrol Activity
3. Extraction of Bacillus Lipopeptides
4. Identification of Bacillus Lipopeptide-Producer Strains
4.1. Phenotype Level
4.2. DNA/RNA Level
4.3. Protein Level
4.4. Metabolic Level
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. Pesticides use, pesticides trade and pesticides indicators. In Global, Regional and Country Trends, 1990–2020; FAOSTAT Analytical Briefs; FAO: Rome, Italy, 2022; pp. 1–13. [Google Scholar]
- Díaz Rodríguez, A.M.; Parra Cota, F.I.; Santoyo, G.; de los Santos Villalobos, S. Chlorothalonil tolerance of indole producing bacteria associated to wheat (Triticum turgidum L.) rhizosphere in the Yaqui Valley, Mexico. Ecotoxicology 2019, 28, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Bhatt, P.; Chen, S. Carbofuran toxicity and its microbial degradation in contaminated environments. Chemosphere 2020, 259, 127419. [Google Scholar] [CrossRef] [PubMed]
- Pang, S.; Lin, Z.; Zhang, Y.; Zhang, W.; Alansary, N.; Mishra, S.; Bhatt, P.; Chen, S. Insights into the Toxicity and Degradation Mechanisms of Imidacloprid Via Physicochemical and Microbial Approaches. Toxics 2020, 8, 65. [Google Scholar] [CrossRef]
- Córdova-Albores, L.C.; Zelaya-Molina, L.X.; Ávila-Alistac, N.; Valenzuela-Ruíz, V.; Cortés-Martínez, N.E.; Parra-Cota, F.I.; Burgos-Canul, Y.Y.; Chávez-Díaz, I.F.; Fajardo-Franco, M.L.; De los Santos-Villalobos, S. Omics sciences potential on bioprospecting of biological control microbial agents: The case of the Mexican agro-biotechnology. Rev. Mex. Fitopatol. 2020, 39, 147–184. [Google Scholar] [CrossRef]
- Hu, J.; Zheng, M.; Dang, S.; Shi, M.; Zhang, J.; Li, Y. Biocontrol Potential of Bacillus amyloliquefaciens LYZ69 against Anthracnose of Alfalfa (Medicago sativa). J. Phytopathol. 2021, 111, 1338–1348. [Google Scholar] [CrossRef]
- Samada, L.H.; Tambunan, U.S.F. Biopesticides as promising alternatives to chemical pesticides: A review of their current and future status. Online J. Biol. Sci. 2020, 20, 66–76. [Google Scholar] [CrossRef]
- Morales-Cedeño, L.R.; Barajas-Barrera, I.A.; Parra-Cota, F.I.; Valenzuela-Ruiz, V.; de los Santos-Villalobos, S.; Loeza-Lara, P.D.; Herrera-Pérez, A.; del Carmen Orozco-Mosqueda, M.; Santoyo, G. Evaluation of Biocontrol Potential of Bacillus spp. and Pseudomonas fluorescens UM270 against Postharvest Fungal Pathogens. Microbiol. Res. 2023, 14, 1511–1523. [Google Scholar] [CrossRef]
- Montoya-Martínez, A.C.; Figueroa-Brambila, K.M.; Escalante-Beltrán, A.; López-Montoya, N.D.; Valenzuela-Ruíz, V.; Parra-Cota, F.I.; Estrada Alvarado, M.I.; de los Santos-Villalobos, S. Biological Control Mechanisms of Bacillus cabrialesii subsp. tritici TSO2T against Fusarium languescens, the Causal Agent of Wilt in Jalapeño Peppers. Horticulturae 2023, 9, 964. [Google Scholar]
- Valenzuela-Ruiz, V.; Parra-Cota, F.I.; Santoyo, G.; de los Santos-Villalobos, S. Potential biocontrol mechanisms of Bacillus sp. TSO2 against Bipolaris sorokiniana, spot blotch in wheat. Rev. Mex. Fitopatol. 2022, 40, 230–239. [Google Scholar] [CrossRef]
- Mishra, R.K.; Bohra, A.; Kamaal, N.; Kumar, K.; Gandhi, K.; Sujayanand, G.K.; Saabale, P.R.; Satheesh Naik, S.J.; Birinchi Kumar, S.; Dharmendra, K.; et al. Utilization of biopesticides as sustainable solutions for management of pests in legume crops: Achievements and prospects. Egypt. J. Biol. Pest Control 2018, 28, 3. [Google Scholar] [CrossRef]
- Ongena, M.; Jacques, P.; Touré, Y.; Destain, J.; Jabrane, A.; Thonart, P. Involvement of fengycin-type lipopeptides in the multifaceted biocontrol potential of Bacillus subtilis. Appl. Microbiol. Biotechnol. 2005, 69, 29–38. [Google Scholar] [CrossRef]
- Tut, G.; Magan, N.; Brain, P.; Xu, X. Critical Evaluation of Two Commercial Biocontrol Agents for Their Efficacy against B. cinerea under In Vitro and In Vivo Conditions in Relation to Different Abiotic Factors. Agronomy 2021, 11, 1868. [Google Scholar] [CrossRef]
- Hashem, A.; Tabassum, B.; Fathi Abd_Allah, E. Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J. Biol. Sci. 2019, 26, 1291–1297. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Ruiz, V.; Gálvez-Gamboa, G.T.; Villla-Rodriguez, E.D.; Parra-Cota, F.I.; Santoyo, G.; de los Santos Villalobos, S. Lipopeptides produced by biological control agents of the genus Bacillus: A review of analytical tools used for their study. Rev. Mex. Cienc. Agríc. 2020, 11, 419–432. [Google Scholar]
- Santoyo, G.; Sánchez-Yáñez, J.; de los Santos-Villalobos, S. Methods for detecting biocontrol and plant growth-promoting traits in rhizobacteria. In Methods in Rhizosphere Biology Research: Rhizosphere Biology; Reinhardt, D., Sharma, A., Eds.; Springer: Singapore, 2019; pp. 133–149. [Google Scholar]
- Salazar, B.; Ortiz, A.; Keswani, C.; Minkina, T.; Mandzhieva, S.; Pratap Singh, S.; Rekadwad, B.; Borriss, R.; Jain, A.; Singh, H.B.; et al. Bacillus spp. as Bio-factories for Antifungal Secondary Metabolites: Innovation Beyond Whole Organism Formulations. Microb. Ecol. 2023, 86, 1–24. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Logan, N.A.; De Vos, P. Bacillus Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc. in Association with Bergey’s Manual Trust: Hoboken, NJ, USA, 2015; 163p. [Google Scholar]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef]
- Jacques, P. Surfactin and Other Lipopeptides from Bacillus spp. In Biosurfactants: From Genes to Applications; Springer: Berlin/Heidelberg, Germany, 2011; pp. 57–91. [Google Scholar]
- Villarreal-Delgado, M.F.; Villa-Rodríguez, E.D.; Cira-Chávez, L.A.; Estrada-Alvarado, M.I.; Parra-Cota, F.I.; De los Santos-Villalobos, S. El género Bacillus como agente de control biológico y sus implicaciones en la bioseguridad agrícola. Rev. Mex. Fitopatol. 2018, 36, 95–130. [Google Scholar] [CrossRef]
- Hamoen, L.W.; Venema, G.; Kuipers, O.P. Controlling competence in Bacillus subtilis: Shared use of regulators. Microbiology 2003, 149, 9–17. [Google Scholar] [CrossRef]
- Villa-Rodriguez, E.; Moreno-Ulloa, A.; Castro-Longoria, E.; Parra-Cota, F.I.; de los Santos-Villalobos, S. Integrated omics approaches for deciphering antifungal metabolites produced by a novel Bacillus species, B. cabrialesii TE3T, against the spot blotch disease of wheat (Triticum turgidum L. subsp. durum). Microbiol. Res. 2021, 251, 126826. [Google Scholar] [CrossRef]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Li, R.; Du, W.; Yang, J.; Liu, Z.; Yousef, A.E. Control of Listeria monocytogenes biofilm by paenibacterin, a natural antimicrobial lipopeptide. Food Control 2018, 84, 529–535. [Google Scholar] [CrossRef]
- Crouzet, J.; Arguelles-Arias, A.; Dhondt-Cordelier, S.; Cordelier, S.; Pršic, J.; Hoff, G.; Mazeyrat-Gourbeyre, F.; Baillieul, F.; Clément, C.; Ongena, M.; et al. Biosurfactants in Plant Protection against Diseases: Rhamnolipids and Lipopeptides Case Study. Front. Bioeng. Biotechnol. 2020, 8, 1014. [Google Scholar] [CrossRef]
- Al-Mutar, D.M.K.; Noman, M.; Abduljaleel Alzawar, N.S.; Azizullah; Li, D.; Song, F. Cyclic Lipopeptides of Bacillus amyloliquefaciens DHA6 Are the Determinants to Suppress Watermelon Fusarium Wilt by Direct Antifungal Activity and Host Defense Modulation. J. Fungi 2023, 9, 687. [Google Scholar] [CrossRef]
- García-Gutiérrez, L.; Zeriouh, H.; Romero, D.; Cubero, J.; de Vicente, A.; Pérez-García, A. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate- and salicylic acid-dependent defence responses. Microb. Biotechnol. 2013, 6, 264–274. [Google Scholar] [CrossRef]
- Andrić, S.; Meyer, T.; Rigolet, A.; Prigent-Combaret, C.; Höfte, M.; Balleux, G.; Steels, S.; Hoff, G.; De Mot, R.; McCann, A.; et al. Lipopeptide Interplay Mediates Molecular Interactions between Soil Bacilli and Pseudomonads. Am. Soc. Microbiol. 2021, 9, e02038-21. [Google Scholar] [CrossRef]
- Ma, Y.; Kong, Q.; Qin, C.; Chen, Y.; Chen, Y.; Lv, R.; Zhou, G. Identification of lipopeptides in Bacillus megaterium by two-step ultrafiltration and LC–ESI–MS/MS. AMB Express 2016, 6, 79. [Google Scholar] [CrossRef]
- Geissler, M.; Heravi, K.M.; Henkel, M.; Hausmann, R. Lipopeptide Biosurfactants from Bacillus Species. In Biobased Surfactants: Synthesis, Properties, and Applications; AOCS Press: Champaign, IL, USA, 2019; pp. 205–240. [Google Scholar]
- Villa-Rodríguez, E.; Parra-Cota, F.; Castro-Longoria, E.; López-Cervantes, J.; de los Santos-Villalobos, S. Bacillus subtilis TE3: A promising biological control agent against Bipolaris sorokiniana, the causal agent of spot blotch in wheat (Triticum turgidum L. subsp. durum). Biol. Control 2019, 132, 135–143. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Zheng, Q.-W.; Wei, T.; Zhang, Z.-Q.; Zhao, C.-F.; Zhong, H.; Xu, Q.-Y.; Lin, J.-F.; Guo, L.-Q. Isolation and Characterization of Fengycins Produced by Bacillus amyloliquefaciens JFL21 and Its Broad-Spectrum Antimicrobial Potential against Multidrug-Resistant Foodborne Pathogens. Front. Microbiol. 2020, 11, 579621. [Google Scholar] [CrossRef] [PubMed]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Nikolić, I.; Berić, T.; Dimkić, I.; Popović, T.; Lozo, J.; Fira, D.; Stanković, S. Biological control of Pseudomonas syringae pv. aptata on sugar beet with Bacillus pumilus SS-10.7 and Bacillus amyloliquefaciens (SS-12.6 and SS-38.4) strains. J. Appl. Microbiol. 2019, 126, 165–176. [Google Scholar]
- Al-Mutar, D.M.K.; Alzawar, N.S.A.; Noman, M.; Li, D.; Song, F. Suppression of fusarium wilt in watermelon by Bacillus amyloliquefaciens DHA55 through extracellular production of antifungal Lipopeptides. J. Fungi 2023, 9, 336. [Google Scholar] [CrossRef]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef]
- Chen, M.; Wang, J.; Liu, B.; Zhu, Y.; Xiao, R.; Yang, W.; Ge, C.; Chen, Z. Biocontrol of tomato bacterial wilt by the new strain Bacillus velezensis FJAT-46737 and its lipopeptides. BMC Microbiol. 2020, 20, 160. [Google Scholar] [CrossRef]
- Preecha, C.; Sadowsky, M.J.; Prathuangwong, S. Lipopeptide surfactin produced by Bacillus amyloliquefaciens KPS46 is required for biocontrol efficacy against Xanthomonas axonopodis pv. glycines. Agric. Nat. Resour. 2010, 44, 84–99. [Google Scholar]
- Alvarez, F.; Castro, M.; Príncipe, A.; Borioli, G.; Fischer, S.; Mori, G.; Jofré, E. The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J. Appl. Microbiol. 2012, 112, 159–174. [Google Scholar] [CrossRef]
- Cuellar-Gaviria, T.Z.; González-Jaramillo, L.M.; Villegas-Escobar, V. Role of Bacillus tequilensis EA-CB0015 cells and lipopeptides in the biological control of black Sigatoka disease. Biol. Control 2021, 155, 104523. [Google Scholar] [CrossRef]
- Ongena, M.; Duby, F.; Jourdan, E.; Beaudry, T.; Jadin, V.; Dommes, J.; Thonart, P. Bacillus subtilis M4 decreases plant susceptibility towards fungal pathogens by increasing host resistance associated with differential gene expression. Appl. Microbiol. Biotechnol. 2005, 67, 692–698. [Google Scholar] [CrossRef]
- Gutiérrez-Chávez, C.; Benaud, N.; Ferrari, B.C. The ecological roles of microbial lipopeptides: Where are we going? Comput. Struct. Biotechnol. J. 2021, 19, 1400–1413. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Vater, J.; Piel, J.; Franke, P.; Scholz, R.; Schneider, K.; Koumoutsi, A.; Hitzeroth, G.; Grammel, N.; Strittmatter, A.W.; et al. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 2006, 188, 4024–4036. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Pila, F.E. Importance of Bacillus subtilis lipopeptides in the biological control of diseases in crops of high economic value. Bionatura 2016, 1, 135–138. [Google Scholar] [CrossRef]
- Sen, R. Surfactin: Biosynthesis, genetics and potential applications. Adv. Exp. Med. Biol. 2010, 672, 316–323. [Google Scholar]
- Mulligan, C.N. Environmental applications for biosurfactants. Environ. Pollut. 2005, 133, 183–198. [Google Scholar] [CrossRef]
- Kalamara, M.; Spacapan, M.; Mandic-Mulec, I.; Stanley-Wall, N.R. Social behaviours by Bacillus subtilis: Quorum sensing, kin discrimination and beyond. Mol. Microbiol. 2018, 110, 863–878. [Google Scholar] [CrossRef]
- Chen, B.; Wen, J.; Zhao, X.; Ding, J.; Qi, G. Surfactin: A Quorum-Sensing Signal Molecule to Relieve CCR in Bacillus amyloliquefaciens. Front Microbiol. 2020, 11, 631. [Google Scholar] [CrossRef]
- López, D.; Kolter, R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 2010, 34, 134–149. [Google Scholar] [CrossRef]
- Spacapan, M.; Danevčič, T.; Mandic-Mulec, I. ComX-induced exoproteases degrade ComX in Bacillus subtilis PS-216. Front. Microbiol. 2018, 9, 105. [Google Scholar]
- Mielich-Süss, B.; Lopez, D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ. Microbiol. 2015, 17, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Boguslawski, K.M.; Hill, P.A.; Griffith, K.L. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis. Mol. Microbiol. 2015, 96, 325–348. [Google Scholar] [CrossRef] [PubMed]
- Piggot, P.J.; Hilbert, D.W. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004, 7, 579–586. [Google Scholar] [CrossRef]
- Errington, J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003, 1, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Griffith, K.L.; Grossman, A.D. A Degenerate Tripartite DNA-Binding Site Required for Activation of ComA-Dependent Quorum Response Gene Expression in Bacillus subtilis. J. Mol. Biol. 2008, 381, 261–275. [Google Scholar] [CrossRef]
- Omer Bendori, S.; Pollak, S.; Hizi, D.; Eldar, A. The RapP-PhrP quorum-sensing system of Bacillus subtilis strain NCIB3610 affects biofilm formation through multiple targets, due to an atypical signal-insensitive allele of RapP. J. Bacteriol. 2015, 197, 592–602. [Google Scholar] [CrossRef] [PubMed]
- Lanigan-Gerdes, S.; Dooley, A.N.; Faull, K.F.; Lazazzera, B.A. Identification of subtilisin, Epr and Vpr as enzymes that produce CSF, an extracellular signalling peptide of Bacillus subtilis. Mol. Microbiol. 2007, 65, 1321–1333. [Google Scholar] [CrossRef]
- Devi, S.N.; Kiehler, B.; Haggett, L.; Fujita, M. Evidence that autophosphorylation of the major sporulation kinase in Bacillus subtilis is able to occur in trans. J. Bacteriol. 2015, 197, 2675–2684. [Google Scholar] [CrossRef] [PubMed]
- Banse, A.V.; Hobbs, E.C.; Losick, R. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein med in Bacillus subtilis. J. Bacteriol. 2011, 193, 3949–3955. [Google Scholar] [CrossRef]
- Shemesh, M.; Kolter, R.; Losick, R. The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J. Bacteriol. 2010, 192, 6352–6356. [Google Scholar] [CrossRef]
- Aguilar, C.; Vlamakis, H.; Guzman, A.; Losick, R.; Kolter, R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio 2010, 1, 10–1128. [Google Scholar] [CrossRef]
- Jiang, M.; Shao, W.; Perego, M.; Hoch, J.A. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000, 38, 535–542. [Google Scholar] [CrossRef]
- Thimon, L.; Peypoux, F.; Wallachb, J.; Michel, G. Ionophorous and sequestering properties of surfactin, a biosurfactant from Bacillus subtilis. Colloids Surf. B Biointerfaces 1993, 1, 57–62. [Google Scholar] [CrossRef]
- Vass, E.; Majer, Z.; Hollósi, M.; Besson, F.; Volpon, L. Ca2+-induced changes of surfactin conformation: AFTIR and circular dichroism study. Biochem. Biophys. Res. Commun. 2001, 282, 361–367. [Google Scholar] [CrossRef]
- Grau, A.; Gómez Fernández, J.C.; Peypoux, F.; Ortiz, A. A study on the interactions of surfactin with phospholipid vesicles. Biochim. Biophys. Acta 1999, 1418, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Cawoy, H.; Debois, D.; Franzil, L.; De Pauw, E.; Thonart, P.; Ongena, M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens. Micro. Biotechnol. 2015, 8, 281–295. [Google Scholar] [CrossRef] [PubMed]
- Le Mire, G.; Siah, A.; Brisset, M.N.; Gaucher, M.; Deleu, M.; Jijakli, M.H. Surfactin protects wheat against Zymoseptoria tritici and activates both salicylic acid- and jasmonic acid-dependent defense responses. Agriculture 2018, 8, 11. [Google Scholar] [CrossRef]
- Bonmatin, J.-M.; Laprévote, O.; Peypoux, F. Diversity Among Microbial Cyclic Lipopeptides: Iturins and Surfactins. Activity-Structure Relationships to Design New Bioactive Agents. Comb. Chem. High Throughput Screen. 2003, 6, 541–556. [Google Scholar] [CrossRef]
- Zhang, F.; Huo, K.; Song, X.; Quan, Y.; Wang, S.; Zhang, Z.; Gao, W.; Yang, C. Engineering of a genome-reduced strain Bacillus amyloliquefaciens for enhancing surfactin production. Microb. Cell Fact. 2020, 19, 223. [Google Scholar] [CrossRef]
- Gao, L.; She, M.; Shi, J.; Cai, D.; Wang, D.; Xiong, M.; Shen, G.; Gao, J.; Zhang, M.; Yang, Z.; et al. Enhanced production of iturin A by strengthening fatty acid synthesis modules in Bacillus amyloliquefaciens. Front. Bioeng. Biotechnol. 2022, 10, 974460. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Liu, G.; Zheng, R.; Sun, C.; Wu, S. Structural and Functional Insights into Iturin W, a Novel Lipopeptide Produced by the Deep-Sea Bacterium Bacillus sp. Strain wsm-1. Appl. Environ. Microbiol. 2020, 86, e01597-20. [Google Scholar] [CrossRef]
- Ranjan, A.; Rajput, V.D.; Prazdnova, E.V.; Gurnani, M.; Bhardwaj, P.; Sharma, S.; Sushkova, S.; Mandzhieva, S.S.; Minkina, T.; Sudan, J.; et al. Nature’s Antimicrobial Arsenal: Non-Ribosomal Peptides from PGPB for Plant Pathogen Biocontrol. Fermentation 2023, 9, 597. [Google Scholar] [CrossRef]
- Seydlová, G.; Svobodová, J. Review of surfactin chemical properties and the potential biomedical applications. Cent. Eur. J. Med. 2008, 3, 123–133. [Google Scholar] [CrossRef]
- Kowalczyk, R.; Harris, P.W.R.; Williams, G.M.; Yang, S.H.; Brimble, M.A. Peptide Lipidation—A Synthetic Strategy to Afford Peptide Based Therapeutics. Adv. Exp. Med. Biol. 2017, 1030, 185–227. [Google Scholar]
- Yue, H.; Zhong, J.; Li, Z.; Zhou, J.; Yang, J.; Wei, H.; Shu, D.; Luo, D.; Tan, H. Optimization of iturin A production from Bacillus subtilis ZK-H2 in submerge fermentation by response surface methodology. 3 Biotech 2021, 11, 36. [Google Scholar] [CrossRef]
- Dang, Y.; Zhao, F.; Liu, X.; Fan, X.; Huang, R.; Gao, W.; Wang, S.; Yang, C. Enhanced production of antifungal lipopeptide iturin A by Bacillus amyloliquefaciens LL3 through metabolic engineering and culture conditions optimization. Microb. Cell Fact. 2019, 18, 68. [Google Scholar] [CrossRef]
- Yu, C.; Qiao, J.; Ali, Q.; Jiang, Q.; Song, Y.; Zhu, L.; Gu, Q.; Borriss, R.; Dong, S.; Gao, X.; et al. degQ associated with the degS/degU two-component system regulates biofilm formation, antimicrobial metabolite production, and biocontrol activity in Bacillus velezensis DMW1. Mol. Plant Pathol. 2023, 24, 1510–1521. [Google Scholar] [CrossRef]
- Vahidinasab, M.; Adiek, I.; Hosseini, B.; Akintayo, S.O.; Abrishamchi, B.; Pfannstiel, J.; Henkel, M.; Lilge, L.; Voegele, R.T.; Hausmann, R. Characterization of Bacillus velezensis UTB96, Demonstrating Improved Lipopeptide Production Compared to the Strain B. velezensis FZB42. Microorganisms 2022, 10, 2225. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Cai, D.; Zhang, H.; Gao, L.; Yang, Y.; Gao, J.; Li, Y.; Yang, C.; Ji, Z.; Yu, J.; et al. Enhanced production of iturin A in Bacillus amyloliquefaciens by genetic engineering and medium optimization. Process Biochem. 2020, 90, 50–57. [Google Scholar] [CrossRef]
- Deleu, M.; Paquot, M.; Nylander, T. Effect of fengycin, a lipopeptide produced by Bacillus subtilis, on model biomembranes. Biophys. J. 2008, 94, 2667–2679. [Google Scholar] [CrossRef]
- Tan, W.; Yin, Y.; Wen, J. Increasing fengycin production by strengthening the fatty acid synthesis pathway and optimizing fermentation conditions. Biochem. Eng. J. 2022, 177, 108235. [Google Scholar] [CrossRef]
- Gimenez, D.; Phelan, A.; Murphy, C.D.; Cobb, S.L. Fengycin A Analogues with Enhanced Chemical Stability and Antifungal Properties. Org. Lett. 2021, 23, 4672–4676. [Google Scholar] [CrossRef]
- Deng, X.; Tian, Y.; Niu, Q.; Xu, X.; Shi, H.; Zhang, H.; Liang, L.; Zhang, K.; Huang, X. The ComP-ComA Quorum System Is Essential For “Trojan horse” Like Pathogenesis in Bacillus nematocida. PLoS ONE 2013, 8, e76920. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Q.; Ma, Y.; Li, S.; Lu, X.; Zhang, X.; Ma, P. DegQ regulates the production of fengycins and biofilm formation of the biocontrol agent Bacillus subtilis NCD-2. Microbiol. Res. 2015, 178, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, Y.; Diop, A.; Gancel, F.; Béchet, M.; Jacques, P.; Drider, D. Polynucleotide phosphorylase is involved in the control of lipopeptide fengycin production in Bacillus subtilis. Arch. Microbiol. 2018, 200, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, R.; Yang, P.; Luo, W.; Chen, S.; Bilal, M.; Xu, H.; Gu, C.; Liu, S.; Zhao, Y.; et al. iTRAQ-BASED Proteomic Analysis of the Mechanism of Fructose on Improving Fengycin Biosynthesis in Bacillus amyloliquefaciens. Molecules 2021, 26, 6309. [Google Scholar] [CrossRef]
- Alarcon, D.A.; Nandi, M.; Carpena, X.; Fita, I.; Loewen, P.C. Structure of glycerol-3-phosphate dehydrogenase (GPD1) from Saccharomyces cerevisiae at 2.45 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012, 68, 1279–1283. [Google Scholar] [CrossRef]
- Desmyttere, H.; Deweer, C.; Muchembled, J.; Sahmer, K.; Jacquin, J.; Coutte, F.; Jacques, P. Antifungal activities of bacillus subtilis lipopeptides to two venturia inaequalis strains possessing different tebuconazole sensitivity. Front. Microbiol. 2019, 10, 2327. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.R.; Hou, Z.J.; Ding, M.Z.; Bai, S.; Wei, S.Y.; Qiao, B.; Xu, Q.M.; Cheng, J.S.; Yuan, Y.J. Improved Production of Fengycin in Bacillus subtilis by Integrated Strain Engineering Strategy. ACS Synth. Biol. 2022, 11, 4065–4076. [Google Scholar] [CrossRef]
- Li, Y.; Wen, J. Metabolomic analysis of the effect glutamate on fengycin-overproducing Bacillus subtilis ATCC 21332 with an enhanced fatty acid synthesis pathway. Biochem. Eng. J. 2023, 196, 108957. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, T.; He, D.; Li, X.Z.; Wu, H.; Liu, W.; Gao, X. Functions of lipopeptides bacillomycin D and fengycin in antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. Microb. Physiol. 2011, 20, 43–52. [Google Scholar] [CrossRef]
- Li, Y.; Héloir, M.C.; Zhang, X.; Geissler, M.; Trouvelot, S.; Jacquens, L.; Henkel, M.; Su, X.; Fang, X.; Wang, Q.; et al. Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 2019, 20, 1037–1050. [Google Scholar] [CrossRef]
- Farzand, A.; Moosa, A.; Zubair, M.; Khan, A.R.; Massawe, V.C.; Tahir, H.A.S.; Sheikh, T.M.M.; Ayaz, M.; Gao, X. Suppression of Sclerotinia sclerotiorum by the Induction of Systemic Resistance and Regulation of Antioxidant Pathways in Tomato Using Fengycin Produced by Bacillus amyloliquefaciens FZB42. Biomolecules 2019, 9, 613. [Google Scholar] [CrossRef]
- Beltran-Gracia, E.; Macedo-Raygoza, G.; Villafaña-Rojas, J.; Martinez-Rodriguez, A.; Chavez-Castrillon, Y.Y.; Espinosa-Escalante, F.M.; Mascio, P.D.; Ogura, T.; Beltran-Garcia, M.J. Production of lipopeptides by fermentation processes: Endophytic bacteria, fermentation strategies and easy methods for bacterial selection. In Fermentation Processes; BoD: Norderstedt, Germany, 2017; pp. 199–222. [Google Scholar]
- Sidorova, T.M.; Asaturova, A.M.; Homyak, A.I.; Zhevnova, N.A.; Shternshis, M.V.; Tomashevich, N.S. Optimization of laboratory cultivation conditions for the synthesis of antifungal metabolites by Bacillus subtilis strains. Saudi J. Biol. Sci. 2020, 27, 1879–1885. [Google Scholar] [CrossRef] [PubMed]
- Cooper, D.G.; Macdonald, C.R.; Duff, S.J.B.; Kosaric, N. Enhanced production of surfactin from Bacillus subtilis by continuous product removal and metal cation additions. Appl. Environ. Microbiol. 1981, 42, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Landy, M.W.G.H.; Warren, G.H.; Rosenman, M.S.B.; Colio, L.G. Bacillomycin: An antibiotic from Bacillus subtilis active against pathogenic fungi. Proc. Soc. Exp. Biol. Med. 1948, 67, 539–541. [Google Scholar] [CrossRef]
- Marcelino, P.R.F.; Gonçalves, F.; Jimenez, I.M.; Carneiro, B.C.; Santos, B.B.; da Silva, S.S. Sustainable production of biosurfactants and their applications. Lignocellul. Biorefining Technol. 2020, 159–183. [Google Scholar]
- Adetunji, C.O.; Jeevanandam, J.; Anani, O.A.; Inobeme, A.; Thangadurai, D.; Islam, S.; Olaniyan, O.T. Strain improvement methodology and genetic engineering that could lead to an increase in the production of biosurfactants. In Green Sustainable Process for Chemical and Environmental Engineering and Science; Elsevier: Amsterdam, The Netherlands, 2021; pp. 299–315. [Google Scholar]
- Barale, S.S.; Ghane, S.G.; Sonawane, K.D. Purification and characterization of antibacteri al surfactin isoforms produced by Bacillus velezensis SK. Amb Express 2022, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Pandey, L.M. Production of biosurfactant by Bacillus subtilis RSL-2 isolated from sludge and biosurfactant mediated degradation of oil. Bioresoure Technol. 2020, 307, 123261. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.J.P.; Perfumo, A.; Marchant, R.; Banat, I.M. Isolation and Analysis of Lipopeptide and high molecular weight biosurfactant. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 3687–3704. [Google Scholar]
- Inès, M.; Dhouha, G. Lipopeptide surfactants: Production, recovery and pore forming capacity. Peptides 2015, 71, 100–112. [Google Scholar] [CrossRef]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathogens 2017, 13, e1006512. [Google Scholar] [CrossRef]
- Gerhardt, H.; Sievers-Engler, A.; Jahanshah, G.; Pataj, Z.; Ianni, F.; Gross, H.; Lindner, W.; Lämmerhofer, M. Methods for the comprehensive structural elucidation of constitution and stereochemistry of lipopeptides. J. Chromatogr. A 2016, 1428, 280–291. [Google Scholar] [CrossRef]
- Lee, S.Y.; Chew, K.W.; Show, P.L. Cell separation and disruption, product recovery, and purification. In Essentials in Fermentation Technology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 237–271. [Google Scholar]
- Venkataraman, S.; Rajendran, D.S.; Kumar, P.S.; Vo, D.V.N.; Vaidyanathan, V.K. Extraction, purification and applications of biosurfactants based on microbial-derived glycolipids and lipopeptides: A review. In Environmental Chemistry Letters; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–22. [Google Scholar]
- Coutte, F.; Lecouturier, D.; Dimitrov, K.; Guez, J.S.; Delvigne, F.; Dhulster, P.; Jacques, P. Microbial lipopeptide production and purification bioprocesses, current progress and future challenges. Biotechnol. J. 2017, 12, 1600566. [Google Scholar] [CrossRef]
- Jauregi, P.; Coutte, F.; Catiau, L.; Lecouturier, D.; Jacques, P. Micelle size characterization of lipopeptides produced by B. subtilis and their recovery by the two-step ultrafiltration process. Sep. Purif. Technol. 2013, 104, 175–182. [Google Scholar] [CrossRef]
- Poole, C.F. New trends in solid-phase extraction. TrAC-Trends Anal Chem. 2003, 22, 362–373. [Google Scholar] [CrossRef]
- Razafindralambo, H.; Paquot, M.; Hbid, C.; Jacques, P.; Destain, J.; Thonart, P. Purification of antifungal lipopeptides by reversed-phase high-performance liquid chromatography. J. Chromatogr. A 1993, 639, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Ciura, K.; Dziomba, S.; Nowakowska, J.; Markuszewski, M.J. Thin layer chromatography in drug discovery process. J. Chromatogr. A 2017, 1520, 9–22. [Google Scholar] [CrossRef]
- Jamshidi-Aidji, M.; Dimkić, I.; Ristivojević, P.; Stanković, S.; Morlock, G.E. Effect-directed screening of Bacillus lipopeptide extracts via hyphenated high-performance thin-layer chromatography. J. Chromatogr. A 2019, 1605, 460366. [Google Scholar] [CrossRef] [PubMed]
- de Souza, C.G.; Martins, F.I.C.C.; Zocolo, G.J.; Figueiredo, J.E.F.; Canuto, K.M.; de Brito, E.S. Simultaneous quantification of lipopeptide isoforms by UPLC-MS in the fermentation broth from Bacillus subtilis CNPMS22. Anal. Bioanal. Chem. 2018, 410, 682–736. [Google Scholar] [CrossRef] [PubMed]
- Hussein, W.; Fahim, S. Detection of synthetases genes involved in non ribosomal lipopeptides (NRLPS) biosynthesis from Bacillus species by bioinformatics and PCR degenerated primers and estimation of their production. Int. J. Pharm. Bio Sci. 2017, 8, 116–125. [Google Scholar]
- Wang, K.; Qin, Z.; Wu, S.; Zhao, P.; Zhen, C.; Gao, H. Antifungal Mechanism of Volatile Organic Compounds Produced by Bacillus subtilis CF-3 on Colletotrichum gloeosporioides Assessed Using Omics Technology. J. Agric. Food Chem. 2021, 69, 5267–5278. [Google Scholar] [CrossRef]
- Burch, A.Y.; Browne, P.J.; Dunlap, C.A.; Price, N.P.; Lindow, S.E. Comparison of biosurfactant detection methods reveals hydrophobic surfactants and contact-regulated production. Microbiology 2011, 13, 2681–2691. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; de Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef]
- Biniarz, P.; Łukaszewicz, M.; Janek, T. Screening concepts, characterization and structural analysis of microbial-derived bioactive lipopeptides: A review. Crit. Rev. Biotechnol. 2016, 37, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Satpute, S.K.; Bhawsar, B.D.; Dhakephalkar, P.K.; Chopade, B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian J. Mar. Sci. 2008, 37, 243–250. [Google Scholar]
- Hernández-Salmerón, J.E.; Prieto-Barajas, C.M.; Valencia-Cantero, E.; Moreno-Hagelsieb, G.; Santoyo, G. Isolation and characterization of genetic variability in bacteria with β-hemolytic and antifungal activity isolated from the rhizosphere of Medicago truncatula plants. Genet. Mol. Res. 2014, 13, 4967–4975. [Google Scholar] [CrossRef]
- Chen, C.Y.; Baker, S.C.; Darton, R.C. The application of a high throughput analysis method for the screening of potential biosurfactants from natural sources. J. Microbiol. Methods 2007, 70, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Burch, A.Y.; Shimada, B.K.; Browne, P.J.; Lindow, S.E. Novel high throughput detection method to assess bacterial surfactant production. Appl. Environ. Microbiol. 2010, 76, 5363–5372. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, I.V.; Blagova, D.K.; Veselova, S.V.; Sorokan, A.V.; Burkhanova, G.F.; Cherepanova, E.A.; Sarvarova, E.R.; Rumyantsev, S.D.; Alekseev, V.Y.; Khayrullin, R.M. Recombinant Bacillus subtilis 26DCryChS line with gene Btcry1Ia encoding Cry1Ia toxin from Bacillus thuringiensis promotes integrated wheat defense against pathogen Stagonospora nodorum Berk. and greenbug Schizaphis graminum Rond. Biol. Control 2020, 144, 104242. [Google Scholar] [CrossRef]
- Afsharmanesh, H.; Perez-Garcia, A.; Zeriouh, H.; Ahmadzadeh, M.; Romero, D. Afatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 2018, 94, 48–55. [Google Scholar] [CrossRef]
- Safari, N.; Mirabzadeh Ardakani, M.; Hemmati, R.; Parroni, A.; Beccaccioli, M.; Reverberi, M. The Potential of Plant-Based Bioactive Compounds on Inhibition of Aflatoxin B1 Biosynthesis and Down-regulation of aflR, aflM and aflP Genes. Antibiotics 2020, 9, 728. [Google Scholar] [CrossRef]
- Djavaheri, M.; Mercado-Blanco, J.; Versluis, C.; Meyer, J.-M.; Van Loon, L.C.; Bakker, P.A.H.M. Ironregulated metabolites produced by Pseudomonas fluorescens WCS374r are not required for eliciting induced systemic resistance (ISR) against Pseudomonas syringae pv. tomato in Arabidopsis. Microbiol. Open 2012, 1, 311–325. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures, and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Chauhan, P.S.; Agrawal, L.; Raj, R.; Srivastava, A.; Gupta, S.; Mishra, S.K.; Yadav, S.; Singh, P.C.; Raj, S.K.; et al. Paenibacillus lentimorbus inoculation enhances tobacco growth and extenuates the virulence of cucumber mosaic virus. PLoS ONE 2016, 11, e0149980. [Google Scholar] [CrossRef] [PubMed]
- Harun-Or-Rashid, M.; Kim, H.J.; Yeom, S.I.; Yu, H.A.; Manir, M.M.; Moon, S.S.; Chung, Y.R. Bacillus velezensis YC7010 enhances plant defenses against brown planthopper through transcriptomic and metabolic changes in rice. Front Plant Sci. 2018, 9, 1904. [Google Scholar] [CrossRef] [PubMed]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.F., Jr. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2015, 172, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, P.; Fan, H.; Liu, L.; Yin, K.; Yang, B.; Li, Y.; Huang, S.M.; Li, X.; Zheng, S.J. A real-time fluorescent reverse transcription quantitative PCR assay for rapid detection of genetic markers’ expression associated with Fusarium wilt of banana biocontrol activities in Bacillus. J. Fungi 2021, 7, 353. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhao, M.; Li, R.; Huang, Q.; Rensing, C.; Shen, Q. Lipopeptides produced by B. amyloliquefaciens NJN-6 altered the soil fungal community and non-ribosomal peptides genes harboring microbial community. Appl. Soil Ecol. 2017, 117, 96–105. [Google Scholar] [CrossRef]
- Moldes, A.B.; Álvarez-Chaver, P.; Vecino, X.; Cruz, J.M. Purification of lipopeptide biosurfactant extracts obtained from a complex residual food stream using Tricine-SDSPAGE electrophoresis. Front. Bioeng. Biotechnol. 2023, 11, 1199103. [Google Scholar] [CrossRef] [PubMed]
- López-Prieto, A.; Rodríguez-López, L.; Rincón-Fontán, M.; Cruz, J.M.; Moldes, A.B. Characterization of extracellular and cell bound biosurfactants produced by Aneurinibacillus aneurinilyticus isolated from commercial corn steep liquor. Microbiol. Res. 2021, 242, 126614. [Google Scholar] [CrossRef]
- Smyth, T.J.; Rudden, M.; Tsaousi, K.; Marchant, R.; Banat, I.M. Protocols for the Isolation and Analysis of Lipopeptides and Bioemulsifiers. In Hydrocarbon and Lipid Microbiology Protocols; McGenity, T., Timmis, K., Nogales, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–28. [Google Scholar]
- Ma, Z.; Hu, J.; Wang, X.; Wang, S. NMR spectroscopic and MS/MS spectrometric characterization of a new lipopeptide antibiotic bacillopeptin B1 produced by a marine sediment-derived Bacillus amyloliquefaciens SH-B74. J. Antibiot. 2014, 67, 175–178. [Google Scholar] [CrossRef][Green Version]
- Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S.S.; Wohlgemuth, G.; Barupal, D.K.; Showalter, M.R.; Arita, M.; et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom. Rev. 2018, 37, 513–532. [Google Scholar] [CrossRef]
- Elkahoui, S.; Djébali, N.; Karkouch, I.; Ibrahim, A.H.; Kalai, L.; Bachkovel, S.; Tabbene, O.; Limam, F. Mass spectrometry identification of antifungal lipopeptides from Bacillus sp. BCLRB2 against Rhizoctonia solani and Sclerotinia sclerotiorum. Prikl. Biokhim. Mikrobiol. 2014, 50, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Olajide, O.E.; Yi, Y.; Zheng, J.; Hamid, A.M. Species-level discrimination of microorganisms by high-resolution paper spray—Ion mobility—Mass spectrometry. Int. J. Mass Spectrom. 2022, 478, 116871. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, X.; Lei, S.; Shao, D.; Jiang, C.; Shi, J.; Zhang, Y.; Liu, L.; Lei, S.; Sun, H.; et al. Iturin A-like lipopeptides from Bacillus subtilis trigger apoptosis, paraptosis, and autophagy in Caco-2 cells. J. Cell. Physiol. 2019, 234, 6414–6427. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, J.B.; de Oliveira Cruz, J.; Geraldo, L.C.; Dias, E.G.; Queiroz, P.R.M.; Monnerat, R.G.; Borges, M.; Blassioli-Moraes, M.C.; Blum, L.E.B. Detection and evaluation of volatile and non-volatile antifungal compounds produced by Bacillus spp. strains. Microbiol. Res. 2023, 275, 127465. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-López, L.; Rincón-Fontán, M.; Vecino, X.; Cruz, J.M.; Moldes, A.B. Extraction, separation and characterization of lipopeptides and phospholipids from corn steep water. Sep. Purif. Technol. 2020, 248, 117076. [Google Scholar] [CrossRef]
- Rathankumar, A.K.; Saikia, K.; Palanisamy, S.; Ahalliya, R.M.; Arasu, M.V. Purification Assessment and Assay of Biosurfactant Efficacy. In Multifunctional Microbial Biosurfactants; Springer: Berlin/Heidelberg, Germany, 2023; pp. 25–50. [Google Scholar]
- Sani, A.; Qin, W.Q.; Li, J.Y.; Liu, Y.F.; Zhou, L.; Yang, S.Z.; Mu, B.Z. Structural diversity and applications of lipopeptide biosurfactants as biocontrol agents against phytopathogens: A review. In Microbiological Research; Elsevier: Amsterdam, The Netherlands, 2023; p. 127518. [Google Scholar]
- Favaro, G.; Bogialli, S.; Di Gangi, I.M.; Nigris, S.; Baldan, E.; Squartini, A.; Pastore, P.; Baldan, B. Characterization of lipopeptides produced by Bacillus licheniformis using liquid chromatography with accurate tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2016, 30, 2237–2252. [Google Scholar] [CrossRef]
- Ho, Y.P.; Reddy, P.M. Advances in mass spectrometry for the identification of pathogens. Mass Spectrum. Rev. 2011, 30, 1203–1224. [Google Scholar] [CrossRef]
- Krásný, L.; Hynek, R.; Hochel, I. Identification of bacteria using mass spectrometry techniques. Int. J. Mass Spectrom. 2013, 353, 67–79. [Google Scholar] [CrossRef]
- Geissler, M.; Oellig, C.; Moss, K.; Schwack, W.; Henkel, M.; Hausmann, R. High-performance thin-layer chromatography (HPTLC) for the simultaneous quantification of the cyclic lipopeptides Surfactin, Iturin A and Fengycin in culture samples of Bacillus species. J. Chrom. B 2017, 1044, 214–224. [Google Scholar] [CrossRef]



| Plant Disease | Phytopathogen | Lipopeptide Producing Specie | Lipopeptide Inhibiting the Phytopathogen | Application Method | Inhibition | Reference |
|---|---|---|---|---|---|---|
| Gray mold disease of apple | Botrytis cinerea | Bacillus subtilis S499 | Fengycin | Cell-free lipopeptide applied directly to the infected apple | 70% | [12] |
| Spot blotch on wheat plants | Bipolaris sorokiniana | Bacillus cabrialesii TE3T | Surfactin and fencgycin | Foliar application of crude lipopeptide extract | 93% | [24] |
| Leaf spot on sugar beet | Pseudomonas syringae | Bacillus pumilus (SS—10.7) and Bacillus amyloliquefaciens (SS—12.6 and SS—38.4) | Surfactin, fengycin and iturin | Foliar application of crude lipopeptide extracts | 92% | [36] |
| Watermelon wilt | Fusarium oxysporum | Bacillus amyloliquefaciens DHA55 | Surfactin, fengycin and iturin | Plants drenched in inoculum suspensions | 71.50% | [37] |
| Arabidopsis root infection | Pseudomonas syringae | Bacillus subtilis 6051 | Surfactin | Bacterial inoculation on plant | 70% | [38] |
| Tomato wilt | Ralstonia solanacearum | Bacillus velezensis FJAT-46737 | Surfactin, fengycin and iturin | Tomato seedling roots were dipped in the crude lipopeptide solution | 96.20% | [39] |
| Root and foliar diseases of soybeans | Xanthomonas axonopodis PV. Glycines | Bacillus amyloliquefaciens KPS46 | Surfactin | Cell-free supernatant treatment of soybean seeds | 30% | [40] |
| Sclerotinia stem rot disease | Sclerotinia sclerotiorum | Bacillus amyloliquefaciens | Surfactin and fengycin | Spray (bacterial cells grown in MOLP) on soybean plants | 100% | [41] |
| Sigatoka disease of banana | Pseudocercospora fijiensis | Bacillus tequilensis EA-CB0015 | Surfactin, fengycin and iturin | Banana plants were sprayed with liquid culture including biomass of bacteria and the lipopeptides | 100% | [42] |
| Damping-off bean | Pythium ultimum | Bacillus subtilis M4 | Iturin and fengycin | Bean seed soaked in cell suspension n of 5 × 108 CFUs | 98% | [43] |
| Method | Description | Reference |
|---|---|---|
| Drop-collapse assay | Each well of a microplate is coated with a layer of oil (i.e., mineral oil) before analysis. Then, a drop of the supernatant is added to the center of a well and observed after 1 min. The drop formed (as it is immiscible) will collapse revealing the presence of biosurfactants, including lipopeptides. | [122,123] |
| Hemolytic assay | The bacteria are cultured on blood agar in a Petri dish. After a number of certain days of growth, the formation of a halo around the colony (β-hemolysis) indicates the production of lipopeptides. | [123] |
| Meniscus formation assay | One volume of supernatant is placed in a 96-well microplate. When biosurfactants (including lipopeptides) are present in the supernatant, the surface of a wellbore liquid forms a concave lens that distorts the view of a grid. | [124] |
| Oil atomization | The bacteria are grown on Petri dish agar. After a number of certain days of growth, a mist of mineral oil is applied to an agar surface with an airbrush. A halo around the colony indicates the production of biosurfactants, including lipopeptides. | [119,125] |
| Technique | Principle | Advantages | Limitations |
|---|---|---|---|
| Liquid chromatography (HPLC) | Separation based on hydrophobicity. | Separates lipopeptides from complex mixtures. | Limited to the separation of intact lipopeptides without detailed structural information. |
| Gas chromatography (GC) | Separation based on volatility. | Quantifies fatty acid chains of lipopeptides. | Requires saponification and derivatization, limited to the analysis of lipid tails. |
| Tandem mass spectrometry (MS/MS) | Fragmentation of ions for structural elucidation. | Provides detailed structural information and identifies specific lipopeptides. | Requires prior chromatographic separation, and database searching for identification. |
| Thin-layer chromatography (TLC) | Separation based on hydrophobicity. | Provides easy visualization of separated compounds. | Limited resolution and sensitivity. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valenzuela Ruiz, V.; Gándara-Ledezma, A.; Villarreal-Delgado, M.F.; Villa-Rodríguez, E.D.; Parra-Cota, F.I.; Santoyo, G.; Gómez-Godínez, L.J.; Cira Chávez, L.A.; de los Santos-Villalobos, S. Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens. Stresses 2024, 4, 107-132. https://doi.org/10.3390/stresses4010007
Valenzuela Ruiz V, Gándara-Ledezma A, Villarreal-Delgado MF, Villa-Rodríguez ED, Parra-Cota FI, Santoyo G, Gómez-Godínez LJ, Cira Chávez LA, de los Santos-Villalobos S. Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens. Stresses. 2024; 4(1):107-132. https://doi.org/10.3390/stresses4010007
Chicago/Turabian StyleValenzuela Ruiz, Valeria, Azucena Gándara-Ledezma, María Fernanda Villarreal-Delgado, Eber Daniel Villa-Rodríguez, Fannie Isela Parra-Cota, Gustavo Santoyo, Lorena Jacqueline Gómez-Godínez, Luis A. Cira Chávez, and Sergio de los Santos-Villalobos. 2024. "Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens" Stresses 4, no. 1: 107-132. https://doi.org/10.3390/stresses4010007
APA StyleValenzuela Ruiz, V., Gándara-Ledezma, A., Villarreal-Delgado, M. F., Villa-Rodríguez, E. D., Parra-Cota, F. I., Santoyo, G., Gómez-Godínez, L. J., Cira Chávez, L. A., & de los Santos-Villalobos, S. (2024). Regulation, Biosynthesis, and Extraction of Bacillus-Derived Lipopeptides and Its Implications in Biological Control of Phytopathogens. Stresses, 4(1), 107-132. https://doi.org/10.3390/stresses4010007







