MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
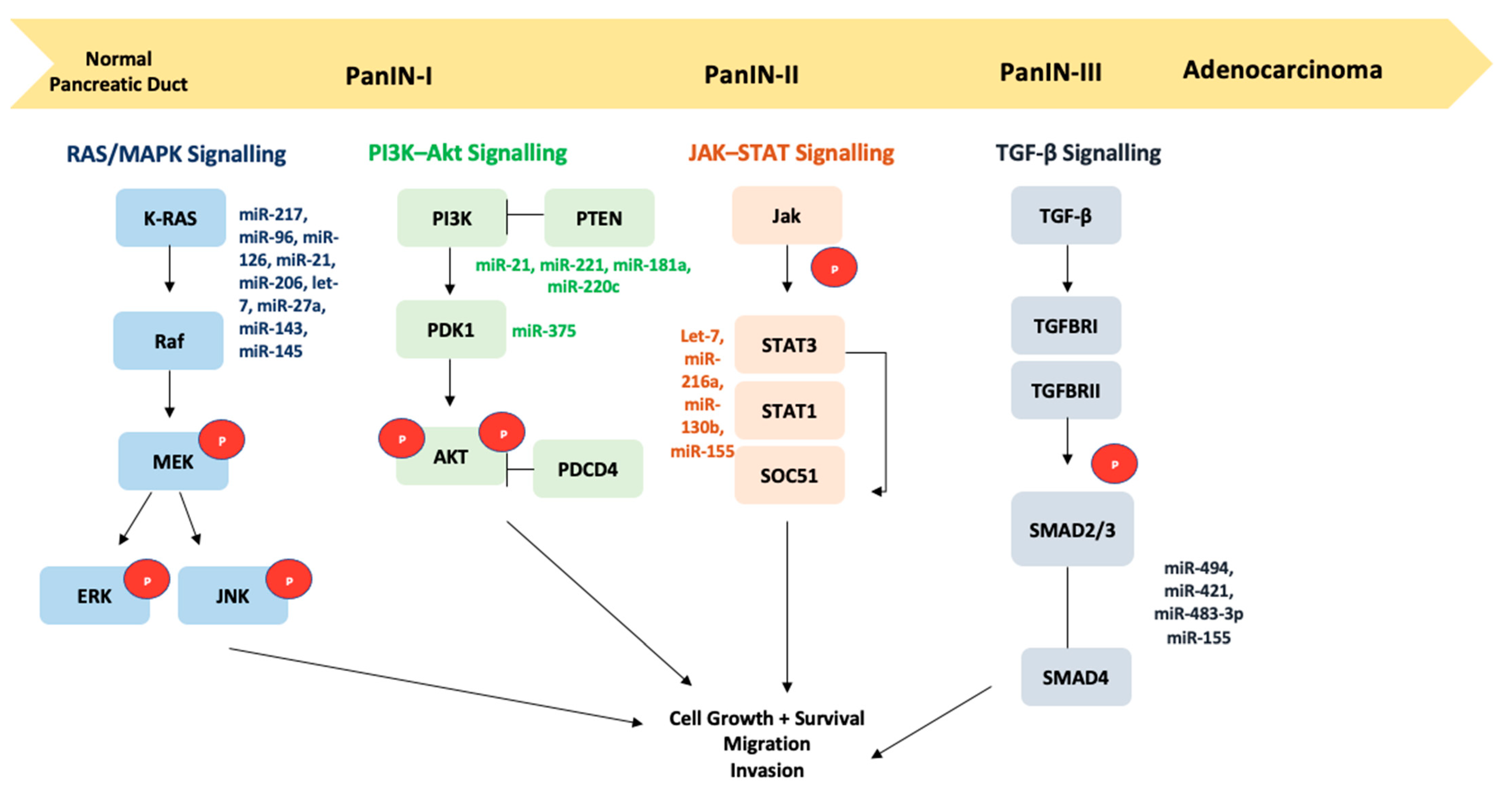
2. Signaling Pathways Associated with PDAC
2.1. TGF-β and HGF-MET Signaling Pathways
2.2. JAK–STAT Signaling Pathway
2.3. PI3K–AKT Signaling Pathway
2.4. TP53 Signaling Pathway and Apoptosis
2.5. KRAS Signaling Pathway
2.6. Epidermal Growth Factor Receptor and HER2/neu Signaling Pathways
2.7. Notch and Hedgehog Signaling Pathways
2.8. Wnt/β-Catenin Signaling Pathway
2.9. Cell Cycle Signaling Pathway and p16/CDKN2A Inactivation
2.10. Transcription Factors and DNA Methylation
3. Involvement of Metabolic Stress in PDAC
4. Angiogenesis and miRs in PDAC
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Von Hoff, D.D.; Korn, R.; Mousses, S. Pancreatic Cancer—Could It Be that Simple? A Different Context of Vulnerability. Cancer Cell 2009, 16, 7–8. [Google Scholar] [CrossRef][Green Version]
- Srivastava, S.K.; Bhardwaj, A.; Singh, S.; Arora, S.; Wang, B.; Grizzle, W.E.; Singh, A.P. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis 2011, 32, 1832–1839. [Google Scholar] [CrossRef]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer statistics, 2014. CA Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Baradaran, B.; Shahbazi, R.; Khordadmehr, M. Dysregulation of key microRNAs in pancreatic cancer development. Biomed. Pharmacother. 2019, 109, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.K.; Arora, S.; Singh, S.; Bhardwaj, A.; Averett, C.; Singh, A.P. MicroRNAs in pancreatic malignancy: Progress and promises. Cancer Lett. 2014, 347, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Blum, R.S.; Kloog, Y. Metabolism addiction in pancreatic cancer. Cell Death Dis. 2014, 5, e1065. [Google Scholar] [CrossRef]
- Abbruzzese, J.L. Adjuvant therapy for surgically resected pancreatic adenocarcinoma. JAMA 2008, 299, 1066–1067. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Kleeff, J.; Michl, P.; Costello, E.; Greenhalf, W.; Palmer, D.H. Therapeutic developments in pancreatic cancer: Current and future perspectives. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 333–348. [Google Scholar] [CrossRef]
- Olive, K.P.; Jacobetz, M.A.; Davidson, C.J.; Gopinathan, A.; McIntyre, D.; Honess, D.; Madhu, B.; Goldgraben, M.A.; Caldwell, M.E.; Allard, D.; et al. Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science 2009, 324, 1457–1461. [Google Scholar] [CrossRef]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; De La Fouchardière, C.; et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Xu, W.; Taranets, L.; Popov, N. Regulating Fbw7 on the road to cancer. Semin. Cancer Biol. 2016, 36, 62–70. [Google Scholar] [CrossRef]
- Maitra, A.; Hruban, R.H. Pancreatic Cancer. Annu. Rev. Pathol. Mech. Dis. 2008, 3, 157–188. [Google Scholar] [CrossRef] [PubMed]
- Hezel, A.F.; Kimmelman, A.C.; Stanger, B.Z.; Bardeesy, N.; Depinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006, 20, 1218–1249. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A.; Velculescu, V.E.; Wolfgang, C.L.; Hruban, R.H. Genetic Basis of Pancreas Cancer Development and Progression: Insights from Whole-Exome and Whole-Genome Sequencing. Clin. Cancer Res. 2012, 18, 4257–4265. [Google Scholar] [CrossRef] [PubMed]
- Bükki, J. Pancreatic Adenocarcinoma. N. Engl. J. Med. 2014, 371, 2139–2141. [Google Scholar] [CrossRef]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Sang, M.; Li, S.; Sun, X.; Yang, C.; Xi, Y.; Wang, L.; Zhang, F.; Bi, Y.; Fu, Y.; et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget 2015, 6, 39756–39792. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.E.; Hernandez, Y.G.; Frucht, H.; Lucas, A.L. Pancreatic ductal adenocarcinoma: Risk factors, screening, and early detection. World J. Gastroenterol. 2014, 20, 11182–11198. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Lowenfels, A.B. Risk factors for pancreatic cancer: A summary review of meta-analytical studies. Int. J. Epidemiol. 2015, 44, 186–198. [Google Scholar] [CrossRef]
- Barone, E.; Corrado, A.; Gemignani, F.; Landi, S. Environmental risk factors for pancreatic cancer: An update. Arch. Toxicol. 2016, 90, 2617–2642. [Google Scholar] [CrossRef] [PubMed]
- Buha, A.; Wallace, D.; Matovic, V.; Schweitzer, A.; Oluic, B.; Micic, D.; Djordjevic, V. Cadmium Exposure as a Putative Risk Factor for the Development of Pancreatic Cancer: Three Different Lines of Evidence. BioMed Res. Int. 2017, 2017, 1–8. [Google Scholar] [CrossRef]
- Djordjevic, V.R.; Wallace, D.R.; Schweitzer, A.; Boricic, N.; Knezevic, D.; Matic, S.; Grubor, N.; Kerkez, M.; Radenkovic, D.; Bulat, Z.; et al. Environmental cadmium exposure and pancreatic cancer: Evidence from case control, animal and in vitro studies. Environ. Int. 2019, 128, 353–361. [Google Scholar] [CrossRef]
- Wallace, D.R.; Djordjevic, A.B. Heavy metal and pesticide exposure: A mixture of potential toxicity and carcinogenicity. Curr. Opin. Toxicol. 2020, 19, 72–79. [Google Scholar] [CrossRef]
- Wallace, D.R.; Spandidos, D.A.; Tsatsakis, A.; Schweitzer, A.; Djordjevic, V.; Djordjevic, A.B. Potential interaction of cadmium chloride with pancreatic mitochondria: Implications for pancreatic cancer. Int. J. Mol. Med. 2019, 44, 145–156. [Google Scholar] [CrossRef]
- Wallace, D.R.; Taalab, Y.M.; Heinze, S.; Lovaković, B.T.; Pizent, A.; Renieri, E.; Tsatsakis, A.; Farooqi, A.A.; Javorac, D.; Andjelkovic, M.; et al. Toxic-Metal-Induced Alteration in miRNA Expression Profile as a Proposed Mechanism for Disease Development. Cells 2020, 9, 901. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.R.; Buha-Đorđević, A.; Benton, A. Toxicity of organic and inorganic nickel in pancreatic cell cultures: Comparison to cadmium. Arh. za Farm. 2020, 70, 344–359. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333. [Google Scholar] [CrossRef]
- Słotwiński, R.; Lech, G.; Słotwińska, S.M. MicroRNAs in pancreatic cancer diagnosis and therapy. Central Eur. J. Immunol. 2018, 43, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, P.S. Small RNAs with big impacts. Nat. Cell Biol. 2005, 435, 745–746. [Google Scholar] [CrossRef]
- Amirkhah, R.; Schmitz, U.; Linnebacher, M.; Wolkenhauer, O.; Farazmand, A. MicroRNA-mRNA interactions in colorectal cancer and their role in tumor progression. Genes Chromosom. Cancer 2015, 54, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Galasso, M.; Sandhu, S.K.; Volinia, S. MicroRNA Expression Signatures in Solid Malignancies. Cancer J. 2012, 18, 238–243. [Google Scholar] [CrossRef]
- Yu, I.S.; Cheung, W.Y. A Contemporary Review of the Treatment Landscape and the Role of Predictive and Prognostic Biomarkers in Pancreatic Adenocarcinoma. Can. J. Gastroenterol. Hepatol. 2018, 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gilles, M.-E.; Hao, L.; Huang, L.; Rupaimoole, R.; López-Casas, P.P.; Pulver, E.; Jeong, J.C.; Muthuswamy, S.K.; Hidalgo, M.; Bhatia, S.N.; et al. Personalized RNA Medicine for Pancreatic Cancer. Clin. Cancer Res. 2018, 24, 1734–1747. [Google Scholar] [CrossRef]
- Jay, C.; Nemunaitis, J.; Chen, P.; Fulgham, P.; Tong, A.W. miRNA Profiling for Diagnosis and Prognosis of Human Cancer. DNA Cell Biol. 2007, 26, 293–300. [Google Scholar] [CrossRef]
- Truty, M.J.; Urrutia, R. Basics of TGF-ß and Pancreatic Cancer. Pancreatology 2007, 7, 423–435. [Google Scholar] [CrossRef]
- Birchenall-Roberts, M.C.; Fu, T.; Bang, O.-S.; Dambach, M.; Resau, J.H.; Sadowski, C.L.; Bertolette, D.C.; Lee, H.-J.; Kim, S.-J.; Ruscetti, F.W. Tuberous Sclerosis Complex 2 Gene Product Interacts with Human SMAD Proteins. J. Biol. Chem. 2004, 279, 25605–25613. [Google Scholar] [CrossRef]
- Ellenrieder, V.; Hendler, S.F.; Boeck, W.; Seufferlein, T.; Menke, A.; Ruhland, C.; Adler, G.; Gress, T.M. Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 2001, 61, 4222–4228. [Google Scholar] [PubMed]
- McCleary-Wheeler, A.L.; McWilliams, R.; Fernandez-Zapico, M.E. Aberrant signaling pathways in pancreatic cancer: A two compartment view. Mol. Carcinog. 2011, 51, 25–39. [Google Scholar] [CrossRef]
- Blackford, A.; Serrano, O.K.; Wolfgang, C.L.; Parmigiani, G.; Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Leary, R.J.; Eshleman, J.R.; et al. SMAD4 Gene Mutations Are Associated with Poor Prognosis in Pancreatic Cancer. Clin. Cancer Res. 2009, 15, 4674–4679. [Google Scholar] [CrossRef] [PubMed]
- Iacobuzio-Donahue, C.A.; Fu, B.; Yachida, S.; Luo, M.; Abe, H.; Henderson, C.M.; Vilardell, F.; Wang, Z.; Keller, J.W.; Banerjee, P.; et al. DPC4 Gene Status of the Primary Carcinoma Correlates with Patterns of Failure in Patients with Pancreatic Cancer. J. Clin. Oncol. 2009, 27, 1806–1813. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Gudey, S.K.; Landström, M. Non-Smad signaling pathways. Cell Tissue Res. 2011, 347, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, Z.; Kong, X.; Xie, D.; Jia, Z.; Jiang, W.; Cui, J.; Du, Y.; Wei, D.; Huang, S.; et al. Down-regulation of MicroRNA-494 via Loss of SMAD4 Increases FOXM1 and β-Catenin Signaling in Pancreatic Ductal Adenocarcinoma Cells. Gastroenterology 2014, 147, 485–497.e18. [Google Scholar] [CrossRef]
- Garajová, I.; Giovannetti, E.; Caponi, S.; Van Zweeden, A.; Peters, G.J. MiRNAs and Their Interference with the Main Molecular Mechanisms Responsible for Drug Resistance in Pancreatic Cancer. Curr. Pharmacol. Rep. 2015, 1, 223–233. [Google Scholar] [CrossRef][Green Version]
- Hao, J.; Zhang, S.; Zhou, Y.; Liu, C.; Hu, X.; Shao, C. MicroRNA 421 suppresses DPC4/Smad4 in pancreatic cancer. Biochem. Biophys. Res. Commun. 2011, 406, 552–557. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Zhang, S.; Zhou, Y.; Hu, X.; Shao, C. MicroRNA 483-3p suppresses the expression of DPC4/Smad4 in pancreatic cancer. FEBS Lett. 2010, 585, 207–213. [Google Scholar] [CrossRef]
- Delitto, D. c-Met signaling in the development of tumorigenesis and chemoresistance: Potential applications in pancreatic cancer. World J. Gastroenterol. 2014, 20, 8458–8470. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.-H.; Huang, C.; Qiu, Z.-J.; Liu, J.; Zhang, Z.-H.; Zhao, N.; Feng, Z.-Z.; Lv, X.-H. Expression and Prognostic Significance of CD151, c-Met, and Integrin alpha3/alpha6 in Pancreatic Ductal Adenocarcinoma. Dig. Dis. Sci. 2010, 56, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.N.; Summy, J.M.; Zhang, J.; Park, S.I.; Parikh, N.U.; Gallick, G.E. Development and Characterization of Gemcitabine-Resistant Pancreatic Tumor Cells. Ann. Surg. Oncol. 2007, 14, 3629–3637. [Google Scholar] [CrossRef]
- Bao, B.; Wang, Z.; Ali, S.; Ahmad, A.; Azmi, A.S.; Sarkar, S.H.; Banerjee, S.; Kong, D.; Li, Y.; Thakur, S.; et al. Metformin Inhibits Cell Proliferation, Migration and Invasion by Attenuating CSC Function Mediated by Deregulating miRNAs in Pancreatic Cancer Cells. Cancer Prev. Res. 2012, 5, 355–364. [Google Scholar] [CrossRef]
- Wu, K.; Hu, G.; He, X.; Zhou, P.; Li, J.; He, B.; Sun, W. MicroRNA-424-5p Suppresses the Expression of SOCS6 in Pancreatic Cancer. Pathol. Oncol. Res. 2013, 19, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.; Kollory, A.; Takashima, A.; Sarkar, S.; Faller, D.V.; Ghosh, S.K. MicroRNA let-7 downregulates STAT3 phosphorylation in pancreatic cancer cells by increasing SOCS3 expression. Cancer Lett. 2014, 347, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 2016, 62, 33–40. [Google Scholar] [CrossRef]
- Hou, B.-H.; Jian, Z.-X.; Cui, P.; Li, S.-J.; Tian, R.-Q.; Ou, J.-R. miR-216a may inhibit pancreatic tumor growth by targeting JAK. FEBS Lett. 2015, 589, 2224–2232. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Zhang, J.-G.; Shi, Y.; Qin, Q.; Liu, Y.; Wang, B.; Tian, K.; Deng, S.-C.; Li, X.; Zhu, S.; et al. MiR-130b Is a Prognostic Marker and Inhibits Cell Proliferation and Invasion in Pancreatic Cancer through Targeting STAT3. PLoS ONE 2013, 8, e73803. [Google Scholar] [CrossRef]
- Huang, C.; Li, H.; Wu, W.; Jiang, T.; Qiu, Z. Regulation of miR-155 affects pancreatic cancer cell invasiveness and migration by modulating the STAT3 signaling pathway through SOCS1. Oncol. Rep. 2013, 30, 1223–1230. [Google Scholar] [CrossRef]
- Ferro, R. Emerging role of the KRAS-PDK1 axis in pancreatic cancer. World J. Gastroenterol. 2014, 20, 10752–10757. [Google Scholar] [CrossRef]
- Park, J.-K.; Lee, E.J.; Esau, C.; Schmittgen, T.D. Antisense Inhibition of microRNA-21 or -221 Arrests Cell Cycle, Induces Apoptosis, and Sensitizes the Effects of Gemcitabine in Pancreatic Adenocarcinoma. Pancreas 2009, 38, e190–e199. [Google Scholar] [CrossRef]
- Sarkar, S.; Dubaybo, H.; Ali, S.; Goncalves, P.; Kollepara, S.L.; Sethi, S.; Philip, P.A.; Li, Y. Down-regulation of miR-221 inhibits prolif-eration of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am. J. Cancer Res. 2013, 3, 465–477. [Google Scholar]
- Liu, J.; Xu, D.; Wang, Q.; Zheng, D.; Jiang, X.; Xu, L. LPS Induced miR-181a Promotes Pancreatic Cancer Cell Migration via Targeting PTEN and MAP2K4. Dig. Dis. Sci. 2014, 59, 1452–1460. [Google Scholar] [CrossRef]
- Arisan, E.; Rencuzogullari, O.; Cieza-Borrella, C.; Arenas, F.M.; Dwek, M.; Lange, S.; Uysal-Onganer, P. MiR-21 is Required for the Epithelial–Mesenchymal Transition in MDA-MB-231 Breast Cancer Cells. Int. J. Mol. Sci. 2021, 22, 1557. [Google Scholar] [CrossRef] [PubMed]
- Uysal-Onganer, P.; D’Alessio, S.; Mortoglou, M.; Kraev, I.; Lange, S. Peptidylarginine Deiminase Inhibitor Application, Using Cl-Amidine, PAD2, PAD3 and PAD4 Isozyme-Specific Inhibitors in Pancreatic Cancer Cells, Reveals Roles for PAD2 and PAD3 in Cancer Invasion and Modulation of Extracellular Vesicle Signatures. Int. J. Mol. Sci. 2021, 22, 1396. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Song, S.; He, S.; Zhu, X.; Zhang, Y.; Yi, B.; Zhang, B.; Qin, G.; Li, D. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int. J. Mol. Med. 2014, 33, 950–956. [Google Scholar] [CrossRef]
- Yu, J.; Ohuchida, K.; Mizumoto, K.; Sato, N.; Kayashima, T.; Fujita, H.; Nakata, K.; Tanaka, M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer 2010, 9, 169. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, P.; Mohr, A.M.; Grandgenett, P.M.; Steele, M.M.; Batra, S.K.; Hollingsworth, M.A. MicroRNA-200c Modulates the Expression of MUC4 and MUC16 by Directly Targeting Their Coding Sequences in Human Pancreatic Cancer. PLoS ONE 2013, 8, e73356. [Google Scholar] [CrossRef]
- Rachagani, S.; Macha, M.A.; Ponnusamy, M.P.; Haridas, D.; Kaur, S.; Jain, M.; Batra, S.K. MUC4 Potentiates Invasion and Metastasis of Pancreatic Cancer Cells through Stabilization of Fibroblast Growth Factor Receptor 1. Carcinogenesis 2012, 33, 1953–1964. [Google Scholar] [CrossRef]
- Kaur, S.; Sharma, N.; Krishn, S.R.; Lakshmanan, I.; Rachagani, S.; Baine, M.J.; Smith, L.M.; Lele, S.M.; Sasson, A.R.; Guha, S.; et al. MUC4-Mediated Regulation of Acute Phase Protein Lipocalin 2 through HER2/AKT/NF-κB Signaling in Pancreatic Cancer. Clin. Cancer Res. 2014, 20, 688–700. [Google Scholar] [CrossRef]
- Bardeesy, N.; Depinho, R.A. Pancreatic Cancer Biology and Genetics. Nat. Rev. Cancer 2002, 2, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Rozenblum, E.; Schutte, M.; Goggins, M.; Hahn, S.A.; Panzer, S.; Zahurak, M.; Goodman, S.N.; Sohn, T.A.; Hruban, R.H.; Yeo, C.J.; et al. Tumour-Suppressive Pathways in Pancreatic Carcinoma. Cancer Res. 1997, 57, 1731–1734. [Google Scholar] [PubMed]
- Nalls, D.; Tang, S.-N.; Rodova, M.; Srivastava, R.K.; Shankar, S. Targeting Epigenetic Regulation of miR-34a for Treatment of Pancreatic Cancer by Inhibition of Pancreatic Cancer Stem Cells. PLoS ONE 2011, 6, e24099. [Google Scholar] [CrossRef]
- Greither, T.; Grochola, L.F.; Udelnow, A.; Lautenschläger, C.; Würl, P.; Taubert, H. Elevated Expression of microRNAs 155, 203, 210 and 222 in Pancreatic Tumors is Associated with Poorer Survival. Int. J. Cancer 2009, 126, 73–80. [Google Scholar] [CrossRef]
- Gironella, M.; Seux, M.; Xie, M.-J.; Cano, C.; Tomasini, R.; Gommeaux, J.; Garcia, S.; Nowak, J.; Yeung, M.L.; Jeang, K.-T.; et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc. Natl. Acad. Sci. USA 2007, 104, 16170–16175. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S. Apoptosis Pathways and Their Therapeutic Exploitation in Pancreatic Cancer. J. Cell. Mol. Med. 2009, 13, 1221–1227. [Google Scholar] [CrossRef]
- Arlt, A.; Müerköster, S.S.; Schäfer, H. Targeting Apoptosis Pathways in Pancreatic Cancer. Cancer Lett. 2013, 332, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, H.; Wang, X.; Zhou, L.; Likun, Z.; Deng, T.; Qu, Y.; Duan, J.; Bai, M.; Ge, S.; et al. The miR-24-Bim Pathway Promotes Tumor Growth and Angiogenesis in Pancreatic Carcinoma. Oncotarget 2015, 6, 43831–43842. [Google Scholar] [CrossRef] [PubMed]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of Somatic Mutations in Most Early-Stage Pancreatic Intraepithelial Neoplasia. Gastroenterology 2012, 142, 730–733.e9. [Google Scholar] [CrossRef]
- Timar, J.; Kashofer, K. Molecular epidemiology and diagnostics of KRAS mutations in human cancer. Cancer Metastasis Rev. 2020, 39, 1029–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.-G.; Yu, S.-N.; Lu, Z.-H.; Ma, Y.-H.; Gu, Y.-M.; Chen, J. The miR-217 microRNA functions as a potential tumor suppressor in pancreatic ductal adenocarcinoma by targeting KRAS. Carcinogenesis 2010, 31, 1726–1733. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lu, Z.; Liu, C.; Meng, Y.; Ma, Y.; Zhao, W.; Liu, J.; Yu, J.; Chen, J. miRNA-96 Suppresses KRAS and Functions as a Tumor Suppressor Gene in Pancreatic Cancer. Cancer Res. 2010, 70, 6015–6025. [Google Scholar] [CrossRef]
- Jiao, L.R.; Frampton, A.E.; Jacob, J.; Pellegrino, L.; Krell, J.; Giamas, G.; Tsim, N.; Vlavianos, P.; Cohen, P.; Ahmad, R.; et al. MicroRNAs Targeting Oncogenes Are Down-Regulated in Pancreatic Malignant Transformation from Benign Tumors. PLoS ONE 2012, 7, e32068. [Google Scholar] [CrossRef]
- Talotta, F.; Cimmino, A.; Matarazzo, M.R.; Casalino, L.; De Vita, G.; D’Esposito, M.; Di Lauro, R.; Verde, P. An autoregulatory loop mediated by miR-21 and PDCD4 controls the AP-1 activity in RAS transformation. Oncogene 2008, 28, 73–84. [Google Scholar] [CrossRef]
- Vorvis, C.; Koutsioumpa, M.; Iliopoulos, D. Developments in miRNA gene signaling pathways in pancreatic cancer. Future Oncol. 2016, 12, 1135–1150. [Google Scholar] [CrossRef]
- Keklikoglou, I.; Hosaka, K.; Bender, C.M.; Bott, A.; Koerner, C.; Mitra, D.; Will, R.G.; Woerner, A.; Muenstermann, E.; Wilhelm, H.E.; et al. MicroRNA-206 functions as a pleiotropic modulator of cell proliferation, invasion and lymphangiogenesis in pancreatic adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene 2015, 34, 4867–4878. [Google Scholar] [CrossRef]
- Ma, Y.; Yu, S.; Zhao, W.; Lu, Z.; Chen, J. miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett. 2010, 298, 150–158. [Google Scholar] [CrossRef]
- Kent, O.A.; Chivukula, R.R.; Mullendore, M.; Wentzel, E.A.; Feldmann, G.; Lee, K.H.; Liu, S.; Leach, S.D.; Maitra, A.; Mendell, J.T. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010, 24, 2754–2759. [Google Scholar] [CrossRef] [PubMed]
- Efurukawa, T. Impacts of Activation of the Mitogen-Activated Protein Kinase Pathway in Pancreatic Cancer. Front. Oncol. 2015, 5, 23. [Google Scholar] [CrossRef]
- Eser, S.; Schnieke, A.; Schneider, G.; Saur, D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer 2014, 111, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Trejo, C.L.; Silva, J.M.; Gu, S.; Korkola, J.E.; Heiser, L.M.; Charles, R.-P.; Rabinovich, B.A.; Hann, B.; Dankort, D.; et al. A Central Role for RAF→MEK→ERK Signaling in the Genesis of Pancreatic Ductal Adenocarcinoma. Cancer Discov. 2012, 2, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Maitra, A.; Ghosh, B.; Zechner, U.; Argani, P.; Iacobuzio-Donahue, C.A.; Sriuranpong, V.; Iso, T.; Meszoely, I.M.; Wolfe, M.S.; et al. Notch mediates TGFα-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell 2003, 3, 565–576. [Google Scholar] [CrossRef]
- Friess, H.; Yamanaka, Y.; Kobrin, M.S.; Do, D.A.; Buchler, M.W.; Korc, M. Enhanced erbB-3 expression in human pancreatic cancer correlates with tumour progression. Clin. Cancer Res. 1995, 1, 1413–1420. [Google Scholar] [PubMed]
- Ueda, S.; Ogata, S.; Tsuda, H.; Kawarabayashi, N.; Kimura, M.; Sugiura, Y.; Tamai, S.; Matsubara, O.; Hatsuse, K.; Mochizuki, H. The Correlation Between Cytoplasmic Overexpression of Epidermal Growth Factor Receptor and Tumor Aggressiveness. Pancreas 2004, 29, e1–e8. [Google Scholar] [CrossRef]
- Ali, S.; Ahmad, A.; Aboukameel, A.; Ahmed, A.; Bao, B.; Banerjee, S.; Philip, P.A.; Sarkar, F.H. Deregulation of miR-146a Expression in a Mouse Model of Pancreatic Cancer Affecting EGFR Signaling. Cancer Lett. 2014, 351, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Du Rieu, M.C.; Torrisani, J.; Selves, J.; Al Saati, T.; Souque, A.; Dufresne, M.; Tsongalis, G.J.; Suriawinata, A.A.; Carrère, N.; Buscail, L.; et al. MicroRNA-21 Is Induced Early in Pancreatic Ductal Adenocarcinoma Precursor Lesions. Clin. Chem. 2010, 56, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Izumchenko, E.; Chang, X.; Michailidi, C.; Kagohara, L.; Ravi, R.; Paz, K.; Brait, M.; Hoque, M.O.; Ling, S.; Bedi, A.; et al. The TGFβ–miR200–MIG6 Pathway Orchestrates the EMT-Associated Kinase Switch That Induces Resistance to EGFR Inhibitors. Cancer Res. 2014, 74, 3995–4005. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, H.; Gu, J.; Zheng, L. Prognostic role of HER2 amplification based on fluorescence in situ hybridization (FISH) in pancreatic ductal adenocarcinoma (PDAC): A meta-analysis. World J. Surg. Oncol. 2016, 14, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aumayr, K.; Soleiman, A.; Sahora, K.; Schindl, M.; Werba, G.; Schoppmann, S.F.; Birner, P. HER2 Gene Amplification and Protein Expression in Pancreatic Ductal Adenocarcinomas. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Radtke, F.; Raj, K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor? Nat. Rev. Cancer 2003, 3, 756–767. [Google Scholar] [CrossRef]
- De Lao, J.-P.; Murtaugh, L.C. Notch and Kras in pancreatic cancer: At the crossroads of mutation, differentiation and signaling. Cell Cycle 2009, 8, 1860–1864. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, Y.; Li, Y.; Banerjee, S.; Liao, J.; Sarkar, F.H. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol. Cancer Ther. 2006, 5, 483–493. [Google Scholar] [CrossRef]
- Ji, Q.; Hao, X.; Zhang, M.; Tang, W.; Yang, M.; Li, L.; Xiang, D.; DeSano, J.T.; Bommer, G.T.; Fan, D.; et al. MicroRNA miR-34 Inhibits Human Pancreatic Cancer Tumor-Initiating Cells. PLoS ONE 2009, 4, e6816. [Google Scholar] [CrossRef]
- Brabletz, S.; Bajdak, K.; Meidhof, S.; Burk, U.; Niedermann, G.; Firat, E.; Wellner, U.; Dimmler, A.; Faller, G.; Schubert, J.; et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011, 30, 770–782. [Google Scholar] [CrossRef]
- Sureban, S.M.; May, R.; Qu, D.; Weygant, N.; Chandrakesan, P.; Ali, N.; Lightfoot, S.A.; Pantazis, P.; Rao, C.V.; Postier, R.G.; et al. DCLK1 Regulates Pluripotency and Angiogenic Factors via microRNA-Dependent Mechanisms in Pancreatic Cancer. PLoS ONE 2013, 8, e73940. [Google Scholar] [CrossRef]
- Altaba, A.R.I.; Sánchez, P.; Dahmane, N. Gli and hedgehog in cancer: Tumours, embryos and stem cells. Nat. Rev. Cancer 2002, 2, 361–372. [Google Scholar] [CrossRef]
- Thayer, S.P.; di Magliano, M.P.; Heiser, P.W.; Nielsen, C.M.; Roberts, D.J.; Lauwers, G.Y.; Qi, Y.P. Hedgehog is an early and late mediator of pancreatic cancer tumourigenesis. Nature 2003, 425, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Dosch, J.S.; Di Magliano, M.P.; Simeone, D.M. Pancreatic Cancer and Hedgehog Pathway Signaling: New Insights. Pancreatology 2010, 10, 151–157. [Google Scholar] [CrossRef]
- Ma, C.; Nong, K.; Wu, B.; Dong, B.; Bai, Y.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. miR-212 promotes pancreatic cancer cell growth and invasion by targeting the hedgehog signaling pathway receptor patched-1. J. Exp. Clin. Cancer Res. 2014, 33, 54. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yu, C.; Chen, M.; Zhang, H.; Tian, S.; Sun, C. Reduction of miR-29c enhances pancreatic cancer cell migration and stem cell-like phenotype. Oncotarget 2014, 6, 2767–2778. [Google Scholar] [CrossRef] [PubMed]
- Nagano, H.; Tomimaru, Y.; Eguchi, H.; Hama, N.; Wada, H.; Kawamoto, K.; Kobayashi, S.; Mori, M.; Doki, Y. MicroRNA-29a induces resistance to gemcitabine through the Wnt/β-catenin signaling pathway in pancreatic cancer cells. Int. J. Oncol. 2013, 43, 1066–1072. [Google Scholar] [CrossRef]
- Zeng, G.; Germinaro, M.; Micsenyi, A.; Monga, N.K.; Bell, A.; Sood, A.; Malhotra, V.; Sood, N.; Midda, V.; Monga, D.K.; et al. Aberrant Wnt/β-Catenin Signaling in Pancreatic Adenocarcinoma. Neoplasia 2006, 8, 279–289. [Google Scholar] [CrossRef]
- Lowy, A.M.; Fenoglio-Preiser, C.; Kim, O.J.; Kordich, J.; Gómez, A.; Knight, J.; James, L.; Groden, J. Dysregulation of β-Catenin Expression Correlates with Tumor Differentiation in Pancreatic Duct Adenocarcinoma. Ann. Surg. Oncol. 2003, 10, 284–290. [Google Scholar] [CrossRef]
- Santo, L.; Siu, K.T.; Raje, N. Targeting Cyclin-Dependent Kinases and Cell Cycle Progression in Human Cancers. Semin. Oncol. 2015, 42, 788–800. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, D.; Killary, A.M.; Sen, S.; Amos, C.I.; Evans, D.B.; Abbruzzese, J.L.; Frazier, M.L. Polymorphisms of p16, p27, p73, and MDM2 Modulate Response and Survival of Pancreatic Cancer Patients Treated with Preoperative Chemoradiation. Ann. Surg. Oncol. 2008, 16, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Lotterman, C.; Karikari, C.; Omura, N.; Feldmann, G.; Habbe, N.; Goggins, M.G.; Mendell, J.T.; Maitra, A. Epigenetic Silencing of MicroRNA miR-107 Regulates Cyclin-Dependent Kinase 6 Expression in Pancreatic Cancer. Pancreatology 2009, 9, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Zhang, J.; Zhang, S.; Yu, D.; Chen, Y.; Liu, Q.; Shi, M.; Ni, C.; Zhu, M. Diagnostic and biological significance of microRNA-192 in pancreatic ductal adenocarcinoma. Oncol. Rep. 2013, 30, 276–284. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, L.-Y.; Dai, H.-Y.; Wang, P.; Gao, S.; Wang, K. miR-301a promotes pancreatic cancer cell proliferation by directly inhibiting bim expression. J. Cell. Biochem. 2012, 113, 3229–3235. [Google Scholar] [CrossRef]
- Willis, S.N.; Chen, L.; Dewson, G.; Wei, A.; Naik, E.; Fletcher, J.I.; Adams, J.M.; Huang, D.C. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005, 19, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Sun, Y.; Yang, H.; Li, J.; Yu, S.; Chang, X.; Lu, Z.; Chen, J. Deregulation of the MiR-193b-KRAS Axis Contributes to Impaired Cell Growth in Pancreatic Cancer. PLoS ONE 2015, 10, e0125515. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Fang, B.; Zeng, F.; Ma, C.; Pang, H.; Cheng, L.; Shi, Y.; Wang, H.; Yin, B.; Xia, J.; et al. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget 2015, 6, 1740–1749. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Y.; Yang, Y.; Liu, J.; Song, Y.; Cao, Y.; Chen, X.; Yang, W.; Wang, F.; Gao, J.; et al. MicroRNA-222 Controls Human Pancreatic Cancer Cell Line Capan-2 Proliferation by P57 Targeting. J. Cancer 2015, 6, 1230–1235. [Google Scholar] [CrossRef][Green Version]
- Boutros, R.; Lobjois, V.; Ducommun, B. CDC25 phosphatases in cancer cells: Key players? Good targets? Nat. Rev. Cancer 2007, 7, 495–507. [Google Scholar] [CrossRef]
- Liffers, S.-T.; Munding, J.B.; Vogt, M.; Kuhlmann, J.D.; Verdoodt, B.; Nambiar, S.; Maghnouj, A.; Mirmohammadsadegh, A.; Hahn, S.A.; Tannapfel, A. MicroRNA-148a is down-regulated in human pancreatic ductal adenocarcinomas and regulates cell survival by targeting CDC25B. Lab. Investig. 2011, 91, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Schutte, M.; Hruban, R.H.; Geradts, J.; Maynard, R.; Hilgers, W.; Rabindran, S.K.; Moskaluk, C.A.; Hahn, S.A.; Schwarte-Waldhoff, I.; Schmiegel, W.; et al. Abrogation of the Rb/p16 tumour-suppressive pathway in virtually all pan-creatic carcinomas. Cancer Res. 1997, 57, 3126–3130. [Google Scholar] [PubMed]
- Sharpless, N.E.; DePinho, R.A. Cancer: Crime and punishment. Nature 2005, 436, 636–637. [Google Scholar] [CrossRef] [PubMed]
- Wilentz, R.E.; Geradts, J.; Maynard, R.; Offerhaus, G.J.A.; Kang, M.; Goggins, M.; Yeo, C.J.; Kern, S.E.; Hruban, R.H. Inactivation of the p16 (INK4A) tumour-suppressor gene in pancreatic duct lesions: Loss of intranuclear expression. Cancer Res. 1998, 58, 4740–4744. [Google Scholar]
- Partensky, C. Toward a Better Understanding of Pancreatic Ductal Adenocarcinoma. Pancreas 2013, 42, 729–739. [Google Scholar] [CrossRef]
- Okamoto, A.; Demetrick, D.J.; Spillare, E.A.; Hagiwara, K.; Hussain, S.P.; Bennett, W.P.; Forrester, K.; Gerwin, B.; Serrano, M.; Beach, D.H. Mutations and altered expression of p16INK4 in human cancer. Proc. Natl. Acad. Sci. USA 1994, 91, 11045–11049. [Google Scholar] [CrossRef]
- Medina, R.F.; Zaidi, S.K.; Liu, C.-G.; Stein, J.L.; Van Wijnen, A.J.; Croce, C.M.; Stein, G.S. MicroRNAs 221 and 222 Bypass Quiescence and Compromise Cell Survival. Cancer Res. 2008, 68, 2773–2780. [Google Scholar] [CrossRef]
- Park, J.-K.; Henry, J.C.; Jiang, J.; Esau, C.; Gusev, Y.; Lerner, M.R.; Postier, R.G.; Brackett, D.J.; Schmittgen, T.D. miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem. Biophys. Res. Commun. 2011, 406, 518–523. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, H.; Yan, H. Gene expression profiling of 1200 pancreatic ductal adenocarcinoma reveals novel subtypes. BMC Cancer 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Nebert, D.W. Transcription factors and cancer: An overview. Toxicology 2002, 181–182, 131–141. [Google Scholar] [CrossRef]
- Lomberk, G.A.; Urrutia, R. The Triple-Code Model for Pancreatic Cancer: Cross Talk Among Genetics, Epigenetics, and Nuclear Structure. Surg. Clin. N. Am. 2015, 95, 935–952. [Google Scholar] [CrossRef] [PubMed]
- Farhan, M.; Wang, H.; Gaur, U.; Little, P.J.; Xu, J.; Zheng, W. FOXO Signaling Pathways as Therapeutic Targets in Cancer. Int. J. Biol. Sci. 2017, 13, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Chen, Q.; Fu, J.; Shankar, S.; Srivastava, R.K. Resveratrol Inhibits Growth of Orthotopic Pancreatic Tumors through Activation of FOXO Transcription Factors. PLoS ONE 2011, 6, e25166. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Liwei, W.; Wang, L.; Wang, L. Modulation of FoxO1 Expression by miR-21 to Promote Growth of Pancreatic Ductal Adenocarcinoma. Cell. Physiol. Biochem. 2015, 35, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhu, C.-F.; Ma, M.-Z.; Chen, G.; Song, M.; Zeng, Z.-L.; Lu, W.-H.; Yang, J.; Wen, S.; Chiao, P.J.; et al. Micro-RNA-155 is induced by K-Ras oncogenic signal and promotes ROS stress in pancreatic cancer. Oncotarget 2015, 6, 21148–21158. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Wang, Z.; Ali, S.; Kong, D.; Banerjee, S.; Ahmad, A.; Li, Y.; Azmi, A.S.; Miele, L.; Sarkar, F.H. Over-expression of FoxM1 leads to epithelial-mesenchymal transition and cancer stem cell phenotype in pancreatic cancer cells. J. Cell. Biochem. 2011, 112, 2296–2306. [Google Scholar] [CrossRef]
- Fujioka, S.; Sclabas, G.M.; Schmidt, C.; Frederick, W.A.; Dong, Q.G.; Abbruzzese, J.L.; Evans, D.B.; Baker, C.; Chiao, P.J. Function of Nuclear Factor kappaB in Pancreatic Cancer Metastasis. Clin. Cancer Res. 2003, 9, 346–354. [Google Scholar] [PubMed]
- Li, Y.; Vandenboom, T.G.; Wang, Z.; Kong, D.; Ali, S.; Philip, P.A.; Sarkar, F.H. miR-146a Suppresses Invasion of Pancreatic Cancer Cells. Cancer Res. 2010, 70, 1486–1495. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Li, Y.; Takwi, A.; Li, B.; Zhang, J.; Conklin, D.J.; Young, K.H.; Martin, R.; Li, Y. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2010, 30, 57–67. [Google Scholar] [CrossRef]
- Denis, H.; Ndlovu, M.N.; Fuks, F. Regulation of Mammalian DNA Methyltransferases: A Route to New Mechanisms. EMBO Rep. 2011, 12, 647–656. [Google Scholar] [CrossRef]
- Dhe-Paganon, S.; Syeda, F.; Park, L. DNA methyl Transferase 1: Regulatory mechanisms and implications in health and disease. Int. J. Biochem. Mol. Boil. 2011, 2, 58–66. [Google Scholar]
- Azizi, M.; Teimoori-Toolabi, L.; Arzanani, M.K.; Azadmanesh, K.; Fard-Esfahani, P.; Zeinali, S. MicroRNA-148b and microRNA-152 reactivate tumor suppressor genes through suppression of DNA methyltransferase-1 gene in pancreatic cancer cell lines. Cancer Biol. Ther. 2014, 15, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Moroishi, T.; Hansen, C.G.; Guan, K.-L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 2015, 15, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Yuan, H.; Su, L. The emerging and diverse roles of sirtuins in cancer: A clinical perspective. OncoTargets Ther. 2013, 6, 1399–1416. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Zhu, S.; Wang, B.; Li, X.; Liu, Y.; Qin, Q.; Gong, Q.; Niu, Y.; Xiang, C.; Chen, J.; et al. Chronic pancreatitis and pancreatic cancer demonstrate active epithelial–mesenchymal transition profile, regulated by miR-217-SIRT1 pathway. Cancer Lett. 2014, 355, 184–191. [Google Scholar] [CrossRef]
- Ye, X.; Wei, X.; Liao, J.; Chen, P.; Li, X.; Chen, Y.; Yang, Y.; Zhao, Q.; Sun, H.; Pan, L.; et al. 4-Hydroxyphenylpyruvate Dioxygenase-Like Protein Promotes Pancreatic Cancer Cell Progression and Is Associated with Glutamine-Mediated Redox Balance. Front. Oncol. 2021, 10, 617190. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Cho, Y.-R.; Lee, S.-Y.; Sung, G.-J.; Shin, D.-M.; Choi, K.-C.; Son, J. TFEB Supports Pancreatic Cancer Growth through the Transcriptional Regulation of Glutaminase. Cancers 2021, 13, 483. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Widmann, S.; Ho, C.; Nguyen, D.; Nguyen, A.; Premaratne, A.; Gustafsson, J.-Å.; Lin, C.-Y. Novel Liver X Receptor Ligand GAC0001E5 Disrupts Glutamine Metabolism and Induces Oxidative Stress in Pancreatic Cancer Cells. Int. J. Mol. Sci. 2020, 21, 9622. [Google Scholar] [CrossRef] [PubMed]
- Ashrafizadeh, M.; Bakhoda, M.R.; Bahmanpour, Z.; Ilkhani, K.; Zarrabi, A.; Makvandi, P.; Khan, H.; Mazaheri, S.; Darvish, M.; Mirzaei, H. Apigenin as Tumor Suppressor in Cancers: Biotherapeutic Activity, Nanodelivery, and Mechanisms with Emphasis on Pancreatic Cancer. Front. Chem. 2020, 8, 829. [Google Scholar] [CrossRef]
- Cinque, G.; Ferino, A.; Pedersen, E.B.; Xodo, L.E. Role of Poly [ADP-ribose] Polymerase 1 in Activating the Kirsten ras (KRAS) Gene in Response to Oxidative Stress. Int. J. Mol. Sci. 2020, 21, 6237. [Google Scholar] [CrossRef]
- Kim, H.-J.; Kim, D.; Yoon, H.; Choi, C.S.; Oh, Y.S.; Jun, H.-S. Prevention of Oxidative Stress-Induced Pancreatic Beta Cell Damage by Broussonetia kazinoki Siebold Fruit Extract via the ERK-Nox4 Pathway. Antioxidants 2020, 9, 406. [Google Scholar] [CrossRef] [PubMed]
- Gibson, A.R.; O’Leary, B.R.; Du, J.; Sarsour, E.H.; Kalen, A.L.; Wagner, B.A.; Stolwijk, J.M.; Falls-Hubert, K.C.; Alexander, M.S.; Carroll, R.S.; et al. Dual Oxidase-Induced Sustained Generation of Hydrogen Peroxide Contributes to Pharmacologic Ascorbate-Induced Cytotoxicity. Cancer Res. 2020, 80, 1401–1413. [Google Scholar] [CrossRef]
- Lee, S.-W.; Commisso, C. Metabolic regulation of EGFR effector and feedback signaling in pancreatic cancer cells requires K-Ras. Biochem. Biophys. Res. Commun. 2020, 533, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Recouvreux, M.V.; Moldenhauer, M.R.; Galenkamp, K.M.; Jung, M.; James, B.; Zhang, Y.; Lowy, A.; Bagchi, A.; Commisso, C. Glutamine depletion regulates Slug to promote EMT and metastasis in pancreatic cancer. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Y.; Han, F.; Zhao, Y.; Tu, M.; Wang, Y.; Huang, C.; Fan, S.; Chen, P.; Yao, X.; et al. A novel miR-1291-ERRα-CPT1C axis modulates tumor cell proliferation, metabolism and tumorigenesis. Theranostics 2020, 10, 7193–7210. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, T.; Zhou, Y.; Wang, S.; Zhou, X.; Wang, L.; Ou, T.; Chen, Y.; Zhou, Y.; Zhang, H.; et al. Carnitine palmitoyltransferase 1C contributes to progressive cellular senescence. Aging 2020, 12, 6733–6755. [Google Scholar] [CrossRef]
- Cannataro, R.; Perri, M.; Gallelli, L.; Caroleo, M.C.; De Sarro, G.; Cione, E. Ketogenic Diet Acts on Body Remodeling and MicroRNAs Expression Profile. MicroRNA 2019, 8, 116–126. [Google Scholar] [CrossRef]
- Chang, T.-C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef]
- Guo, R.; Gu, J.; Zhang, Z.; Wang, Y.; Gu, C. MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life 2015, 67, 42–53. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ali, S.; Ahmad, A.; Azmi, A.S.; Li, Y.; Banerjee, S.; Kong, D.; Sethi, S.; Aboukameel, A.; Padhye, S.B.; et al. Hypoxia-Induced Aggressiveness of Pancreatic Cancer Cells Is Due to Increased Expression of VEGF, IL-6 and miR-21, Which Can Be Attenuated by CDF Treatment. PLoS ONE 2012, 7, e50165. [Google Scholar] [CrossRef]
- Khan, S.; Ansarullah; Kumar, D.; Jaggi, M.; Chauhan, S.C. Targeting microRNAs in pancreatic cancer: Microplayers in the big game. Cancer Res. 2013, 73, 6541–6547. [Google Scholar] [CrossRef] [PubMed]
- Mace, T.A.; Collins, A.L.; Wojcik, S.E.; Croce, C.M.; Lesinski, G.B.; Bloomston, M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J. Surg. Res. 2013, 184, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Yang, Y.; Xia, L.; Yang, Y.; Wang, F.; Song, M.; Chen, X.; Liu, J.; Song, Y.; Zhao, Y.; et al. MiR-221 Promotes Capan-2 Pancreatic Ductal Adenocarcinoma Cells Proliferation by Targeting PTEN-Akt. Cell. Physiol. Biochem. 2016, 38, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
| miRs | Regulation in PDAC | Signaling Pathways Involved | Target Genes | Functional Involvement in PDAC | References |
|---|---|---|---|---|---|
| let-7 | Down | JAK–STAT K-RAS | STAT3, SOCS3, N-cadherin, ZEB1 | Tumor growth, migration | [53] |
| let-7a | Up | NOTCH | JAK, STAT | EMT | [103] |
| let-7d | Up | K-RAS | KRAS | Cell proliferation, migration, invasion, apoptosis | [81] |
| miR-21 | Up | PI3K–AKT K-RAS EGFR Cell cycle Apoptosis | G12D, p27, p57, FOXO1, Bcl-2, FasL, PI3K, AKT, PTEN, RECK, SPRY2 | Cell cycle arrest, apoptosis, gemcitabine resistance, aggressiveness | [59,82,94] |
| miR-23a | Up | Apoptosis | APAF1 | Apoptosis | [76] |
| miR-24 | Up | Apoptosis | BIM | Apoptosis, cell cycle | [76] |
| miR-26a-5p | Down | HGF–MET | ARMTL2, Cyclin E2, MMP12 | Cell proliferation, migration, survival, EMT | [51] |
| miR-27a | Up | K-RAS | Sprouty2 | Cell proliferation, migration, invasion, apoptosis | [85] |
| miR-29a | Up | Wnt | MUC1 | Cell proliferation, differentiation, invasion, migration, gemcitabine resistance | [109] |
| miR-29c | Down | Wnt | ZD4, FZD5, FRAT2, LRP-6 | Cell proliferation, differentiation, invasion, migration | [108] |
| miR-34a | Down | Tp53 NOTCH | NOTCH1/2CDK6, SIRT1 | Cell proliferation, migration, invasion, apoptosis | [71,83] |
| miR-96b | Down | K-RAS | KRAS, AKT | Cell proliferation, migration, invasion, apoptosis | [80] |
| miR-107 | Up | Cell cycle | Cyclin D1-dependent kinase | Cell proliferation | [114] |
| miR-126 | Down | K-RAS | KRAS | Cell proliferation, migration, invasion, apoptosis | [81] |
| miR-130b | Down | JAK–STAT | STAT3 | Cell proliferation, invasion | [54,56] |
| miR-132 | Up | P16 | Rb1 | G2/M cell cycle arrest, cell proliferation | [129] |
| miR-141 | Down | DNA methylation | YAP1, MAP4K4 | Tissue homeostasis, organ size, regeneration | [144] |
| miR-143 | Down | K-RAS | KRAS, MMP-2, MMP1 | Tumor growth | [86] |
| miR-144 | Down | NOTCH | PRR11 | EMT | [103] |
| miR-145 | Down | K-RAS NOTCH | KRAS | Tumor growth, EMT | [86,103] |
| miR-146a | Down | EGFR | EGFR, NF-κB, IRAK-1 | Increased tumor growth | [93] |
| miR-148a | Down | Cell cycle | CDC25B | Cell proliferation, differentiation, invasion, migration | [121] |
| miR-148b | Down | DNA methylation | DNMT1, AMPKa1 | Cell proliferation, differentiation, invasion, migration | [143] |
| miR-150 | Up | HER-2/neu | MUC4, IGF-1R | Invasion, migration | [45] |
| miR-152 | Down | DNA methylation | DNMT1 | Cell proliferation, differentiation, invasion, migration | [143] |
| miR-155 | Up | JAK–STAT Tp53 | SOCS1 TP53-induced nuclear protein 1 gene, FOXO3a, RHOA, SMAD1/5, ZNF652 | Cell invasion, migration, metastasis, generation of reactive oxygen species | [54,57,73] |
| miR-181a | Up | PI3K–AKT | PTEN | Migration | [61] |
| miR-192 | Up | Cell Cycle | Cyclin D1, cyclin D2, CDK4 | Cell proliferation | [115] |
| miR-193b | Down | Cell Cycle | KRAS | G1-phase arrest | [118] |
| miR-200 | Up | NOTCH | Jagged1, Maml2,Maml3 | EMT | [102] |
| miR-200c | Down | EGFR | MIG6, EP300 | Invasion, migration | [95] |
| miR-203 | Up | Tp53 | p53 | Cell proliferation, migration, invasion, apoptosis | [72] |
| miR-206 | Down | K-RAS | KRAS, ANXA2 | Decrease in proangiogenic and proinflammatory mediators, tumor growth, progression | [83,84] |
| miR-212 | Up | P16 Hedgehog | Rb1 | G2/M cell cycle arrest, cell proliferation | [107,129] |
| miR-216a | Down | JAK–STAT | JAK2 | Cell proliferation, invasion | [54,55] |
| miR-217 | Down | K-RAS, DNA methylation | KRAS, SIRT1 | Cell proliferation, migration, invasion, apoptosis, metabolism, DNA damage, stress responses, genome stability, cell survival | [79,145] |
| miR-220c | Up | PI3K–AKT | MUC4 | EMT | [54] |
| miR-221 | Up | PI3K–AKT | Cdk4, p16, E2F, CDKN1B, MMP-2, MMP-9, PUMA, PTEN, MDM2, ICAM-1, P27, BIM, SOD2, STAT5A | Proliferation, migration | [60] |
| miR-222 | Up | Tp53 P16 Cell cycle | p53, p27, p57, MMP2, MMP9, PUMA, PTEN, BIM, MMP1, SOD2, STAT5 | Cell proliferation, migration, invasion, apoptosis | [72,120,128] |
| miR-223 | Up | Cell Cycle | FBXW7, Cyclin E2 | Cell proliferation, differentiation, invasion, migration | [11,119] |
| miR-301a | Up | Cell Cycle | Bim, Bak, Bax, NF-κB | Cell proliferation | [116] |
| miR-375 | Down | PI3K–AKT | PDK1 | Cell proliferation, invasion, migration | [54,64] |
| miR-421 | Up | TGF-β | SMAD4 | Cell proliferation, migration, invasion | [46] |
| miR-424-5p | Up | HGF–MET | SOCS6 | Cell proliferation, migration, survival, EMT | [52] |
| miR-483-3p | Up | TGF-β | SMAD4 | Cell proliferation, migration, invasion | [47] |
| miR-494 | Down | TGF-β | FOXM1 | Cell proliferation, migration, invasion, increased resistance to gemcitabine | [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mortoglou, M.; Wallace, D.; Buha Djordjevic, A.; Djordjevic, V.; Arisan, E.D.; Uysal-Onganer, P. MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma. Stresses 2021, 1, 30-47. https://doi.org/10.3390/stresses1010004
Mortoglou M, Wallace D, Buha Djordjevic A, Djordjevic V, Arisan ED, Uysal-Onganer P. MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma. Stresses. 2021; 1(1):30-47. https://doi.org/10.3390/stresses1010004
Chicago/Turabian StyleMortoglou, Maria, David Wallace, Aleksandra Buha Djordjevic, Vladimir Djordjevic, E. Damla Arisan, and Pinar Uysal-Onganer. 2021. "MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma" Stresses 1, no. 1: 30-47. https://doi.org/10.3390/stresses1010004
APA StyleMortoglou, M., Wallace, D., Buha Djordjevic, A., Djordjevic, V., Arisan, E. D., & Uysal-Onganer, P. (2021). MicroRNA-Regulated Signaling Pathways: Potential Biomarkers for Pancreatic Ductal Adenocarcinoma. Stresses, 1(1), 30-47. https://doi.org/10.3390/stresses1010004








