Personalized Prediction of Total Knee Arthroplasty Mechanics Based on Sparse Input Data—Model Validation Using In Vivo Force Data
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vivo Validation Data
| Patient | Height [cm] | Weight [kg] | Age [Years] | Gender | Side |
|---|---|---|---|---|---|
| GCC2 | 172 | 67 | 83 | Male | Right |
| GCC3 | 167 | 78.4 | 68 | Female | Left |
| GCC5 | 180 | 75 | 86 | Male | Left |
| GCC6 | 172 | 70 | ~86 | Male | Right |
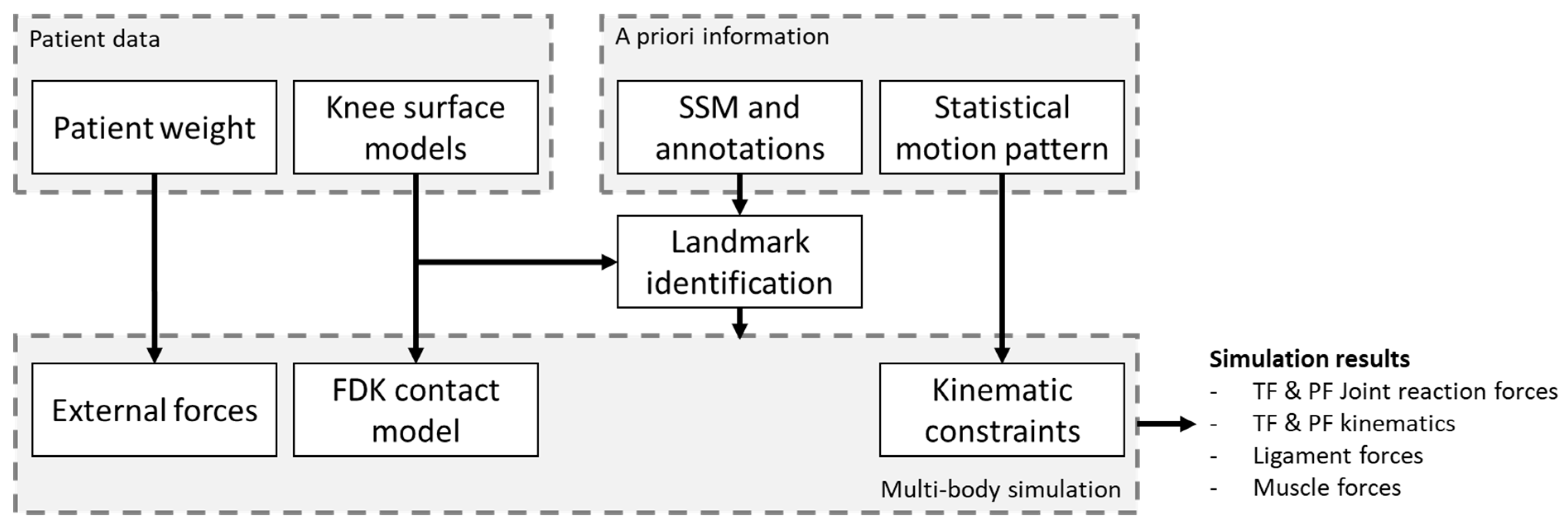
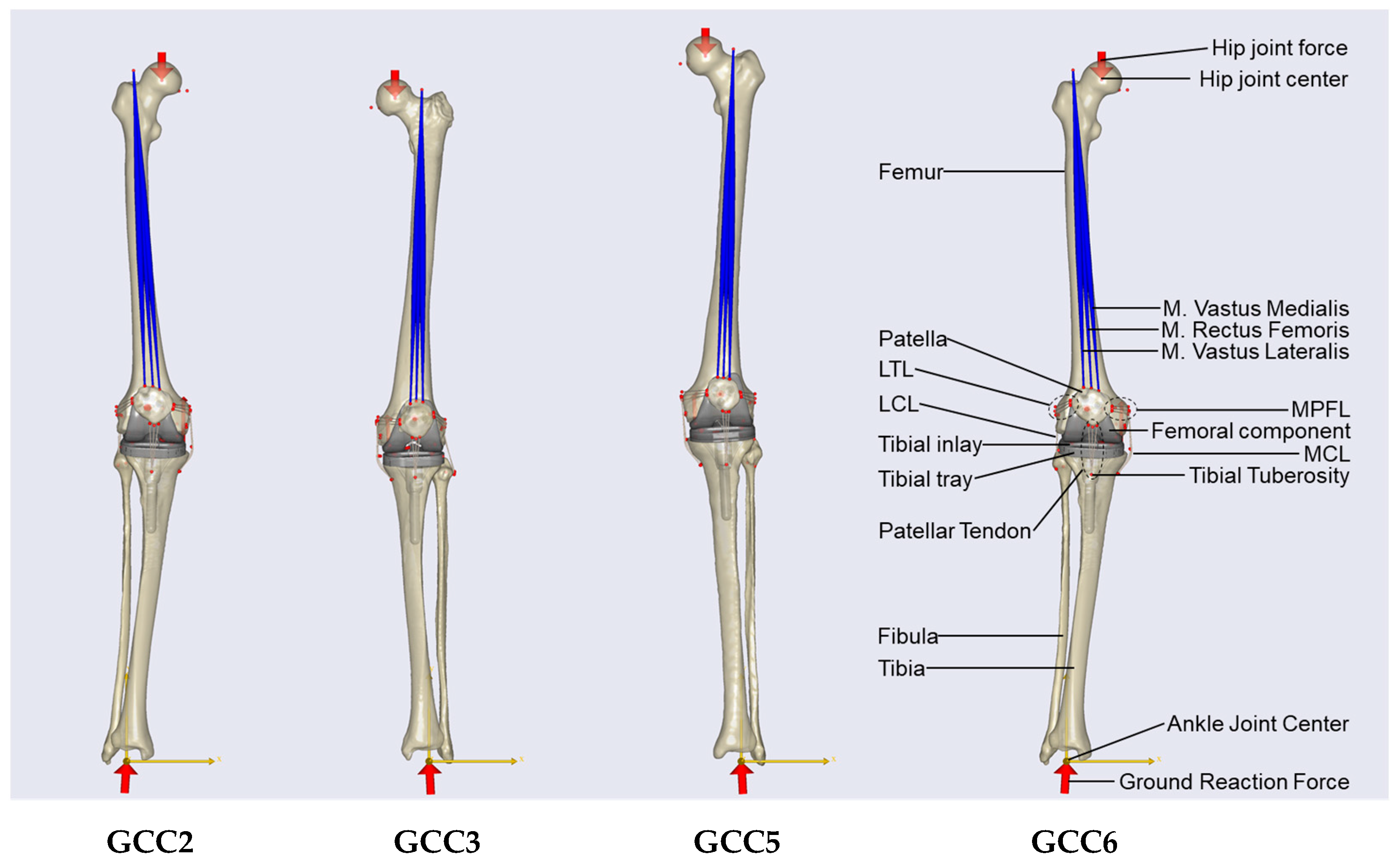
2.2. Multi-Body Simulation Model
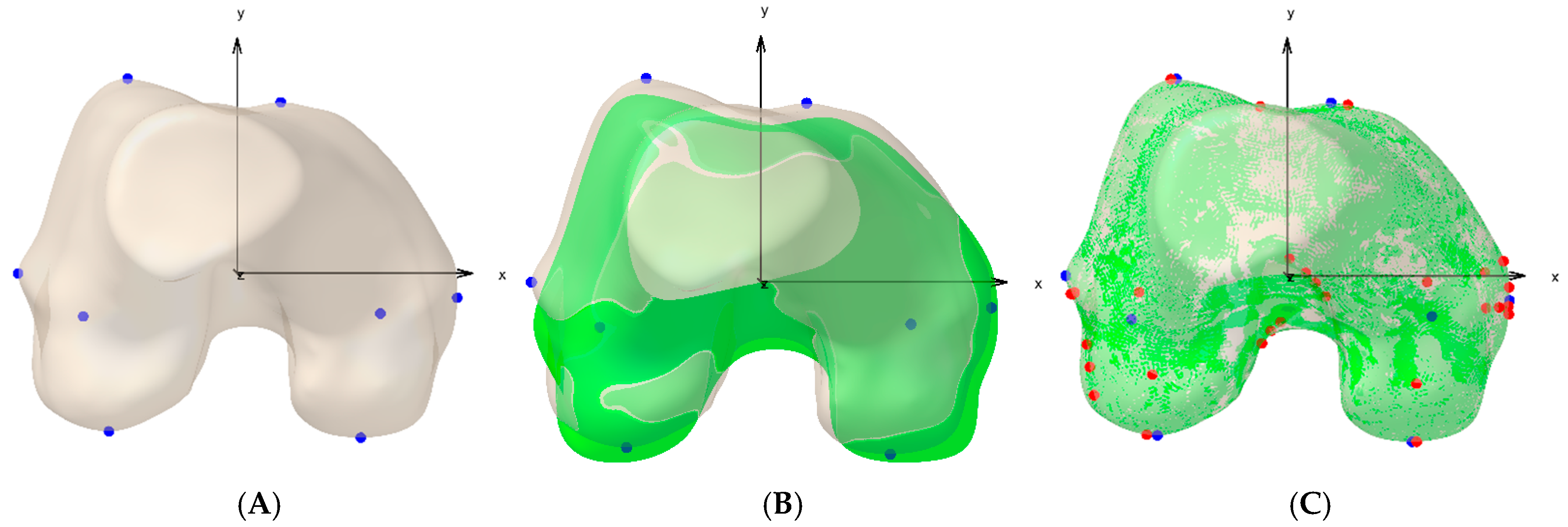
2.3. Biomechanical Modeling Using a Priori Knowledge
2.4. Statistical Analysis and Evaluation
3. Results
4. Discussion
| Study | Year | Activity | No. of Patients | RMSE Total [%BW], Mean (Range) | RMSE Medial [%BW], Mean (Range) | RMSE Lateral [%BW], Mean (Range) | Simulation Time [min] |
|---|---|---|---|---|---|---|---|
| Stylianou et al. [5] | 2013 | Squat | 1 | 42.3 (only superior-inferior) | / | / | n.a. |
| Thelen et al. [23] | 2014 | Gait | 1 | 51.0 | 26 | 42 | 100 |
| Chen et al. [4] | 2014 | Gait | 1 | 44.7 | 28.0 | 23.3 | n.a. |
| Marra et al. [3] | 2015 | Gait | 1 | 26.0 | 26.0 | 35.0 | 540 |
| Chen et al. [19] | 2016 | Gait | 3 | 36.3 (29–44) | 29.0 (26–34) | 27.3 (18–36) | n.a. |
| Ding et al. [8] | 2016 | Squat | 3 | 77.8 (46.3–101.0) | 50.2 (25.2–66.0) | 45.8 (22.0–74.1) | n.a. |
| Kebbach et al. [6] | 2020 | Gait | 1 | 39.0 | 35.0 | 10.0 | n.a. |
| Present study | 2024 | Squat | 4 | 41.7 (19.9–66.1) | 32.4 (15.9–52.6) | 18.1 (8.2–26.1) | <4 |
4.1. Limitations
4.2. Outlook
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mehta, S.; Tyler, A.; Hast, M. Understanding the Basics of Computational Models in Orthopaedics: A Nonnumeric Review for Surgeons. J. Am. Acad. Orthop. Surg. 2017, 25, 684–692. [Google Scholar] [CrossRef]
- Innocenti, B.; Bori, E.; Armaroli, F.; Schlager, B.; Jonas, R.; Wilke, H.-J.; Galbusera, F. The use of computational models in orthopedic biomechanical research. In Human Orthopaedic Biomechanics; Elsevier: Amsterdam, The Netherlands, 2022; pp. 681–712. ISBN 9780128244814. [Google Scholar]
- Marra, M.A.; Vanheule, V.; Fluit, R.; Koopman, B.H.; Rasmussen, J.; Verdonschot, N.; Andersen, M.S. A subject-specific musculoskeletal modeling framework to predict in vivo mechanics of total knee arthroplasty. J. Biomech. Eng. 2015, 137, 20904. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, X.; Ardestani, M.M.; Wang, L.; Liu, Y.; Lian, Q.; He, J.; Li, D.; Jin, Z. Prediction of in vivo joint mechanics of an artificial knee implant using rigid multi-body dynamics with elastic contacts. Proc. Inst. Mech. Eng. H 2014, 228, 564–575. [Google Scholar] [CrossRef]
- Stylianou, A.P.; Guess, T.M.; Kia, M. Multibody muscle driven model of an instrumented prosthetic knee during squat and toe rise motions. J. Biomech. Eng. 2013, 135, 41008. [Google Scholar] [CrossRef]
- Kebbach, M.; Darowski, M.; Krueger, S.; Schilling, C.; Grupp, T.M.; Bader, R.; Geier, A. Musculoskeletal Multibody Simulation Analysis on the Impact of Patellar Component Design and Positioning on Joint Dynamics after Unconstrained Total Knee Arthroplasty. Materials 2020, 13, 2365. [Google Scholar] [CrossRef]
- Otten, E. Inverse and forward dynamics: Models of multi-body systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003, 358, 1493–1500. [Google Scholar] [CrossRef][Green Version]
- Ding, Z.; Nolte, D.; Kit Tsang, C.; Cleather, D.J.; Kedgley, A.E.; Bull, A.M.J. In Vivo Knee Contact Force Prediction Using Patient-Specific Musculoskeletal Geometry in a Segment-Based Computational Model. J. Biomech. Eng. 2016, 138, 21018. [Google Scholar] [CrossRef]
- Krueger, C.A.; Austin, M.S.; Levicoff, E.A.; Saxena, A.; Nazarian, D.G.; Courtney, P.M. Substantial Preoperative Work Is Unaccounted for in Total Hip and Knee Arthroplasty. J. Arthroplast. 2020, 35, 2318–2322. [Google Scholar] [CrossRef]
- Tzanetis, P.; Fluit, R.; de Souza, K.; Robertson, S.; Koopman, B.; Verdonschot, N. Pre-Planning the Surgical Target for Optimal Implant Positioning in Robotic-Assisted Total Knee Arthroplasty. Bioengineering 2023, 10, 543. [Google Scholar] [CrossRef]
- Grosso, M.J.; Courtney, P.M.; Kerr, J.M.; Della Valle, C.J.; Huddleston, J.I. Surgeons’ Preoperative Work Burden Has Increased Before Total Joint Arthroplasty: A Survey of AAHKS Members. J. Arthroplast. 2020, 35, 1453–1457. [Google Scholar] [CrossRef]
- Wasterlain, A.S.; Courtney, P.M.; Yayac, M.F.; Nazarian, D.G.; Austin, M.S. Quantifying the Perioperative Work Associated with Total Hip and Knee Arthroplasty: The Burden Has Increased with Contemporary Care Pathways. J. Arthroplast. 2019, 34, 2528–2531. [Google Scholar] [CrossRef] [PubMed]
- Fregly, B.J.; Besier, T.F.; Lloyd, D.G.; Delp, S.L.; Banks, S.A.; Pandy, M.G.; D’Lima, D.D. Grand challenge competition to predict in vivo knee loads. J. Orthop. Res. 2012, 30, 503–513. [Google Scholar] [CrossRef]
- D’Lima, D.D.; Patil, S.; Steklov, N.; Slamin, J.E.; Colwell, C.W. Tibial forces measured in vivo after total knee arthroplasty. J. Arthroplast. 2006, 21, 255–262. [Google Scholar] [CrossRef] [PubMed]
- Wong, P.D.; Callewaert, B.; Desloovere, K.; Labey, L.; Innocenti, B. Determination of In Vivo Three-Dimensional Lower Limb Kinematics for Simulation of High-Flexion Squats. In XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010, Chalkidiki, Greece, 27–30 May 2010; Magjarevic, R., Bamidis, P.D., Pallikarakis, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 483–486. ISBN 978-3-642-13038-0. [Google Scholar]
- Asseln, M.; Hänisch, C.; Quack, V.; Radermacher, K. A Mesh Morphing Based Method to Estimate Cruciate Ligament Attachments Based on CT-Data. In Proceedings of the 15th Annual Meeting of the International Society for Computer Assisted Orthopaedic Surgery, Vancouver, BC, Canada, 17–20 June 2015. [Google Scholar]
- Amberg, B.; Romdhani, S.; Vetter, T. Optimal Step Nonrigid ICP Algorithms for Surface Registration. In Proceedings of the 2007 IEEE Conference on Computer Vision and Pattern Recognition, Minneapolis, MN, USA, 17–22 June 2007; IEEE: Piscataway, NJ, USA, 2007; pp. 1–8, ISBN 1-4244-1179-3. [Google Scholar]
- Zavatsky, A.B. A kinematic-freedom analysis of a flexed-knee-stance testing rig. J. Biomech. 1997, 30, 277–280. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Wang, L.; Li, D.; Zhang, Y.; Jin, Z. Evaluation of a subject-specific musculoskeletal modelling framework for load prediction in total knee arthroplasty. Med. Eng. Phys. 2016, 38, 708–716. [Google Scholar] [CrossRef]
- Raikova, R.T.; Prilutsky, B.I. Sensitivity of predicted muscle forces to parameters of the optimization-based human leg model revealed by analytical and numerical analyses. J. Biomech. 2001, 34, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Cockburn, R.A.; Hemmerich, A.; Li, R.M.; Wyss, U.P. Tibiofemoral joint contact forces and knee kinematics during squatting. Gait Posture 2008, 27, 376–386. [Google Scholar] [CrossRef]
- Hsu, A.T.; Perry, J.; Gronley, J.K.; Hislop, H.J. Quadriceps force and myoelectric activity during flexed knee stance. Clin. Orthop. Relat. Res. 1993, 288, 254–262. [Google Scholar] [CrossRef]
- Thelen, D.G.; Won Choi, K.; Schmitz, A.M. Co-simulation of neuromuscular dynamics and knee mechanics during human walking. J. Biomech. Eng. 2014, 136, 21033. [Google Scholar] [CrossRef] [PubMed]
- Lachiewicz, P.F.; Henderson, R.A. Patient-specific instruments for total knee arthroplasty. J. Am. Acad. Orthop. Surg. 2013, 21, 513–518. [Google Scholar] [CrossRef]
- Dartus, J.; Jacques, T.; Martinot, P.; Pasquier, G.; Cotten, A.; Migaud, H.; Morel, V.; Putman, S. The advantages of cone-beam computerised tomography (CT) in pain management following total knee arthroplasty, in comparison with conventional multi-detector CT. Orthop. Traumatol. Surg. Res. 2021, 107, 102874. [Google Scholar] [CrossRef]
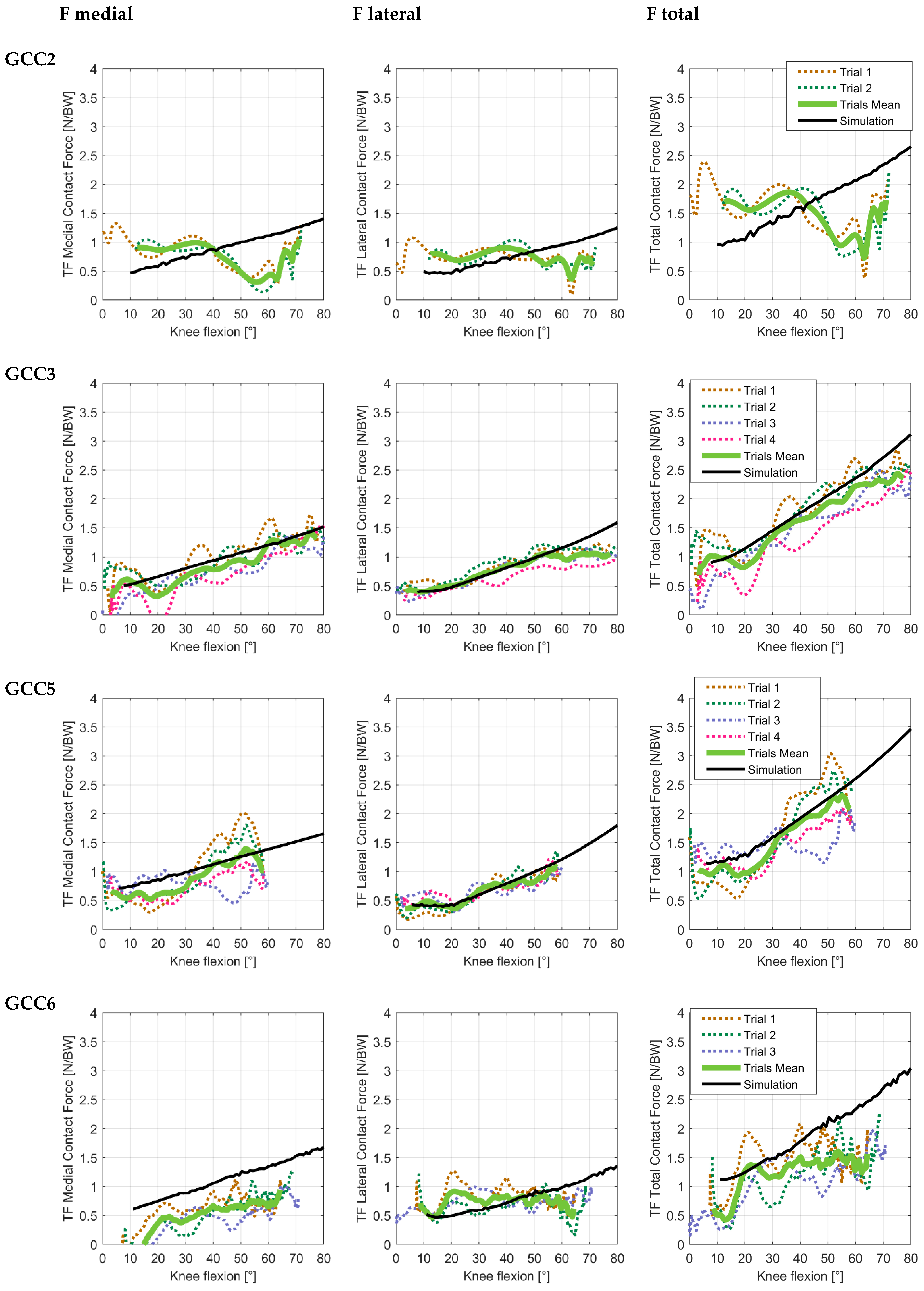
| Subject | Trial/Mean | Medial Contact Force | Lateral Contact Force | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAD [%BW] | RMSE [%BW] | r | M | P | C | MAD [%BW] | RMSE [%BW] | r | M | P | C | ||
| GCC2 | Trial 1 | 35.13 | 39.49 | 0.35 | 0.11 | 0.14 | 0.18 | 24.08 | 28.25 | 0.37 | 0.01 | 0.12 | 0.12 |
| Trial 2 | 39.07 | 48.52 | 0.66 | 0.19 | 0.18 | 0.26 | 25.31 | 28.20 | 0.30 | 0.01 | 0.12 | 0.12 | |
| Mean | 36.89 | 42.73 | 0.61 | 0.18 | 0.15 | 0.24 | 23.20 | 26.09 | 0.42 | 0.03 | 0.11 | 0.11 | |
| GCC3 | Trial 1 | 16.68 | 20.09 | 0.84 | −0.06 | 0.06 | 0.09 | 11.41 | 14.50 | 0.94 | 0.06 | 0.05 | 0.08 |
| Trial 2 | 16.38 | 19.31 | 0.85 | 0.04 | 0.06 | 0.07 | 16.61 | 20.97 | 0.85 | 0.05 | 0.07 | 0.08 | |
| Trial 3 | 20.58 | 23.43 | 0.93 | 0.19 | 0.05 | 0.20 | 14.20 | 22.04 | 0.92 | 0.20 | 0.05 | 0.20 | |
| Trial 4 | 25.85 | 31.94 | 0.92 | 0.21 | 0.09 | 0.23 | 25.63 | 33.13 | 0.94 | 0.42 | 0.05 | 0.43 | |
| Mean | 13.68 | 15.94 | 0.95 | 0.12 | 0.04 | 0.12 | 9.97 | 16.44 | 0.94 | 0.13 | 0.05 | 0.14 | |
| GCC5 | Trial 1 | 35.82 | 39.83 | 0.90 | −0.12 | 0.11 | 0.16 | 11.17 | 13.40 | 0.90 | 0.06 | 0.06 | 0.09 |
| Trial 2 | 24.71 | 26.44 | 0.92 | 0.01 | 0.08 | 0.08 | 8.88 | 10.07 | 0.94 | 0.01 | 0.04 | 0.04 | |
| Trial 3 | 27.56 | 36.67 | 0.14 | 0.30 | 0.10 | 0.31 | 15.15 | 16.91 | 0.77 | 0.08 | 0.07 | 0.11 | |
| Trial 4 | 28.48 | 30.98 | 0.84 | 0.35 | 0.05 | 0.36 | 10.89 | 13.10 | 0.85 | 0.03 | 0.06 | 0.07 | |
| Mean | 15.65 | 18.50 | 0.95 | 0.13 | 0.05 | 0.14 | 6.80 | 8.15 | 0.96 | 0.05 | 0.03 | 0.06 | |
| GCC6 | Trial 1 | 36.49 | 40.60 | 0.69 | 0.53 | 0.07 | 0.53 | 25.53 | 32.42 | 0.20 | −0.15 | 0.12 | 0.19 |
| Trial 2 | 53.31 | 54.96 | 0.93 | 0.76 | 0.12 | 0.77 | 17.11 | 26.45 | 0.12 | 0.16 | 0.11 | 0.19 | |
| Trial 3 | 63.56 | 66.25 | 0.80 | 1.08 | 0.15 | 1.09 | 20.72 | 23.62 | 0.12 | 0.01 | 0.10 | 0.10 | |
| Mean | 51.61 | 52.61 | 0.91 | 0.90 | 0.08 | 0.90 | 17.38 | 21.87 | 0.01 | −0.03 | 0.09 | 0.10 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehreiser, S.; Asseln, M.; Radermacher, K. Personalized Prediction of Total Knee Arthroplasty Mechanics Based on Sparse Input Data—Model Validation Using In Vivo Force Data. Biomechanics 2025, 5, 8. https://doi.org/10.3390/biomechanics5010008
Ehreiser S, Asseln M, Radermacher K. Personalized Prediction of Total Knee Arthroplasty Mechanics Based on Sparse Input Data—Model Validation Using In Vivo Force Data. Biomechanics. 2025; 5(1):8. https://doi.org/10.3390/biomechanics5010008
Chicago/Turabian StyleEhreiser, Sonja, Malte Asseln, and Klaus Radermacher. 2025. "Personalized Prediction of Total Knee Arthroplasty Mechanics Based on Sparse Input Data—Model Validation Using In Vivo Force Data" Biomechanics 5, no. 1: 8. https://doi.org/10.3390/biomechanics5010008
APA StyleEhreiser, S., Asseln, M., & Radermacher, K. (2025). Personalized Prediction of Total Knee Arthroplasty Mechanics Based on Sparse Input Data—Model Validation Using In Vivo Force Data. Biomechanics, 5(1), 8. https://doi.org/10.3390/biomechanics5010008








