A Proteomics Method for Presumptive Identification of Human Tissue
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Samples
2.2. Proteomics Analysis
2.2.1. Questioned and Known Human Samples
2.2.2. Formalin-Fixed Paraffin-Embedded (FFPE) Tissue
2.2.3. Optimal Cutting Temperature (OCT) Compound-Embedded Tissue
2.2.4. Dried Blood Spots
2.2.5. Saliva and Blood from Countertops
2.3. Tandem Mass Spectrometry
2.4. Database Search
2.5. Development of Data Filtration Rules for Human Tissue and Organ Identification
- (a)
- Step 1: Use high-confidence peptides to search a “mammalian” database with Proteome Discoverer and the Sequest HT algorithm. Export the result to MS Excel and delete any protein annotated as keratin.
- (b)
- Step 2: Delete proteins identified with less than 2 unique peptides.
- (c)
- Step 3: Delete identified proteins with less than 5% protein sequence coverage.
- (d)
- Step 4: Examine the remaining proteins on the list to determine the percentage annotated as human.
2.6. Validation of Bottom-Up Proteomics-Based Human Tissue Identification
3. Results and Discussion
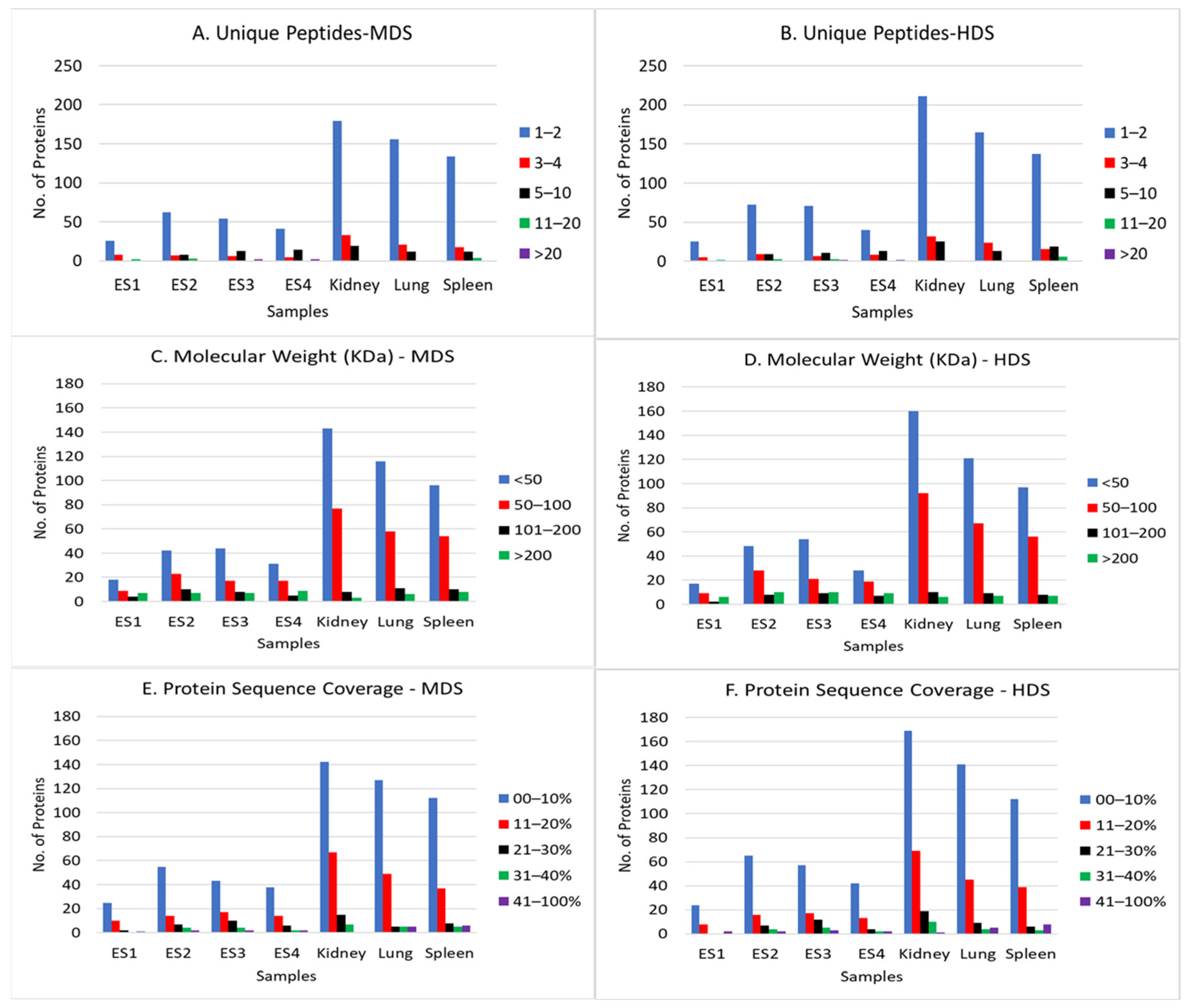
3.1. Proteomic Analysis of Known Human and Questioned Tissue
3.2. Identification of Human and Non-Human Tissue Using Proteomic Data
4. Limitations and Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Negri, S. Transplant ethics and the international crime or organ trafficking. Int. Crim. Law Rev. 2016, 16, 287–303. [Google Scholar] [CrossRef]
- O’Neill, M. Trafficking and Organs, Tissues, and Cells: An Examination of European and UK Legislation and Gaps European. J. Crime Crim. Law Crim. Justice 2024, 32, 32–57. [Google Scholar] [CrossRef]
- Meshelemiah, J.C.A.; Lynch, R.E. The Cause and Consequence of Human Trafficking: Human Rights Violations; The Ohio State University Pressbook: Columbus, OH, USA, 2019. [Google Scholar]
- Parker, G.J.; Leppert, T.; Anex, D.S.; Hilmer, J.K.; Matsunami, N.; Baird, L.; Stevens, J.; Parsawar, K.; Durbin-Johnson, B.P.; Rocke, D.M.; et al. Demonstration of Protein-Based Human Identification Using the Hair Shaft Proteome. PLoS ONE 2016, 11, e0160653. [Google Scholar] [CrossRef]
- Somiari, R.I.; Sullivan, A.; Russell, S.; Somiari, S.; Hu, H.; Jordan, R.; George, A.; Katenhusen, R.; Buchowiecka, A.; Arciero, C.; et al. High-throughput proteomic analysis of human infiltrating ductal carcinoma of the breast. Proteomics 2003, 3, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Duong, V.-A.; Park, J.-M.; Lim, H.-J. Proteomics in Forensic Analysis: Applications for Human Samples. Appl. Sci. 2021, 11, 3393. [Google Scholar] [CrossRef]
- Oonk, S.; Schuurmans, T.; Pabst, M.; de Smet, L.C.P.M.; de Put, M. Proteomics as a new tool to study fingermark ageing in forensics. Sci. Rep. 2018, 8, 16425. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.M.; Zissler, A.; Kim, E.; Ehrenfellner, B.; Cho, E.; Lee, S.I.; Steinbacher, P.; Yun, K.N.; Shin, J.H.; Kim, J.Y.; et al. Postmortem proteomics to discover biomarkers for forensic PMI estimation. Int. J. Leg. Med. 2019, 133, 899–908. [Google Scholar] [CrossRef]
- Boroumand, M.; Grassi, V.M.; Castagnola, F.; De-Giorgio, F.; D’aLoja, E.; Vetrugno, G.; Pascali, V.L.; Vincenzoni, F.; Iavarone, F.; Faa, G.; et al. Estimation of postmortem interval using top-down HPLC–MS analysis of peptide fragments in vitreous humour: A pilot study. Int. J. Mass Spectrom. 2023, 483, 116952. [Google Scholar] [CrossRef]
- Alex, S.; Shehata, T.P.; Gergely, A.I.; de Puit, M. Proteomics in forensics: From source attribution to reconstruction of events. Sci. Justice 2025, 65, 101320. [Google Scholar] [CrossRef]
- Parker, G.J.; McKiernan, H.E.; Legg, K.M.; Goecker, Z.C. Forensic proteomics. Forensic Sci. Int. Genet. 2021, 54, 102529. [Google Scholar] [CrossRef]
- Wadsworth, C.; Buckley, M. Proteome degradation in fossils: Investigating the longevity of protein survival in ancient bone. Rapid Commun. Mass Spectrom. RCM 2014, 28, 605–615. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bunger, M.K.; Cargile, B.J.; Sevinsky, J.R.; Deyanova, E.; Yates, N.A.; Hendrickson, R.C.; Stephenson, J.J.L. Detection and validation of non-synonymous coding SNPs from orthogonal analysis of shotgun proteomics data. J. Proteome Res. 2007, 6, 2331–2340. [Google Scholar] [CrossRef] [PubMed]
- Sheynkman, G.M.; Shortreed, M.R.; Frey, B.L.; Scalf, M.; Smith, L.M. Large-scale mass spectrometric detection of variant peptides resulting from nonsynonymous nucleotide differences. J. Proteome Res. 2014, 13, 228–240. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mason, K.E.; Anex, D.; Grey, T.; Hart, B.; Parker, G. Protein-based forensic identification using genetically variant peptides in human bone. Forensic Sci. Int. 2018, 288, 89–96. [Google Scholar] [CrossRef]
- Pastwa, E.; Somiari, S.B.; Czyz, M.; Somiari, R.I. Proteomics in human cancer research. Proteom. Clin. Appl. 2007, 1, 4–17. [Google Scholar] [CrossRef]
- Somiari, R.I.; Renganathan, K.; Russell, S.; Wolfe, S.; Mayko, F.; Somiari, S.B. A Colorimetric Method for Monitoring Tryptic Digestion Prior to Shotgun Proteomics. Int. J. Proteom. 2014, 2014, 125482. [Google Scholar] [CrossRef] [PubMed]
- Rispoli, L.A.; Edwards, J.L.; Pohler, K.G.; Russell, S.; Somiari, R.I.; Payton, R.R.; Schrick, F.N. Heat-induced hyperthermia impacts the follicular fluid proteome of the periovulatory follicle in lactating dairy cows. PLoS ONE 2019, 14, e0227095. [Google Scholar] [CrossRef] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 11, 923–925. [Google Scholar] [CrossRef] [PubMed]
- The, M.; MacCoss, M.J.; Noble, W.S.; Käll, L. Fast and Accurate Protein False Discovery Rates on Large-Scale Proteomics Data Sets with Percolator 3.0. J. Am. Soc. Mass Spectrom. 2016, 27, 1719–1727. [Google Scholar] [CrossRef] [PubMed]
- Käll, L.; Storey, J.D.; MacCoss, M.J.; Noble, W.S. Posterior Error Probabilities False Discovery Rates: Two Sides of the Same Coin. J. Proteome Res. 2008, 7, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Pontén, F.; Gry, M.; Fagerberg, L.; Lundberg, E.; Asplund, A.; Berglund, L.; Oksvold, P.; Björling, E.; Hober, S.; Kampf, C.; et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 2009, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Turingan, R.S.; Brown, J.; Kaplun, L.; Smith, J.; Watson, J.; Boyd, D.A.; Steadman, D.W.; Selden, R.F. Identification of human remains using Rapid DNA analysis. Int. J. Leg. Med. 2019, 134, 863–872. [Google Scholar] [CrossRef]
- Raj, T.A.; Aravind, G.B.; Arun, M.; Aneesh, E.M. Mass spectrometry-based proteomics in forensic investigations: A focused review of LC MS applications. Egypt. J. Forensic Sci. 2025, 15, 75. [Google Scholar] [CrossRef]

| Tissue Samples | Number of Proteins Identified | Average | % of Proteins Annotated as Human in MDS | |
|---|---|---|---|---|
| HDS | MDS | |||
| ||||
| Kidney | 268 | 224 | 246 | 74 |
| Lung | 204 | 187 | 196 | 70 |
| Spleen | 168 | 161 | 165 | 62 |
| ||||
| ES1 | 34 | 35 | 35 | 71 |
| ES2 | 94 | 74 | 84 | 72 |
| ES3 | 94 | 72 | 83 | 70 |
| ES4 | 63 | 55 | 59 | 74 |
| Sample | Proteins Identified | Proteins with >1 UP 1 | Proteins with >1 UP and >4% SC 2 | % of Human Proteins with >1 UP and >4% SC 3 | ||
|---|---|---|---|---|---|---|
| Total | Human | % Human | ||||
| Kidney | 224 | 166 | 74% | 92 | 84 | 83% |
| Lung | 187 | 128 | 68% | 60 | 51 | 76% |
| Spleen | 161 | 98 | 61% | 63 | 57 | 65% |
| ES1 | 35 | 24 | 69% | 17 | 14 | 86% |
| ES2 | 74 | 53 | 72% | 23 | 20 | 85% |
| ES3 | 72 | 49 | 68% | 30 | 24 | 67% |
| ES4 | 55 | 40 | 73% | 26 | 23 | 78% |
| S/N | Origin | CODE | Condition 1 | Identified Proteins | Classification 6 | |||
|---|---|---|---|---|---|---|---|---|
| Total 2 | Human 3 | Human A 4 | Human B 5 | |||||
| 1 | HUMAN | GPB1 | A | 13 | 12 | 92% | 100% | H |
| 2 | HUMAN | GPB2 | A | 16 | 14 | 88% | 100% | H |
| 3 | HUMAN | GPB3 | A | 21 | 15 | 71% | 100% | H |
| 4 | HUMAN | FFPE1 | B | 29 | 15 | 51% | 60% | H |
| 5 | HUMAN | FFPE2 | B | 26 | 17 | 65% | 67% | H |
| 6 | HUMAN | FFPE3 | B | 30 | 15 | 50% | 83% | H |
| 7 | HUMAN | DHS1 | C | 24 | 22 | 92% | 100% | H |
| 8 | HUMAN | DHB1 | D | 29 | 26 | 90% | 100% | H |
| 9 | HUMAN | OCT1 | E | 376 | 233 | 62% | 71% | H |
| 10 | HUMAN | OCT2 | F | 68 | 38 | 56% | 59% | H |
| 11 | Bovine | B1 | G | 14 | 2 | 14% | 0% | NH |
| 12 | Bovine | B2 | G | 86 | 17 | 20% | 6.1% | NH |
| 13 | Mouse | M1 | H | 15 | 2 | 13% | 0% | NH |
| 14 | Mouse | M2 | H | 15 | 3 | 20% | 20% | NH |
| 15 | Mouse | M3 | H | 26 | 6 | 23% | 14.3% | NH |
| 16 | Mouse | M4 | H | 86 | 20 | 23% | 21% | NH |
| 17 | Mouse | M5 | H | 20 | 5 | 25% | 25% | NH |
| 18 | Mouse | M6 | H | 385 | 79 | 21% | 18.5% | NH |
| 19 | Mouse | M7 | H | 50 | 9 | 18% | 5.5% | NH |
| 20 | Rat | R1 | I | 39 | 8 | 21% | 43% | NH |
| 21 | Rat | R2 | I | 76 | 12 | 16% | 13% | NH |
| 22 | Rat | R3 | I | 106 | 21 | 20% | 12% | NH |
| 23 | Rat | R4 | I | 1133 | 220 | 19% | 15% | NH |
| 24 | Rat | R5 | I | 16 | 3 | 19% | 0% | NH |
| 25 | Rat | R6 | I | 107 | 21 | 20% | 23% | NH |
| 26 | Sheep | S1 | J | 104 | 30 | 29% | 16.6% | NH |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Somiari, R.I.; Russell, S.J.; Feeley, J.; Somiari, S.B. A Proteomics Method for Presumptive Identification of Human Tissue. Forensic Sci. 2025, 5, 75. https://doi.org/10.3390/forensicsci5040075
Somiari RI, Russell SJ, Feeley J, Somiari SB. A Proteomics Method for Presumptive Identification of Human Tissue. Forensic Sciences. 2025; 5(4):75. https://doi.org/10.3390/forensicsci5040075
Chicago/Turabian StyleSomiari, Richard Idem, Stephen J. Russell, John Feeley, and Stella B. Somiari. 2025. "A Proteomics Method for Presumptive Identification of Human Tissue" Forensic Sciences 5, no. 4: 75. https://doi.org/10.3390/forensicsci5040075
APA StyleSomiari, R. I., Russell, S. J., Feeley, J., & Somiari, S. B. (2025). A Proteomics Method for Presumptive Identification of Human Tissue. Forensic Sciences, 5(4), 75. https://doi.org/10.3390/forensicsci5040075






