1. Introduction
Using a swab to collect forensically relevant samples, including buccal swabs for reference, is a common method for recovering biological material from a crime scene. A swab can be defined as an absorbent pad with a shaft. Originally designed as ‘ear pickers’, swabs are now used for various purposes, including sample collection (e.g., microorganisms, DNA/RNA, and forensic traces) [
1,
2,
3].
The use and selection of the most appropriate swab type received much attention during the COVID-19 pandemic, since swabs are a fundamental tool in collecting samples from individuals to diagnose the presence of the SARS-CoV-2 virus [
4].
In this review, the performance of swabs is examined closely. The overall performance or efficiency is composed of the extraction efficiency and the recovery efficiency, and the absorption capacity plays a role. These terms are explained in the next section, along with the various factors influencing these efficiencies, such as the swab material and the type of substrate. Subsequently, a critical reflection of swab performance as reported in the literature is provided. Since swabs generally exhibit low overall efficiency, various available alternatives are discussed. In the Discussion, the performance of swabs and their alternatives is analyzed in light of the various factors that influence efficiency. Moreover, a critical reflection is given on the other criteria that determine the selection of a swab, such as the usability of a swab or alternatives.
2. Swab Efficiency
The performance of a swab can be expressed in terms of the extraction and recovery efficiencies. The extraction efficiency refers to the material transfer effectiveness from the collection medium to the extraction solution, while the recovery (or overall) efficiency is defined as the transfer effectiveness from a sampled surface to the extraction solution. Most of the literature focuses solely on the recovery efficiency, whereas a swab may exhibit excellent collection properties but bad performance upon release of the sample [
5,
6].
Although not widely reported, the absorption capacity can have a substantial influence on the extraction and recovery efficiency of swabs. This absorption capacity mostly depends on the swab tip’s (also called the swab head) dimensions and morphology rather than on swab material composition. The amount of DNA collected is inversely proportional to the density of the fibers in the swab tip. Accounting for absorption capacity allows for the determination of maximum theoretical efficiency, assuming uniform distribution of DNA across the liquid phases on the swab and in the extraction vial [
2,
6,
7].
The exact recovery efficiency of a swab is influenced by various factors, including the swab material, the type of sample, the type of substrate and the swabbing protocol. It should be noted that the recovery of more DNA does not necessarily result in good or complete DNA profiles [
3].
2.1. Swab Material
The existing types of swabs can be categorized into three main design categories: (1) a wound swab is made up of either many fibers or one long fiber wound around a shaft; (2) a flocked swab, i.e., a swab that has small fibers glued onto a shaft; and (3) a pad swab, which contains a sleeve of foam (or other porous material) attached to a shaft.
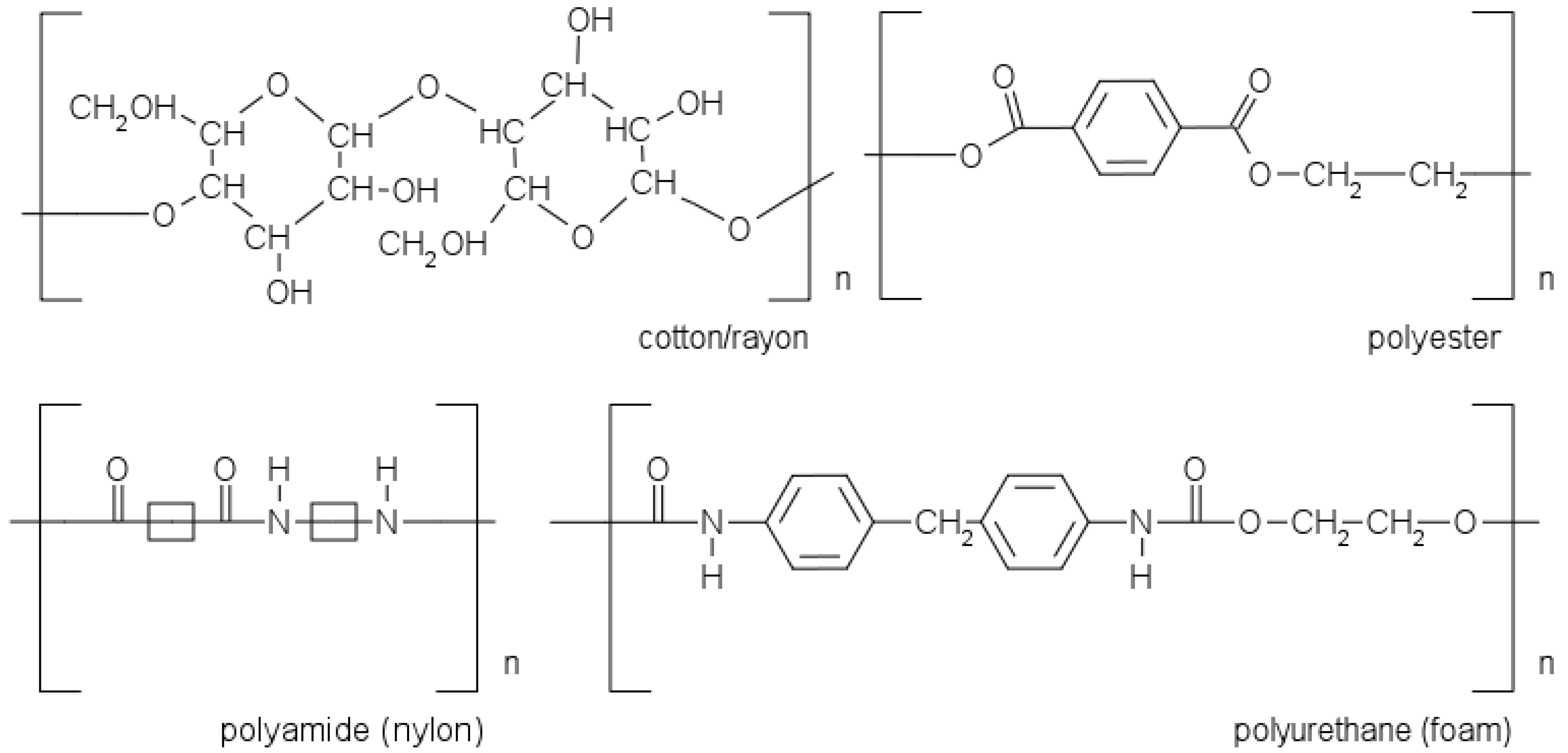
For forensic use, cotton and nylon-flocked are the most commonly used; alternatives include foam, polyester, and rayon. The molecular structures of the different swabs can be seen in
Figure 1. An ideal swab material should exhibit strong DNA binding during swabbing while allowing for easy DNA release during the extraction process [
3,
6].
2.1.1. Cotton and Rayon
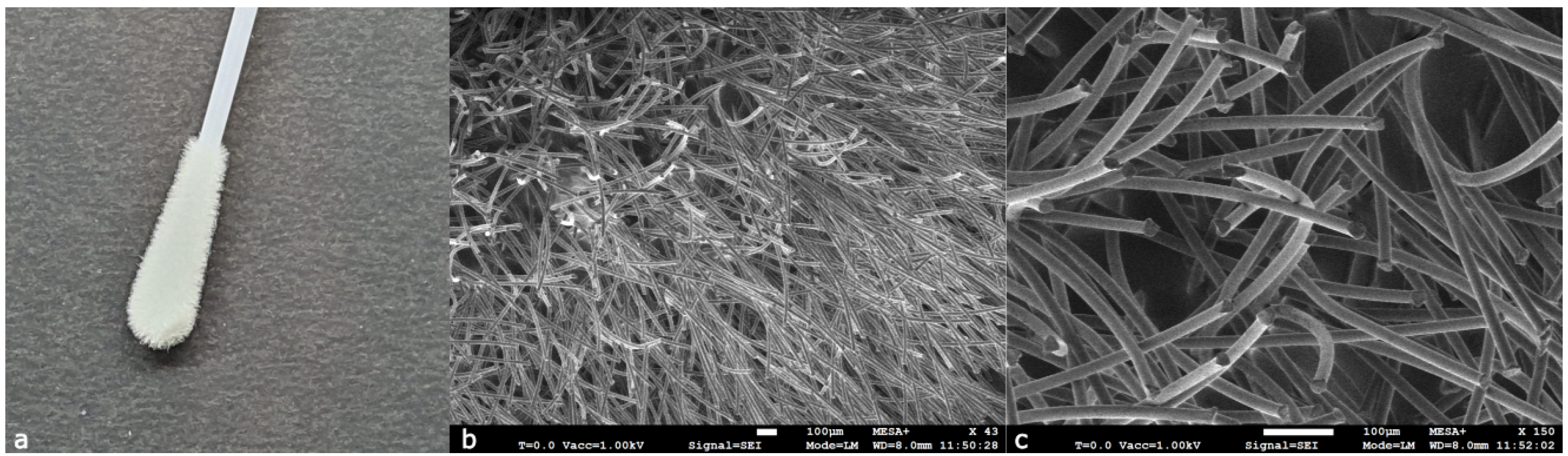
Cotton, one of the oldest materials cultivated, is currently considered the state of the art within the forensic field as swab material, falling in the first category of swabs. However, cotton swabs may leave fibers or other impurities in a reaction mixture, potentially negatively affecting the PCR, for example, through inhibition. Another drawback of tightly wound swabs, such as cotton or rayon, is their limited material retrieval and releasing performance. Rayon, also known as viscose, is a spun cellulose fiber made from wood pulp and is mainly designed for the recovery of micro-organisms. The texture and absorbance characteristics of rayon are similar to those of cotton, since cotton and rayon have the same chemical structure but differ in the degree of polymerization with values of 400–700 and 5000 cellulose units for rayon and cotton, respectively. Cellulose contains hydroxyl (O–H) groups; they form hydrogen bonds with nucleic acid chains and carbohydrates in cell membranes, which is beneficial for sample pick-up but not for sample extraction from the swab. In comparison to cotton, rayon is somewhat softer and less prone to leaving fibers behind [
3,
7,
8]. A photograph and SEM images of a cotton and rayon swab can be seen in
Figure 2 and
Figure 3, respectively.
2.1.2. Nylon-Flocked Swabs
Nylon-flocked swabs are composed of short nylon fiber strands attached to a (plastic) shaft. The hydrophilic open-fiber morphology is specially designed to enhance sampling, ensuring the efficient collection and release of the sample. The swabs are suitable for the sampling of saliva, blood, and skin epithelial cells from various substrates. Nylon-flocked swabs show a better sample release and no sample entrapment, as is the case with conventional cotton swabs. However, flocked swabs might leave swab material on the surface, particularly on rough ones. Nylon, a polyamide, contains N–H groups, which are known to form hydrogen bonds with nucleic acids. While these bonds are advantageous for sample collection, they are not suitable for sample extraction from the swab [
3,
9]. A photograph and SEM images of two types of nylon-flocked swabs can be seen in
Figure 4 and
Figure 5.
2.1.3. Polyester
Polyester, also known as dacron, is a synthetic polymer (PET) fiber. Polyester swabs are known for their high collection and release characteristics. Swabs with (knitted) polyester tips are mainly used for cleaning purposes rather than forensic applications, although they perform quite well on nonporous substrates. Polyester possesses polar ester (C=O part) groups, which form only weak dipole–dipole interactions, having less influence on extraction efficiency than strong interactions. However, compared to cotton and rayon, the long and irregular fibers leave fewer open spaces, making polyester less efficient for extraction [
3,
8].
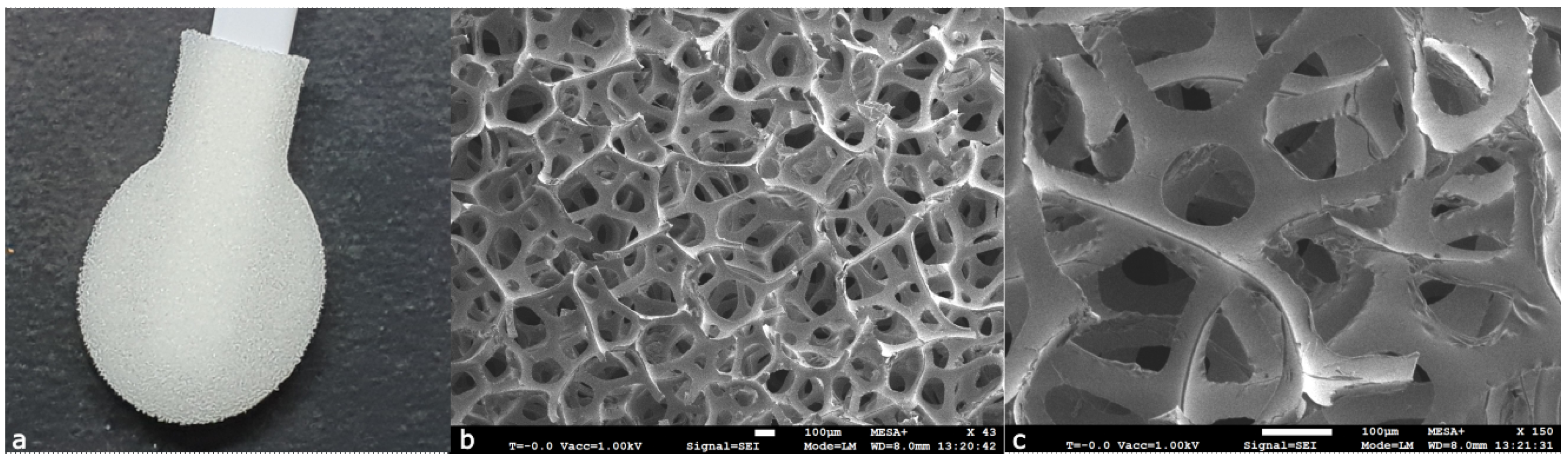
2.1.4. Foam
Foam (polyurethane) swabs are elastomers produced through a condensation reaction of isocyanates with polyols. Swabs constructed from foam have a more open structure than polyester, rayon, and cotton swabs, making them work as a sponge. However, due to the hydrophobic nature of foam, aqueous solutions tend to stay on the outer surface of the swab tip (rather than penetrating in the foam). It has been suggested that foam swabs, due to the flexible nature of the material, can penetrate into porous substrates, which makes them suitable for, e.g., wooden surfaces. Polyurethane, just as polyester, has polar C=O groups, which possess only weak dipole–dipole interactions. Weaker interactions have a relatively minor effect on extraction efficiency compared to strong interactions [
3,
8]. A photograph and SEM images of a foam swab can be seen in
Figure 6.
2.2. Type of Sample
At a crime scene, various types of samples can potentially be found and sampled for forensic DNA analysis. The types of samples can vary depending on the nature of the crime and the circumstances. Blood and saliva, for instance, are frequently encountered in violent crimes, while touch DNA and cell-free DNA underscore the significance of trace evidence, enabling forensic experts to investigate even subtle contact interactions that may otherwise be unnoticed.
2.2.1. Biological Fluids
DNA profiling is a powerful tool within forensic science because every cell type possesses identical DNA, enabling identification of the donor of biological material regardless of the type of sample. Blood is the most common biological material found at a crime scene, followed by saliva. Additionally, other body fluids can be used for DNA profiling, including semen, urine, sweat, and vaginal secretions. As reference sample, usually a buccal swab is taken, since swabbing the inside of the cheek from a person is a noninvasive procedure, and the amount of DNA collected is sufficient for DNA profiling [
10,
11].
While the DNA is the same for each cell type, the amount of DNA varies substantially. Blood and saliva (on, e.g., drinking cans or cigarette butts) show a high success rate of DNA profiling. Mapes et al. showed that the success ratio of DNA profiling is directly related to the DNA concentration in a sample. Essentially, any traces with a concentration exceeding 100 pg/
L yield usable DNA profiles suitable for inclusion in a DNA database. Consequently, the outcome hinges on the quantity of DNA present, irrespective of the nature of the sampled items. Cigarettes, bloodstains, and headwear exhibit relatively high DNA profiling success rates. Conversely, cartridge cases, crowbars, and tie-wraps fall at the opposite end of the spectrum with lower success rates [
12].
2.2.2. Touch DNA
Touch DNA refers to DNA left behind on an object or surface through direct physical contact. This type of DNA is often collected from skin cells, sweat, or other biological materials that individuals deposit when they touch items like doorknobs, glass, or clothing. Since touch DNA is not visible (e.g., blood), the collection is carried out based on speculations. Additionally, touch DNA samples are often small (and may be subject to degradation over time), making their analysis and interpretation challenging for forensic scientists. As DNA analysis techniques have become more sensitive, touch DNA has become a substantial part of the traces sent to the laboratory for DNA profiling. Consequently, due to the limited amount of DNA available, the majority of the literature on improving DNA recovery focuses on touch DNA [
2,
13,
14,
15,
16].
2.2.3. Cell-Free DNA
Cell-free DNA refers to fragments of DNA found in bodily fluids and tissues outside of cells. Cell-free DNA has become of interest to forensic scientists, since DNA profiles can be generated from cell-free DNA. This DNA can originate from various cell types throughout the body and can be present as extracellular DNA but can also originate from cell membrane rupture due to osmotic lysis caused by soaking off the stain with water [
17,
18]. In a study conducted by Vandewoestyne et al., cell-free DNA was found in 90% of 100 forensic case samples, originating from different sources such as blood, saliva, vomit, and contact traces [
17].
2.3. Type of Substrate
The effectiveness of DNA collection techniques greatly hinges on the physical attributes of the sampled substrate. The dimensions, absorbent qualities, irregular configuration, and coarse texture are parameters that must be taken into account. For example, substrates with irregular shapes or contours can present challenges during swabbing, since DNA can be trapped in crevices, folds, or textured surfaces. The choice of swab and collection technique should align with the specific characteristics of the substrate encountered at a crime scene.
2.3.1. Nonporous Substrates
Nonporous (or nonabsorbing) surfaces can be further categorized in smooth and ridged surfaces. Smooth nonporous surfaces like glass are typically amenable to swab-based sample collection using materials such as cotton or nylon-flocked swabs. However, a challenge arises with ridged nonporous substrates like ridged plastic, where the swab fibers may encounter difficulties due to the uneven surface. The ridges can cause swab fibers to fray, potentially hindering effective DNA collection.
2.3.2. Porous Substrates
Also, porous substrates can be further divided into smooth (e.g., untreated wood) and rough (e.g., textile) substrates. Porous surfaces, such as fabrics or paper, may readily absorb bodily fluids containing DNA, making it crucial to sample thoroughly to capture the maximum amount of biological material. In cases involving porous substrates, tape-lifting emerges as a popular and effective method for sample collection.
2.3.3. Metal Substrates
Within forensics, metal substrates are usually considered a distinct category of substrates. Although metal surfaces are usually nonporous, it is challenging to recover a sufficient amount of DNA from metals. The phosphate backbone of DNA is negatively charged and interacts with the array of ionization and electron affinities of metals, which makes it hard to remove DNA from metallic substrates. Additionally, nitrogen or positive atoms of the nucleobase can form ionic bonds with metals. Commonly encountered in forensic cases, metal substrates such as doorknobs, jewelry, knives, and firearms (including ammunition) necessitate the development of specialized protocols for DNA recovery due to the distinctive properties of these surfaces [
13,
19]. Bonsu et al. concluded that Isohelix swabs (material type unknown) with isopropyl alcohol outperformed rayon swabs with water for DNA recovery from metal surfaces when swabbing touch DNA [
20,
21].
2.4. Protocol
To achieve the highest DNA recovery, the entire protocol is crucial, including the swabbing technique, buffer solution, extraction method, and swab transport/storage. The swabbing technique used impacts the DNA quality and quantity, requiring proper methods to minimize contamination and maximize sample viability. Buffer solutions preserve and stabilize DNA, preventing degradation during transport and storage. The exact composition of the buffer solution also influences the amount of DNA that can be recovered from a substrate. In fact, the selection of the extraction method selection is key for optimal DNA retrieval, minimizing losses and maximizing yield. Well-designed storage containers and conditions protect samples from environmental factors, contamination, and degradation. All of these aspects are discussed in the following subsections.
2.4.1. Swabbing Technique
The single swabbing technique involves a (pre-)wetted swab to recover biological material from a surface. With the double-swabbing technique, a wet swab is followed by a dry swab, which are individually extracted. Especially in the case of touch DNA/epithelial cells, double swabbing can be beneficial, since there is consensus that only a wet swab might not recover this type of sample efficiently. Pooling/combing both swabs can further enhance DNA profiling results. Although only used occasionally, dry–dry swabbing is also possible. Both swab heads are pooled for further analysis. This method is used on substrates that are affected and/or destroyed by wetting them, such as paper [
5,
10,
22,
23].
Three types of double-swabbing techniques were investigated by Adbullah et al. to determine the best technique for optimal DNA recovery. In addition to the wet–wet and wet–dry double-swabbing techniques, wet–moist was also used. This technique, by using a wet swab and then a moist swab, has been used for clothing and fabric substrates. Additionally, a single wet and moist–dry swab were used in the study. For the latter, one side is moistened and applied first to the substrate; after, the dry side of the same swab is used. Using cotton swabs to recover DNA from cotton, the double wet–moist swab with sampling for 30 s gave the highest recovery efficiency. When tile is used as the substrate, either a single wet or moist–dry swab is recommended to be used, in combination with a 15 s swabbing time [
24]. Hedman et al. compared the double-swabbing technique with single swabbing and concluded that double swabbing is especially beneficial for complex surfaces (e.g., very porous surfaces), but that, as a general approach, single wet swabs are better, in particular on nonabsorbing surfaces [
25].
2.4.2. Buffer Solution
A buffer solution (and prewetting agent) can be as little as only water (note, where water is mentioned, nuclease-free water is meant). Detergents, such as SDS, Tween 20, and Triton X-100, can be added to the buffer solution, as well as EDTA or PBS. Due to the amphiphilic nature of detergents, cell components like fats, lipids, and proteins become dissolved in the buffer solution. SDS can denature tertiary structures, aiding in the release of bound DNA. To avoid the osmotic lysis of the cells, PBS can be added to the buffer solution. In term of cell/DNA recovery, Tween 20 is more efficient than PBS. Moreover, detergents can aid cell lysis, but higher concentrations result in precipitation, and these detergents might interfere with commercial DNA extraction kits. EDTA can be added to inactivate DNase to prevent degradation of the DNA. Also, NaCl can be added, but (high concentrations of) salt can interfere with PCR [
1,
5,
13].
Various detergents that can be added to the swabbing solution were investigated by Thomasma et al. to determine their influence on DNA recovery from touch samples. Especially, the addition of Triton X-100 and SDS showed a significantly greater DNA recovery than using only water [
26]. Schulte et al. concluded that the prewetting volume has no significant effect on DNA yield and that 70% ethanol and 0.10%
v/
v Triton X-100 are both suitable as moistening solutions for the collection of touch DNA. Also, SDS can be used but tends to foam when higher concentrations are used, and Triton X-100 is extremely viscous, which makes its handling more difficult [
27]. Canfield et al. recommend the use of AutoMACS running buffer (containing BSA, EDTA, and azide) for the recovery of intact leukocytes over PBS or water in combination with nylon-flocked swabs (instead of cotton or dissolvable swabs) [
28].
2.4.3. Extraction
The overall efficiency of swabs also depends on the extraction method used, since the tight winding and high absorption capacity of swabs necessitates an efficient extraction method that is capable of overcoming the strong DNA binding to the swab. The tip of a cotton swab is sometimes shaved off from the shaft. For tightly wound materials, teasing loose the swab head before extraction is beneficial for DNA recovery.
It is worth mentioning that in forensic case research, so-called spin baskets are commonly used for the extraction (and purification) of DNA from cells. These baskets (see
Figure 7) are developed and recommended for use in combination with a lysis step. However, the basket protocols involve an intensive set of washing and purification steps, which may result in additional DNA loss [
6,
14].
2.4.4. Transport/Storage
In addition to the swabbing technique, buffer solution, and extraction protocol used, the manner of transport to the laboratory is also important. Biological samples are susceptible to degradation from the moment of collection until their analysis in the laboratory. Factors like biological contaminants (e.g., bacteria, fungi, and enzymes) and environmental elements (e.g., sunlight, heat, and humidity) can accelerate DNA degradation [
1].
To prevent fungal and/or bacterial growth, moist samples should be dried as quickly as possible. Therefore, some swabs are available in combination with a preperforated container or a desiccant. In case the swab container is not equipped with this, a hole needs to be made manually [
1].
The performance of two commercially available swab and drying tube combinations was investigated by Garvin et al. It was concluded that faster drying is beneficial for the DNA yield, with 95% versus 12% for fast sample drying (with about 0.64%/min during the first 2 h and completely dry within 4 h for the nylon-flocked swab and 10% of water remaining within 6 h for the cotton swab) and slow sample drying (with about 0.08%/min and completely dry within 29 h), respectively [
29].
3. Comparison of Swab Efficiency
Several research groups have compared the (overall) performance of various swab types. Parameters including substrate and composition of the extraction buffer were in some cases also taken into account. An overview of the parameters tested can be found in
Table 1.
Multiple swab types were tested on porous (wood and brick) and nonporous (glass and pitted plastic) substrates to determine the extraction and recovery efficiency of sampling blood, saliva, and touch DNA by Verdon et al. To determine the best swab, the various swab types were ranked according to their efficiency based on the quantity of DNA extracted (in ng/
L) from different biological fluids. Swabs with the highest recovery efficiency often did not have the highest extraction efficiency. For most combinations of sample type and type of substrate, different swabs gave the best results (see also
Table 2) [
2].
Brownlow et al. compared the efficiency of cotton and nylon-flocked swabs with saliva samples spotted in a petri dish. The efficiency was expressed in percentage recovered DNA for high-quantity DNA (>500 ng) and decreased-quantity DNA (10–100 ng). Three different extraction methods were used; overall, the cotton swabs performed better (with the highest recovery value being 64.5%) than the nylon-flocked swabs. It must be noted that the extraction method had a bigger influence on the result than the type of swab (with the lowest recovery value being 15.09%) [
7].
The absorption capacity and swab morphology were also taken into account by Bruijns et al., who tested various swabs with pure DNA (simulating cell-free DNA). The absorption capacity (>100
L for the tested swabs) mainly depended on the tip dimensions and winding of the fibers rather than on the swab material itself. However, both the extraction and recovery efficiency stayed below 50%. Overall, a nylon-flocked swab performed the best and, by also taking the maximum theoretical efficiency into account, a nylon-flocked and a polyester swab showed the highest efficiency [
6].
Cotton and nylon-flocked swabs were tested on various substrates by Dadhania et al. Two kits based on magnetic bead technology were investigated, whether or not in combination with a spin basket, with the latter increasing the DNA recovery to 48%. The nylon-flocked swabs showed substantially higher recovery rates in terms of total DNA recovered in ng (with an increase of 50% or 480%, dependent on the extraction method used). However, an adaption of the protocol was required, since the swabs were not compatible with one of the lysis buffers used (reason unknown) [
14].
Comment et al. compared five swabs from various materials in combination with touch DNA samples from real case work. Not only was the performance investigated: the crime scene investigators were asked about their experience with the different swabs. The investigators could choose the sampling method, although the recommended method was wet–dry swabbing with one swab by prewetting one side of the swab head and subsequently sampling again with the dry side. They concluded that the user-friendliness or handiness of a swab influences the results and should also be taken into account during swab selection. In terms of performance, expressed as the fraction (%) of suitable profiles/DNA profiling success rate, a cotton swab showed the best results in this study, at ∼39% [
30].
The extraction efficiency of cotton swabs was determined by Adamowicz et al. via the direct deposit of blood and buccal cells on the swabs. A commercially available extraction kit, containing prot K, was used. The extraction efficiency for the buccal suspension was lower than 50%; for blood, more than 80% of the DNA was not recovered with the standard extraction protocol. Increasing the incubation time from one hour to three hours gave better results, while increasing the time even further did not yield in significantly more DNA or even resulted in DNA degradation. The amount of recovered DNA could also be increased by sequentially washing and separating the extraction buffer from the swab [
31].
Mawlood et al. tested cotton and nylon-flocked swabs with saliva samples deposited on glass. The amount of DNA recovered (in ng/
L) was reported and used to determine the best swab type. Compared to cotton, the nylon-flocked swab gave better results. It must be noted that the shaft of the nylon swab was also made of nylon, while for the two other cotton swab types, the shaft was made of wood, possibly absorbing biological material [
32].
Three different cotton swabs and one nylon-flocked swab were tested by Comte et al. Some of the swabs were prewetted (prewetting one side of the swab head and subsequently sampling again with the dry side), according to the protocol of the manufacturer. Smooth and nonporous (screwdriver handles), a rough and nonporous (steering wheels and screwdriver handles), and porous (collars) substrates were investigated. The ratios of the concentrations of DNA released were calculated, whereby one of the cotton swabs was the challenger swab. Also, DNA profiles were used to determine the performance of the swabs, using the peak height (RFU) to calculate the integrity index to determine the stability/degradation of the DNA. Overall, the nylon-flocked swab performed best, both in terms of DNA recovery and in practicality. The DNA also remained stable with this swabs up to 12 months of preservation at room temperature in an air-conditioned room. However, severe DNA degradation was observed when the nylon-flocked swab was treated with an antimicrobial agent [
15].
Not only the type of swab but also the composition of the extraction buffer influence the recovery efficiency. Gray et al. used cell-free DNA (rainbow trout DNA) and cellular DNA (mouse embryonic fibroblasts) samples that were directly deposited onto the cotton swabs heads (and glass and drinking cans were used as substrate to determine the recovery efficiency). Having BSA, Tween 20, or dNTP mix in the extraction buffer had a positive effect on the efficiency (expressed in % recovery), while sodium chloride had a negative influence, since high salt concentrations can increase the binding ability of DNA to cellulose and can interfere or even completely inhibit the PCR as a result. BSA gave the best results but cannot be used in combination with prot K as it destroys the BSA. Therefore, PVP was used instead of BSA, ending up with the following optimal buffer composition: 1% PVP, 1% Tween 20, and 20 mM Tris HCl (with 20
g/mL prot K to remove proteins that are bound to the DNA preventing access for primers and polymerase) [
33].
Zasada et al. investigated several types of buffers, including PBS, water, commercial lysis buffers, and Tween 20. The DNA recovery efficiency was determined by placing the various swab types in a bacterial suspension. The highest amount of DNA (in ng/
L) was recovered using the nylon-flocked swabs, especially in combination with AL buffer. For the other swab types, other buffers gave the best results, whereas 0.5% Tween 20 was for all swabs the worst-performing buffer solution. Zasada et al. concluded that the absorption capacity of a swab was poorly related to the extraction efficiency [
34].
In a study by Wood et al., the efficiency of cotton and nylon-flocked swabs was investigated on nonporous surfaces (knives with a plastic handle, plastic piping, metal cable, firearm metal, and glass). Mini-tapes were also included in their research in combination with glass, firearm metal, and plastic piping as substrates but showed poor recovery (<17%), which also widely varied. The efficiency of both swab types was best on the plastic knife handles (55%) and poor on metal and glass (<30%). They also concluded that the efficiency depends more on the type of substrate and the experience of the investigator than on the type of swab [
35].
The performance of cotton, nylon-flocked, and rayon swabs was compared by Seiberle et al. in a study in which various parameters, such as type of swab, type of surface, and swabbing solution volume, were tested. The rayon swab was excluded in subsequent experiments, since the overall performance of this swab upon generating a DNA profile was too poor. In addition to various substrates and swabbing techniques (i.e., dry or wet swabbing), the handling and packaging of the swabs was taken into account. The swabs with a cardboard box or tube as a passive drying storage system were found to be not ideal due to the proneness to environmental influences. Investigators had doubts about the contamination risk of a swab with an attached protective cap as a storage system. In the experiment in which the swab type was compared, two cotton and two nylon-flocked swabs were used. All swabs performed proportionally similar for each type of sample in terms of DNA recovery (reported in ng/
L) [
16].
Hedman et al. tested various swab types on several substrates and indicated that the recovery efficiency, which was expressed in ng/
L recovered DNA, not only depends on the combination of swab type and substrate but also on the skills of the investigator. At first the recovery of saliva as a sample in combination with cotton, nylon-flocked and foam swabs were tested on window glass, ridged plastic, and wood. Additional surfaces were added for the best-performing swab of each type. The swabbing technique (including angle, amount of wetting agent, rotation, and pressure, which were all not investigated extensively) was investigated in combination with only cotton swabs on window glass, ridged plastic, and wood in the second part of their study. In the last part, a closer look was taken at the swabbing protocol by comparing swabbing from experience with swabbing from a predefined protocol. Overall, the large foam swab resulted in the highest DNA recovery (but required cutting of the large pad into smaller pieces for further processing), while the cotton swab gave sufficient results for a range of substrates. The most effective sampling technique (i.e., retrieving the most DNA) varied somewhat for each substrate. It is expected that using protocols will result in enhanced DNA recovery efficiencies and less interpersonal variation [
36].
Ip et al. studied the preservation of blood on various swab types with a background of microbes by swabbing a tile prior to pipetting blood directly onto the swab. The appearance of the swabs with bloodstains was observed, and the DNA profiling results were reported. It was concluded that in dry conditions, all swab types gave full DNA profiles, while excessive moisture had a negative influence on DNA preservation. Therefore, the drying method rather than swab type is critical for DNA preservation, as well as the antimicrobial properties of the swab [
37].
Several research groups have attempted to determine the best swab type. However, comparing the results is challenging, sinc the initial conditions are often not known, or different parameters were tested. For instance, some studies only report the amount of retrieved DNA (e.g., in ng or ng/
L) but do not specify the amount of DNA within the sample itself; therefore, no recovery rate can be determined. Others only tested a single sample type and/or substrate type. Other parameters are simply not given in most articles, like swabbing angle, pressure, duration, and amount of wetting agent.
Table 2 is an attempt to compare the various types of swabs by indicating the optimal swab type for a variety of substrate and samples types based on the literature data. This table reveals that there is no universal best type of swab for every combination of substrate and sample type. Note that ‘the best’ swab is often not giving a high recovery rate. While most research focuses on improving recovery rates, literature that discusses extraction efficiency indicates that the challenge lies in extracting the sample from the swab. In conclusion, swabs are effective for sample uptake but are not sufficient when it comes to sample extraction.
4. Alternatives
Since swabs generally exhibit low overall efficiency due to poor extraction (i.e., it can be challenging to remove biological material from a swab), various alternatives are available. Tapes are designed especially for porous surfaces, while hydrogels and vacuum techniques can be used for more complex substrates, such as porous or ridged substrates. Additionally, alternatives such as direct extraction and dissolvable materials are discussed below.
4.1. Tapes
Although a swab can be used, tapes/tape lifting is recommended to collect biological material from porous surfaces, like fabrics. The tape must be placed with the adhesive side onto the substrate, while pressing firmly against the tape’s back. This step must be repeated along the whole target area. A more adhesive tape collects more DNA (and of a higher quality) than a less sticky tape. In general tapes with a stronger adhesion layer give a higher (trace) DNA yield than less adhesive tapes or swabs [
1,
20].
Four substrates and two tapes (with different adhesive strengths) were investigated by Verdon et al. The results of sampling touch DNA were also compared to double swabbing with cotton swabs. Overall, the tape with the strongest adhesion gave the best results, and sampling the area multiple times was beneficial for the recovery yield. For fabrics that have loose fibers, which are easily removed with a tape, swabbing is the preferred sampling method [
38].
Hansson et al. compared the performance of three swabs with mini-tape. This tape is completely soluble in water and shows higher DNA concentrations and full profiles when used on textiles. However, a drawback is that the extract becomes gel-like, making pipetting impossible [
39].
4.2. Hydrogels
A hydrogel is a three-dimensional network of hydrophilic polymer chains that has the ability to absorb and retain a significant amount of water or other aqueous solutions. Therefore, this material has been investigated as an alternative to the conventional swab. Due to the flexible nature of these gels, they are potentially more suitable for sampling complex three-dimensional surfaces.
Hydrogels need to be rehydrated before use, which involves a vortexing step for 2 min and heating at 55 °C for 30 min. Recovering Jurkat cell DNA from polycarbonate surfaces showed comparable results for hydrogels and foam swabs. Moreover, collecting cells with a hydrogel was compatible with the extraction process and subsequent qPCR and STR analysis [
40]. Hydrogels were used by van Helmond et al. for collecting amino acids from fingerprints. Also, DNA could be recovered with the hydrogels, but with only 20–60% of the DNA quantity compared to a cotton swab. Only for unequal surfaces (e.g., gun grips) did a hydrogel seem to be beneficial compared with swabs [
41].
4.3. Direct Extraction and Direct PCR
In the case of paper or textiles as the substrate, direct extraction can be performed without the use of swabs. The substrate is cut or punched, and the small piece of paper or textile is placed directly in a tube for extraction and further processing. However, for paper cuttings, the recovery is often too poor to obtain a DNA profile. Another method of direct extraction is the soaking method, whereby a piece of evidence is placed in a lysis buffer. This method is limited to small pieces of substrates to limit the amount of lysis buffer required. Another drawback is the risk of metal ions and contaminants leaching in the buffer, having a negative influence on the subsequent PCR step.
As an alternative to recovering DNA with a swab, direct PCR is possible, whereby the DNA extraction, purification, and quantification steps are omitted. A drawback of direct extraction and direct PCR is the high chance of obtaining PCR inhibitors in the PCR mixture. Additionally, retesting of the sample is not possible with this technique, although only part of a sample can be used. On the other hand, the risks of contamination and loss of DNA are reduced by removing the extraction and quantification steps.
DNA on ammunition, such as casings, can be recovered by placing the casing directly in a lysis buffer. However, since the casings slowly oxidize (and not only releasing metal ions in the buffer), this may cause partial dissolution of the surface, which is destructive for the striations or filings on casings. To minimize such oxidation-related problems, this step must be reduced to 30 min [
19].
Govindarajan et al. pipetted a range of blood dilutions directly onto rayon swabs to investigate the possibility of direct PCR. Small sections were taken from the swabs by means of a scalpel and placed directly in the PCR tubes. A punch was used to obtain small samples from a swab but led to contamination issues despite an extensive cleaning protocol. The number of alleles recovered was comparable to that of conventional DNA profiling. In the case of a mixture, the authors recommended reanalyzing the sample with conventional DNA profiling [
42]. Sherier et al. investigated a direct PCR method whereby a small nylon-flocked swab was used in combination with blood, semen, and saliva on a cotton cloth. With this small swab, a small area of the stain was swabbed and used as input for direct PCR by placing the complete swab head in a well of a 96-well PCR plate. Compared to stains swabbed with a normal cotton or nylon-flocked swab, the profile completeness and total RFU were improved, indicating the suitability of the small swabs in combination with direct PCR for the prescreening methodology/subsampling of a stain (leaving the rest of the stain for further analysis) [
43].
4.4. Vacuuming Device
There are two types of vacuum techniques used to recover cells and/or cell-free DNA from a surface: dry vacuuming and wet vacuuming.
4.4.1. Wet Vacuuming
Bricks are relatively often used in crimes (e.g., burglaries and riots), but recovering (touch) DNA from this material is challenging due to the porous and rough nature of this substrate. Using swabs might cause fraying of the swab material, and tape may lose adhesiveness. With the M-Vac device (
Figure 8), a wet-vacuum system, large and porous surfaces can be sampled. It operates by applying a sterile buffer onto a surface and, at the same time, extracting the liquid along with any cells or DNA that may be present into a sterile container. The resultant solution is subsequently passed through a vacuum filter, which captures the biological material on the filter. This filter can then undergo standard procedures for DNA profiling. While the collection process with the M-Vac is more intricate and time-intensive compared to the double-swabbing technique, the system itself is relatively uncomplicated and demands only minimal training. It must be noted that cell-free DNA might be lost during filtration, and the chances of obtaining a mixed profile increase. Moreover, swabbing is less expensive than wet vacuuming.
The performance of a system on tiles and bricks in combination with diluted saliva to mimic touch DNA was investigated by Vickar et al. Compared to the double-swabbing method, 75% more DNA was recovered from bricks. However, for tiles, the double-swabbing method showed better results [
44]. McLamb et al. used 22 substrates with various porosities in a study in which the performance of the M-Vac was compared to wet swabbing. Overall, more DNA was recovered using the M-Vac, especially from porous substrates. However, the collection efficiency was similar for both methods, meaning that the recovery from the filter or swab had the strongest influence on the amount of DNA obtained [
45].
4.4.2. Dry Vacuuming
In order to circumvent the drawbacks of a wet-vacuuming system (e.g., large buffer volume and spraying a buffer solution on the surface), a dry-vacuuming system was developed: the DNA-Buster. The DNA-Buster makes use of a pump (with an airflow of 3.75 L/min); within the hose, an inverted filter tip is used for DNA collection, whereby the collected material adheres to the filter itself. The filter is subsequently transferred to a test tube for further analysis. The tube can be used in confined spaces and corners. The DNA-Buster was compared to the M-Vac system, and both systems produced (complex) mixed DNA profiles. Carpet, cotton sweater, stone, tile, and wood were used as the sampling surfaces in combination with touch DNA as a sample, but only for textiles did the DNA-Buster show good performance. For wood and tile, a swab and tape performed better, respectively. For stone, swabbing obtained the highest amount of recovered touch DNA, but the cotton was frayed [
46].
Dry vacuuming can also be performed by putting a (prewetted) swab tip in a glass Pasteur pipette or a plastic pipette tip connected to a vacuum machine (e.g., a M-Vac machine). While having the vacuum on, the pipette with swab can be pulled across the surface to be sampled. Subsequently, the swab can be processed further with conventional techniques. Since the method is nondestructive, other evidence types can also be obtained from the piece of evidence. Morgan et al. concluded that, compared to the Pasteur pipette, the plastic pipette tip had stronger suction, which made it harder to move the tip over their paper substrate, and the DNA recovery of both vacuum methods was low (as well as for cutting/direct extraction and wet–dry and dry-dry swabbing) [
23].
4.5. Dissoluble Materials
Alternative options for sample collection that are nowadays available include swabs made of a material that collects high amounts of cells/DNA and subsequently dissolves in the extraction buffer, thereby theoretically releasing more DNA compared to (cotton) swabs.
The Pinpoint DNA Isolation System from Zymo Research was investigated by Verdon et al. as sampling method for forensic applications. A thin layer of polymer must be applied on a nonporous surface, which forms a film upon drying. By dissolving the dried polymer into the extraction solution, more DNA should be released into the solution. While the method is convenient for uneven and unpredictable surfaces (e.g., gun handles), significantly less DNA was recovered compared to the wet–dry swabbing method. Another drawback is the relatively long drying time of the polymer [
47].
The X-Swab from Diomics Corporation dissolves during incubation at 56 °C for 1 h. The reported results for blood and saliva samples with this swab showed a higher yield and average peak heights after profiling compared to the 4N6FLOQSwab, whereas there was an indication that the dissolved swab material enhanced the PCR yield [
48]. Note that this swab is not commercially available anymore.
5. Discussion
The efficiency of swabs for collecting biological samples is pivotal for DNA profiling. However, collecting samples from a crime scene with swabs is not optimal due to their low extraction efficiency. Although ISO 18385 [
49] is a standard that specifies the requirements for the production of products used in the collection, storage, and analysis of biological material for forensic DNA purposes and the guidelines (
https://enfsi.eu/about-enfsi/structure/working-groups/documents-page/documents/best-practice-manuals/, accessed on 18 December 2023) of the ENSFI (European Network of Forensic Science Institutes) cover various topics related to DNA analysis (e.g., a best practice manual on human forensic biology and DNA profiling), there are still no universal guidelines for a sampling protocol [
50]. Most forensic investigators commonly use cotton or nylon-flocked swabs for all sample and substrate types, as this is common practice, and typically only one type of swab is available.
A range of swab materials have been examined in the literature, ranging from cotton to nylon-flocked, polyester, and foam. Each material type has a unique molecular structure, bringing its strengths and limitations to the forensic toolkit. Furthermore, the type of substrate has a significant influence on the swabbing efficiency. Both cotton and nylon-flocked swabs are suitable for smooth nonporous substrates. However, for ridged surfaces, the issue of fraying becomes more prominent, making cotton swabs less suitable. For porous surfaces, like textiles, foam swabs perform better or an alternative like tape. Due to metal–DNA interactions, it is challenging to recover enough DNA for profiling from metal surfaces. Direct extraction or direct PCR seem good alternatives, although leached metal ions could inhibit subsequent PCR.
Buffer solutions and extraction methods have emerged as crucial factors influencing swab efficiency. The choice of the appropriate buffer solution and extraction protocol significantly affects the quality and quantity of DNA recovered. Fore example, NaCl can be added to enhance the recovery efficiency, but a too-high concentration interferes with PCR. Also, detergents like Triton X-100 or SDS can be part of the buffer, although Triton X-100 is hard to handle due to its high viscosity, while SDS tends to foam. BSA can also improve the recovery efficiency, though it deactivates prot K (a compound usually present in lysis buffers). Regarding the extraction, the before-mentioned spin baskets can be used, which is also reflected in
Table 1.
Table 1 also shows several (commercially available) extraction kits, for which the exact content is often unknown. These different extraction chemistries conceivably influence the overall efficiency.
The recovery efficiency is also affected by the lack of uniformity in swabbing techniques. There is no clear indication of how wet or moist such a swab needs to be, and there is no consensus regarding other parameters, including swabbing angle, swabbing time, and applied pressure on the swab. For touch DNA and/or complex substrates, it is recommended to use double swabbing with a wet and subsequent dry swab, whereas for less complex cases, a single wet or moist–dry swab is the best option. Note that the swab glue, fibers, and shaft can negatively interfere with the subsequent DNA profiling process due to fraying or leaching inhibitory chemicals into the PCR mixture [
6].
While there is no standardized international protocol for the swabbing procedure, by combining the available literature, the following general protocol can be proposed to ensure the collection of as much material as possible [
1,
22]:
Premoisten the swab (nuclease-free water or other wetting agent);
Swab the total target area, while applying medium pressure, at an angle relative to the substrate (to assure that a large area of the swab is in contact with the substrate) and while rotating the swab continually;
These steps can be repeated with another dry (if the substrate is still relatively moist) swab, which is coextracted with the first.
Note that there is no clear indication of, for instance, an exact numerical value for the swabbing angle, how moist the swab must be, or for how long the area must be swabbed.
Not only is the efficiency of importance: other criteria also influence the selection of a swab (or alternative sample collection method). Examples include transport to and from the crime scene, stability and length of the shaft, storage and labeling system, and waste management [
15,
30]. For instance, small swabs do not have enough space for proper labeling. Moreover, having a proper breaking point is beneficial to ensure the controlled removal of the swab head from the shaft, reducing the risk of contamination. This method is also less labor-intensive compared to shaving the swab head from the shaft.
However, in addition to the recovery efficiency of the used swab itself, the sampling skills (including swabbing technique) of the investigator substantially influence the obtained overall efficiency. Surprisingly, the influence of the investigator’s skills is frequently underestimated, with only a limited number of articles addressing this crucial aspect [
16,
35,
36].
There are some swab alternatives available, each with its own set of advantages and disadvantages. Tapes, for instance, are well suited for DNA recovery from porous substrates (e.g., fabrics), but they are less efficient on fabrics with very loose fibers or nonporous surfaces. Applying and pressing the tape onto the substrate is somewhat more labor-intensive and time-consuming compared to swabbing. Hydrogels also offer a solution for more complex surfaces, such as three-dimensional objects (e.g., guns). Their drawbacks are the required rehydration of the gels prior to use, which is both labor-intensive and time-consuming; less DNA is recovered with hydrogels than with cotton swabs. Omitting steps like DNA extraction, purification, and quantification with direct PCR reduces the amount of steps, the risk of contamination, and DNA loss. On the other hand, there is a higher chance of PCR inhibitors being present in the PCR mixture, especially with metal substrates (leaching of metal ions in the PCR mixture). Another alternative is vacuuming: an interesting method for the recovery of DNA from porous surfaces. These vacuum methods are, however, relatively expensive, more labor-intensive and time-consuming and the obtained efficiency is variable, since the recovery of the DNA from the filter is challenging, and mixed profiles are more commonly generated. In theory, dissoluble materials enhance the recovery efficiency; however, in practice, the recovery efficiency is lower compared to swabbing.