1. Introduction
The magnetic bead-based PrepFiler™ Bone, Tooth and Adhesive (BTA) Forensic DNA Extraction Kit (Thermo Fisher Scientific, Waltham, MA, USA), as its name suggests, has been developed for the processing of challenging forensic samples such as bone, tooth and samples containing adhesive materials including tape-lifts, chewing gum and cigarette butts. It is different from the PrepFiler™ Forensic DNA Extraction kit for common forensic samples such as swabs or stains of body fluids, as it utilizes the proprietary BTA lysis buffer that has been formulated for more efficient digestion of complex matrices. The PrepFiler™ BTA Forensic DNA Extraction chemistry has been widely validated in the literature [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
11,
12], demonstrating its ability to efficiently extract DNA from challenging forensic sample types. The PrepFiler
Express™ BTA and the PrepFiler™ BTA
Express Forensic DNA Extraction kits are versions of the PrepFiler™ BTA chemistry to be used with the Automate
Express™ instrument or high throughput liquid handling platforms, respectively, for automated magnetic bead-based DNA purification.
For all PrepFiler™ BTA DNA extractions regardless of kit type, samples are first manually lysed using PrepFiler™ BTA lysis buffer supplemented with Proteinase K and DTT, after which the lysate collected by centrifugation. A 300 µL volume of PrepFiler™ lysis buffer is then added to the spun PrepFiler™ BTA lysate, and the following steps of the DNA purification proceeds identically to the PrepFiler™ Forensic DNA Extraction protocol. In the PrepFiler
Express™ BTA Forensic DNA Extraction kit, centrifuged lysates are loaded onto the AutoMate
Express™ Instrument using the pre-programmed “PF Express BTA” protocol, where the AutoMate
Express™ instrument adds 300 µL of PrepFiler™ lysis buffer to the spun lysate, and continues the DNA purification as per the protocol for PrepFiler
Express™ (“PF Express” protocol) [
13].
The PrepFiler™ BTA Forensic DNA Extraction Kit user guides [
13,
14,
15] state that DNA extraction should proceed directly following lysis. This process, while efficient, does not allow contingency for unexpected delays in processing. Earlier versions of the user guide for the PrepFiler™ BTA Forensic DNA Extraction kit [
16] and the PrepFiler™ Automated Forensic DNA Extraction kits [
14,
16,
17], but not in the current version [
15], state that PrepFiler™ BTA lysates, when topped up with 300 µL of PrepFiler™ lysis buffer (not PrepFiler™ BTA lysis buffer) are stable at ambient temperature for up to 24 h. This allows flexibility if DNA extraction could not proceed directly after the initial lysis.
For operational laboratory workflows, there is benefit in ensuring the stability of sample lysates and the resultant DNA quantity and quality, over time to allow for contingencies that could lead to processing delays. Additionally, the added flexibility of being able to combine PrepFiler™ BTA or PrepFiler™ lysis buffer processed lysates in the same DNA extraction batches to facilitate high throughput applications with the addition of PrepFiler™ lysis buffer to samples lysed with PrepFiler™ BTA lysis buffer is advantageous. Furthermore, there is no information regarding the effect on DNA yield and resultant DNA quality when the PrepFiler™ lysis buffer is added over PrepFiler™ BTA-lysed sample remnants and centrifuged (hereafter referred to as the top-up method) in contrast to adding the PrepFiler™ lysis buffer to the spun lysate (standard method) for different sample types commonly processed with PrepFiler™ BTA. Using the PrepFiler Express™ BTA Forensic DNA Extraction kit and the AutoMate Express™ DNA extraction system, this study evaluated the effects of the PrepFiler™ lysis buffer top-up over PrepFiler™ BTA lysed sample remnants on DNA yield and STR profile quality of different sample types processed by the PrepFiler™ BTA Forensic DNA Extraction kit.
2. Materials and Methods
2.1. Sample Preparation
All biological material used in this study was donated by volunteers with written informed consent. Whole blood from a single male donor was collected in EDTA vacutainers and stored at 4 °C. Fresh semen samples provided by a single donor were aliquoted upon receipt, stored at −20 °C, and thawed when required. A third male donor was supplied with rayon swabs (Copan Diagnostics Incorporated, Murrieta, CA, USA) for self-collection of buccal cells. Buccal cells were eluted by manually agitating excised buccal swabs (collected from the same donor) with sterile forceps, in Tris-EDTA (TE) buffer (10 mM Tris-HCl, 0.1 mM EDTA pH 8.0), followed by centrifugation to pellet cells and removal of excess supernatant to create a concentrated, homogenous buccal cell suspension for the preparation of spiked adhesive tape samples.
A 10 µL aliquot of the stock solutions of whole blood, semen and buccal cells were first extracted and quantified to ascertain the amount of DNA per unit volume of the same sample type and subsequently diluted with TE buffer. The diluted stock solutions of blood, semen and buccal cell suspension (between 10 and 50 μL, depending on sample type) was then added onto the adhesive surface of 25 × 19 mm pieces of adhesive tape (Scotch® Brand Transparent Tape 600, currently used in the laboratory for tape-lifting) to generate experimental replicate samples that yield a target expected DNA concentration of ~0.2–1.0 ng/µL, and allowed to air dry overnight. Ten adhesive tape samples (25 × 19 mm) containing each sample type (blood, semen, buccal cells) were excised and placed in a PrepFiler™ LySep™ column, with five samples each directed for processing with either the standard or top-up BTA method.
Human muscle tissue samples from two male cadaveric donors, A and B, were obtained from the Perth Bone and Tissue Bank. A single tooth donated by five individuals were cryogenically pulverized in a SPEX® 6875D bone mill (SPEX® SamplePrep LLC, Metuchen, NJ, USA) to generate ground tooth powder for DNA extraction. Five volunteers were each provided with one piece of chewing gum and instructed to chew the gum for ~10 min.
Chewed gum, human tissue and ground tooth samples were stored frozen at −20 °C until required. The distribution of DNA over a single piece of tissue, gum, tooth and cigarette butt can be very different over different parts of the same sample. In order to ensure that any observed differences in results is attributed to the method, rather than confounded by the variable nature of the sample type tested, tissue, chewing gum and tooth samples were matched by donor and also by their measured weights (~50 mg) as closely as possible in pairs, with each sample directed for processing either by the standard or top-up method for comparison. Filter paper of smoked cigarette butts of the same brand obtained from a single female donor were cut into halves lengthwise (approximately 15 × 20 mm), with each half directed for PrepFiler™ BTA lysis and processing using either the standard or top-up BTA method.
2.2. Sample Lysis and DNA Extraction
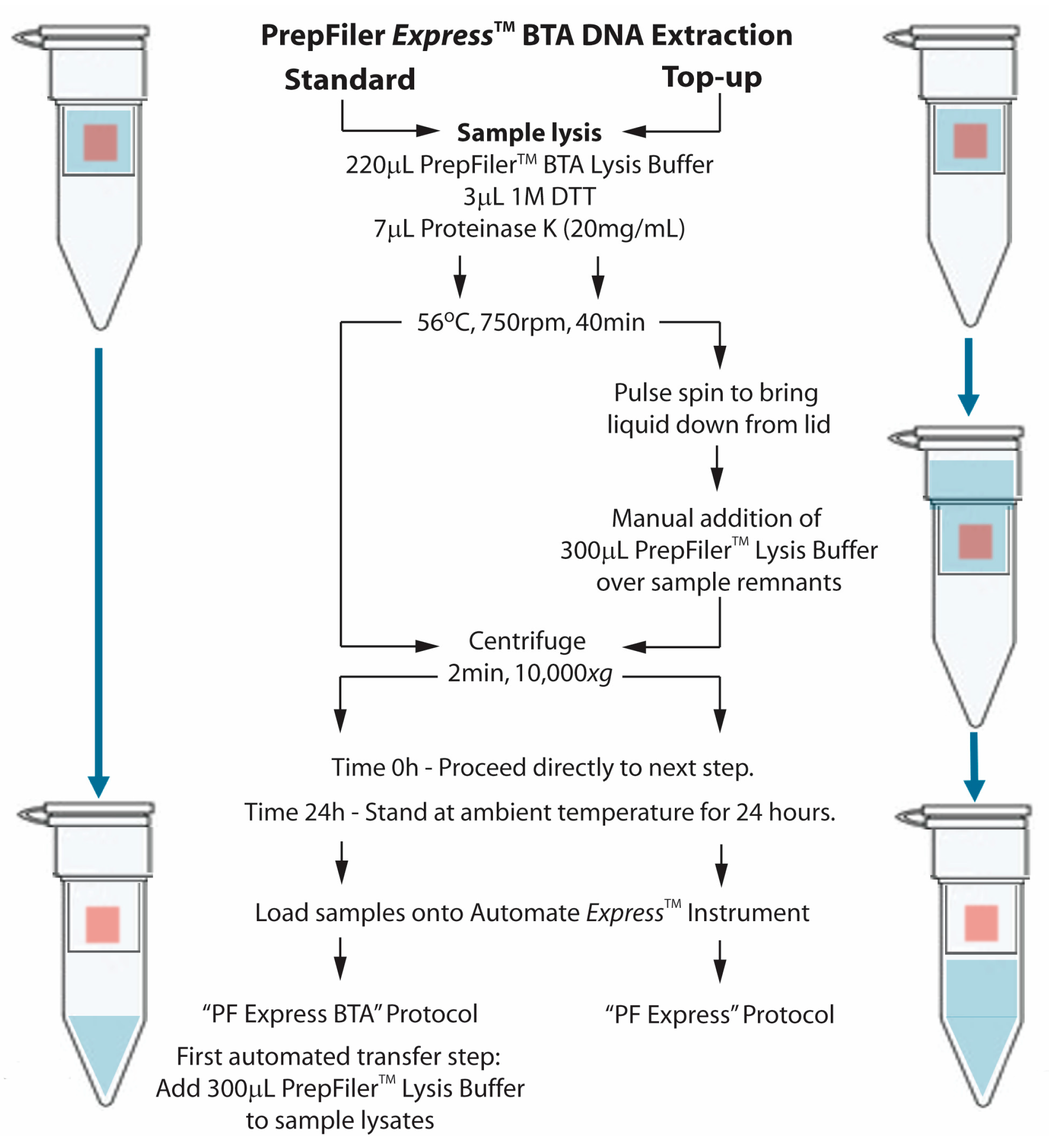
As per the kit user guide, adhesive tape, muscle tissue and cigarette butt samples were incubated in a BTA lysis buffer mix containing 220 µL PrepFiler™ BTA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 7 µL Proteinase K (20 mg/mL) and 3 µL DTT (1M), at 56 °C for 40 min with agitation at 750 rpm. Ground tooth samples were incubated in the same BTA lysis buffer mix at 56 °C for 2 h with agitation at 1100 rpm. Upon completion of the lysis step, samples were processed for DNA extraction with either the standard BTA method (standard), or top-up BTA method (top-up) (
Figure 1).
To determine if the addition of 300 µL of PrepFiler™ lysis buffer (without further supplementation of DTT) over the sample remnants in the top-up method had an effect on DNA recovery and maintained sample lysate stability for 24 h, adhesive tape containing diluted whole blood, semen and buccal cells were processed with the standard and top-up method and DNA extracted on the AutoMate Express™ instrument either immediately (Time 0 h) or after 24 h (Time 24 h) at ambient temperature.
To determine if the top-up method had an impact on DNA recovery and resultant STR profile quality of challenging samples, donor and weight-matched pairs of human muscle tissue, ground tooth powder and halves of cigarette butt paper samples were processed using the standard and top-up methods directly after the lysis step.
For the standard method, the incubated samples were centrifuged for 2 min at 10,000×
g, and loaded on the AutoMate
Express™ instrument (Thermo Fisher Scientific, Waltham, MA, USA) for DNA extraction using the “PF Express BTA” protocol, where the first automated step transfers 300 μL of PrepFiler™ lysis buffer to the spun lysate. For the top-up method, the samples were first centrifuged briefly (to remove any liquid off the lid of the PrepFiler™ LySep™ Column), after which 300 µL PrepFiler™ lysis buffer was added into the LySep™ Column containing the sample remnant and centrifuged for 2 min at 10,000×
g before being processed for DNA extraction on the AutoMate
Express™ instrument using the “PF Express” protocol (
Figure 1). Both the “PF Express BTA” and the “PF Express” protocols generated a 50 μL final elution volume of purified DNA.
2.3. DNA Quantification and STR Profiling
Samples were quantified using the Quantifiler™ Trio DNA Quantification Kit (Thermo Fisher Scientific, Waltham, MA, USA) on the QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) and analyzed using HID real-time PCR analysis software v1.3 (Thermo Fisher Scientific, Waltham, MA, USA). For STR profiling, 0.5 ng DNA, or a maximum DNA volume of 15 µL (if there was <0.5 ng DNA) was amplified using the PowerPlex® 21 System (PP21) (Promega Corporation, Madison, WI, USA) in a 25 µL reaction volume for 30 cycles on the ProFlex™ PCR System (Thermo Fisher Scientific, Waltham, MA, USA). For each sample, 1 µL of amplified PCR product underwent capillary electrophoresis on the 3500xL Genetic Analyser (Thermo Fisher Scientific, Waltham, MA, USA) at injection parameters of 1.2 kV for 24 s. The STR profiles were analyzed using GeneMapper™ ID-X software v1.6 (Thermo Fisher Scientific, Waltham, MA, USA) with the level of detection (LOD) set at 100 RFU and an analytical threshold (AT) of 250 RFU as per internal laboratory validated thresholds. Loci were deemed passing if alleles were above AT, met a homozygote threshold of 1200 RFU or a heterozygote peak height ratio of 50%.
Graphing of results and statistical analyses by Student’s t-test were performed using GraphPad Prism software v9.2.0 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. Evaluation of PrepFiler™ Lysis Buffer Top-Up on DNA Quality of Blood, Semen and Buccal Cell Dried on Adhesive Tape Processed Directly or 24 h after PrepFiler™ BTA Cell Lysis
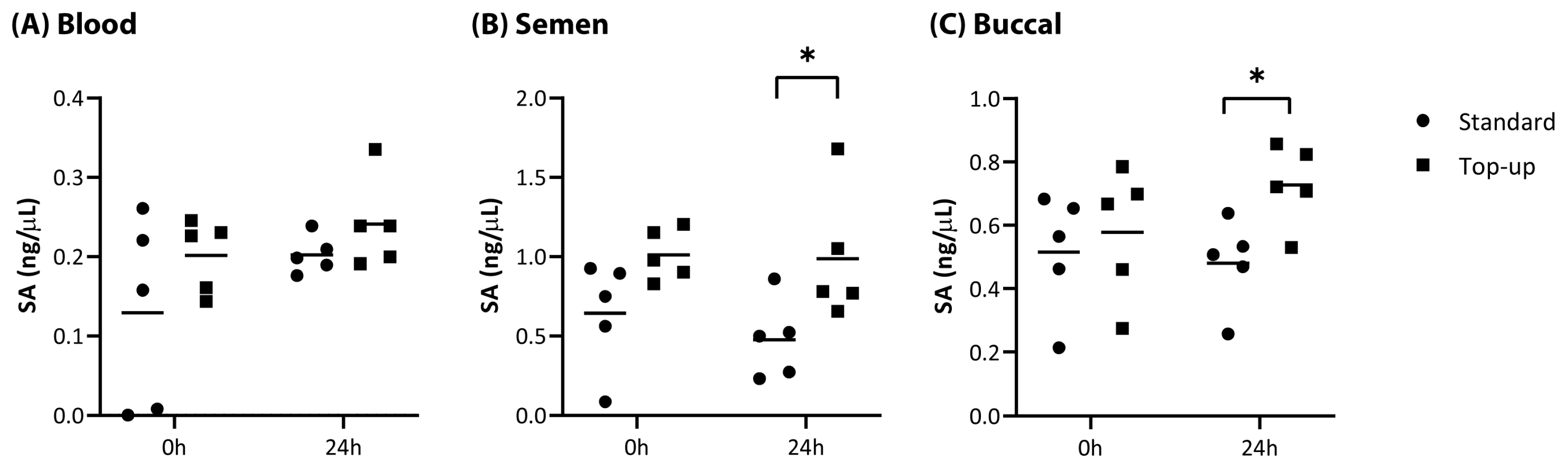
Adhesive tape (containing blood, semen or buccal cells) processed with the standard BTA method, generated comparable quantification results when extracted at Time 0 h and Time 24 h (
Table 1). The Small Autosomal (SA) DNA concentration values obtained were not significantly different between the two time points tested for blood, semen and buccal samples using the standard method (Student’s
t-test,
p > 0.5). Comparison of the degradation index of samples processed at Time 0 h and Time 24 h also showed no significant difference, demonstrating that the lysates remained stable at ambient temperature up to 24 h without the immediate addition of PrepFiler™ lysis buffer following the cell lysis step. Although variable between replicates of the same sample type (
Appendix A,
Figure A1), the top-up method returned higher average DNA quantitation values compared to the standard method for all adhesive tape samples at both time points tested (
Table 1). The improvement in DNA concentration observed with the top-up method was statistically significant for semen and buccal samples at the 24 h timepoint (Student’s
t-test;
p < 0.05), and most marked for diluted semen, where average DNA quantitation values were greater by 57% and 107% at Time 0 h and Time 24 h, respectively.
3.2. Evaluation of PrepFiler™ Lysis Buffer Top-Up on DNA Quantity and Quality for Matched Chewing Gum, Tissue, Ground Tooth and Cigarette Butt Samples
Chewing gum, tissue, ground tooth material and cigarette butt are sample types which are highly variable and often exhibit uneven DNA distribution across the sample. As such, the results were compared for each sample pair which have been matched by donor and weight to evaluate the effect of the top-up method. Overall, for gum, tissue and tooth samples, the top-up method returned higher SA DNA concentration values and marginally higher degradation index than the standard method. The PP21 profiles were assessed qualitatively (number of passing loci, peak height ratio, ski slope effect, etc.), were comparable between the standard and top-up method, demonstrating that there is no detrimental effect of performing the top-up method for these sample types (
Table 2). This is still advantageous as it allows for batching of these samples with those processed with PrepFiler™ lysis buffer for high throughput extraction workflows.
Analysis of the SA DNA concentration and PP21 profile quality of cigarette samples, however, demonstrated that cigarette butts do not benefit from having the additional PrepFiler™ lysis buffer added and centrifuged over the remnants using the top-up method. Cigarette butt samples 1, 2 and 3 processed with the top-up method returned DNA samples with markedly higher degradation index values compared to the standard method, despite relatively comparable SA DNA concentration (
Table 2). More DNA was recovered from the half of cigarette butt sample 4 processed with the top-up method, however, the degradation index was much greater than that obtained using the standard method. The degradation and poorer DNA quality obtained from the cigarette butt samples processed by the top-up method was reflected in the PP21 profiles, where the samples exhibited a stronger ski-slope pattern, leading to fewer passing loci compared to the standard method. All PP21 DNA profiles obtained were concordant with the expected donor reference profile.
3.3. Evaluation of Modified PrepFiler™ Lysis Buffer Top-Up on DNA Quantity and Quality for Cigarette Butt Sample Processing
A further ten smoked cigarette butts of the same brand smoked by the same donor were cut into thirds lengthways (approximately 10 × 20 mm) and each third processed using the standard, top-up or a modified top-up method which involved addition and centrifugation of the PrepFiler™ lysis buffer through the Lysep™ column after the lysed cigarette butt remnants are removed. A comparison of the SA DNA concentration and degradation index showed that cigarette butt samples processed by the modified top-up method was comparable to the standard method, and not significantly different (Student’s
t-test,
p > 0.05) (
Table 3). Notably, DNA quantitation values of cigarette butt samples processed using the top-up method were significantly lower compared to samples obtained using the standard and modified top-up methods (Student’s
t-test,
p < 0.001). The degradation index was also markedly higher in the top-up samples, indicating that poorer quality DNA was generated, which was reflected in the resultant PP21 STR profiles obtained (
Table 3). All PP21 DNA profiles obtained were concordant with the expected donor reference profile. The results showed that the additional 300 μL of PrepFiler™ lysis buffer must be added after the cigarette butt remnants are removed to maintain resultant DNA quality.
4. Discussion
In this study, the primary difference between the PrepFiler™ BTA standard and top-up methods is the timing of the addition of PrepFiler™ lysis buffer either centrifuged over the sample remnant, or added directly to the centrifuged lysate, immediately after the heated lysis step. While the supplementation of PrepFiler™ lysis buffer serves to ensure optimal salt concentration and pH for effective DNA binding to the magnetic beads, the physical addition of PrepFiler™ lysis buffer and centrifugation appeared to act as an additional ‘rinse’ over the already lysed substrate remnants, facilitating the release and collection of more DNA for downstream DNA purification on the AutoMate Express™ instrument. The degradation index and quality of PP21 profiles generated were comparable between all tape samples processed by the standard and top-up methods, with single loci peak height imbalances noted where a full 21/21 profile was not obtained, demonstrating that the DNA released in the lysates were not subject to detectable DNA degradation over the 24 h before DNA was purified.
All adhesive tape sample lysates were separated from the sample remnants by centrifugation after the 40 min lysis step. The median pore size of the Prepfiler™ LySep™ column is 7–12 microns, which will filter out unlysed epithelial cells and white blood cells, which are ~25 and 15 microns in size, respectively. However, unlysed sperm heads (~4 microns) are small enough to be released from the adhesive tape and be spun through with the lysate. As such, at the 24 h timepoint, any unlysed sperm heads would be incubating in lysis buffer at ambient temperature until the lysate is presented to the Automate Express™ instrument for DNA extraction. This may contribute in part, to the greater increase in DNA quantitation values obtained for the semen samples compared to that observed for blood and buccal cell samples. The results for semen samples (
Table 1) shows that the SA DNA concentrations for semen samples were not significantly different at 0 h and 24 h using the standard method, which suggests that the top-up method was the major contributor to the improvement in DNA recovery.
Furthermore, previous work in the developmental validation of the i-sep
® differential separation method showed no quantifiable sperm cell DNA following incubation at 56 °C for 2 h in a Tris-EDTA lysis buffer supplemented with Proteinase K for epithelial cell lysis [
18]. Additionally, Hennekens et al. (2013) [
19] also reported no indication of sperm cell lysis in the absence of the reducing agent, DTT, when comparing various differential lysis protocols. In this study, DTT was only added to the initial PrepFiler™ BTA lysis buffer and not added to the PrepFiler™ lysis buffer used for the top-up. This demonstrates that the higher DNA quantitation values obtained for semen on adhesive tape processed with the top-up method, is not attributed to additional DTT, but likely due to a greater release of DNA with the addition and centrifugation of PrepFiler™ lysis buffer over the sample remnant, into the spun PrepFiler™ BTA lysate. It also demonstrates that amount of DTT present in the lysis step was sufficient to lyse cells including spermatozoa in semen. These results indicate that under circumstances where semen may be present on a tape-lift, the top-up should be performed over the sample remnants to ensure maximal DNA recovery when using PrepFiler™ BTA.
While the results obtained for adhesive tape samples demonstrate that BTA lysates are stable for 24 h without the addition of PrepFiler™ lysis buffer, it should be noted that the biological material used in this study was not subjected to degradative treatments and therefore, expected to produce good quality purified DNA. PCR inhibition was not observed in the samples tested in this study. However, forensic casework samples are inherently unpredictable in the quality and quantity of recoverable DNA, which often also contain PCR inhibitors. While the addition of PrepFiler™ lysis buffer to PrepFiler™ BTA lysates ensures optimal buffer conditions for DNA binding to the magnetic beads, it may also prevent further damage and degradation of samples of substandard DNA quality, post-lysis. The results obtained with good quality DNA do not substantiate the stability of degraded or substandard DNA samples, nor is it indicative that no further deterioration occurs over time, without the addition of PrepFiler™ lysis buffer. Environmental damage and the natural aging of samples is difficult to mimic and replicate in the laboratory to generate experimental samples and therefore, was not evaluated in this study. As such, further testing with degraded or inhibited samples is required. It is therefore recommended to that the top-up method is performed in circumstances where the sample cannot proceed directly to DNA extraction.
It is well-documented in the literature that cigarettes contain PCR inhibitors from the chemical additives and tobacco plant material [
20,
21,
22,
23,
24]. It is likely that the addition of PrepFiler™ lysis buffer over the cigarette butt remnants resulted in the release of more inhibitory substances from the smoked cigarette butt paper. Interestingly, the lysates generated using the top-up method also appeared cloudy, but distinct from the appearance of lysis buffer salt precipitation, suggesting carryover of substances that could not be removed by the magnetic bead-based purification process. The magnetic bead-based DNA purification method employed by the PrepFiler™ Forensic DNA Extraction kit has been demonstrated to be effective in the removal of PCR inhibitors from different biological samples [
2,
3,
5,
6]. However, that the results here demonstrate that the added PrepFiler™ lysis buffer over the cigarette butt remnants had a negative effect, with significant downstream effects which could not be overcome by the PrepFiler™ DNA purification protocol.
Using the modified top-up method, cigarette butt samples could be re-integrated into the workflow by removing the substrate remnant prior to adding PrepFiler™ lysis buffer, facilitating the batching of samples to maintain workflow efficiency.
5. Conclusions
There are currently no published data evaluating PrepFiler™ BTA sample lysate stability over time, and the effects of centrifuging additional PrepFiler™ lysis buffer over the sample remnant or adding it directly to the centrifuged lysate, on DNA quantity and quality. This study demonstrated that DNA recovery from adhesive tape containing diluted whole blood, semen and buccal cells, was improved by the addition and centrifugation of 300 μL PrepFiler™ lysis buffer over the substrate remnant. The improvement in DNA recovery is greatest for adhesive tape samples that contain dried semen, which suggests that the success rates of DNA recovery from tape-lifts of vaginal gauzes or clothing if semen is present can be enhanced with the top-up method. Overall, the results obtained in this study indicate that released DNA in BTA lysates for up to 24 h, with or without the addition of PrepFiler™ lysis buffer after the heated lysis step, however the latter is not recommended for potentially degraded and poor-quality samples.
The top-up method was suitable for the processing of other challenging sample types such as chewing gum, teeth and tissue, where resultant PP21 STR profiles were comparable to those obtained using the standard method. The addition of PrepFiler™ lysis buffer to the lysed sample remnants is not suitable for the processing of cigarette butt sample as it resulted markedly poorer DNA yields and STR profile quality. The removal of the cigarette butt paper remnant prior to the addition of PrepFiler™ lysis buffer demonstrated comparable results to the standard method, standardises the sample lysate required for processing on the AutoMate Express™ instrument using the “PF Express” protocol, allowing for the batching of samples lysed with PrepFiler™ BTA and PrepFiler™ lysis buffer. This also allows for all samples processed with either PrepFiler™ BTA and the PrepFiler™ chemistry to be processed concurrently on high-throughput liquid handling robotic platforms. The results obtained in this study demonstrate that BTA lysates are stable for up to 24 h prior to extraction, allowing for unexpected delays, and with the top-up method facilitating the batching of samples for high throughout processing, and also standardizing workflows, thereby increasing efficiency, turn-around times, and maximizing the quality and quantity of DNA results obtained for challenging samples.
Author Contributions
Conceptualization, C.J.C., C.M.H. and J.W.T.; methodology, C.J.C., C.M.H., K.Q., S.J.B. and J.W.T.; formal analysis, C.J.C., C.M.H., K.Q., S.J.B. and J.W.T.; writing—original draft preparation, C.J.C., C.M.H. and J.W.T.; writing—review and editing, C.J.C., C.M.H., K.Q., S.J.B. and J.W.T.; supervision, J.W.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This study was exempt from review by the Sir Charles Gairdner Osborne Park Health Care Group (SCGOPHCG) Human Research Ethics Committee (HREC). This activity has been assessed as meeting the requirements of the National Statement on Ethical Conduct in Human Research and is endorsed.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
Figure A1.
Comparison of SA DNA concentration values obtained from from PrepFiler™ BTA lysis buffer lysed samples (A) blood, (B) semen and (C) buccal cells obtained on adhesive tape at 0 h and 24 h processed with standard or top-up methods. * Student’s t-test; p < 0.05.
Figure A1.
Comparison of SA DNA concentration values obtained from from PrepFiler™ BTA lysis buffer lysed samples (A) blood, (B) semen and (C) buccal cells obtained on adhesive tape at 0 h and 24 h processed with standard or top-up methods. * Student’s t-test; p < 0.05.
References
- Almeida, M.; Betancor, E.; Fregel, R.; Suárez, N.; Pestano, J. Efficient DNA extraction from hair shafts. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e319–e320. [Google Scholar] [CrossRef]
- Balsa, F.; Bogas, V.; Cunha, P.; Brito, P.; Serra, A.; Lopes, V.; Carvalho, M.; Andrade, L.; Bento, A.; Bento, M.S.; et al. Preliminary validation of Prepfiler Express™ Extraction kit in AutoMate Express DNA Extraction System. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e377–e378. [Google Scholar] [CrossRef]
- Barbaro, A.; Cormaci, P.; Agostino, A. Validation of PrepFiler™ forensic DNA extraction kit (Applied Biosystems). Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 176–177. [Google Scholar] [CrossRef]
- Barbaro, A.; Cormaci, P.; Falcone, G. Validation of BTA™ lysis buffer for DNA extraction from challenged forensic samples. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e61–e62. [Google Scholar] [CrossRef]
- Brevnov, M.; Mundt, J.; Benfield, J.; Treat-Clemons, L.; Kalusche, G.; Meredith, J.; Porter, G.; Furtado, M.R.; Shewale, J.G. Automated Extraction of DNA from Forensic Sample Types Using the PrepFiler Automated Forensic DNA Extraction Kit. JALA J. Assoc. Lab. Autom. 2009, 14, 294–302. [Google Scholar] [CrossRef]
- Brevnov, M.G.; Pawar, H.S.; Mundt, J.; Calandro, L.M.; Furtado, M.R.; Shewale, J.G. Developmental validation of the PrepFiler Forensic DNA Extraction Kit for extraction of genomic DNA from biological samples. J. Forensic Sci. 2009, 54, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Iyavoo, S.; Hadi, S.; Goodwin, W. Evaluation of five DNA extraction systems for recovery of DNA from bone. Forensic Sci. Int. Genet. Suppl. Ser. 2013, 4, e174–e175. [Google Scholar] [CrossRef]
- Liu, J.Y.; Zhong, C.; Holt, A.; Lagacé, R.; Harrold, M.; Dixon, A.B.; Brevnov, M.G.; Shewale, J.G.; Hennessy, L.K. AutoMate Express™ Forensic DNA Extraction System for the Extraction of Genomic DNA from Biological Samples. J. Forensic Sci. 2012, 57, 1022–1030. [Google Scholar] [CrossRef] [PubMed]
- Stray, J.; Holt, H.; Brevnov, M.G.; Calandro, L.M.; Furtado, M.R.; Shewale, J.G. Extraction of high quality DNA from biological materials and calcified tissues. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 159–160. [Google Scholar] [CrossRef]
- Joël, J.; Glanzmann, B.; Germann, U.; Cossu, C. DNA extraction of forensic adhesive tapes—A comparison of two different methods. Forensic Sci. Int. Genet. Suppl. Ser. 2015, 5, e579–e581. [Google Scholar] [CrossRef]
- Rubenstein, L. Comparing two automated methods of DNA extraction from degraded skeletal remains. Forensic Med. Anat. Res. 2021, 9, 24–30. [Google Scholar] [CrossRef]
- Stoop, B.; Defaux, P.M.; Utz, S.; Zieger, M. Touch DNA sampling with SceneSafe Fast™ minitapes. Leg. Med. 2017, 29, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Applied Biosystems. PrepFiler Express™ and PrepFiler Express™ BTA Forensic DNA Extraction Kits User Guide. Publication Part Number 4442699 Revision D. Thermofisher Scientific. 2017. Available online: https://tools.thermofisher.com/content/sfs/manuals/cms_081933.pdf (accessed on 29 June 2018).
- Applied Biosystems. PrepFiler™ and PrepFiler™ BTA Forensic DNA Extraction Kits Quick Reference. Publication Part Number 4468126 Revision B. Thermofisher Scientific. 2012. Available online: https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_099065.pdf (accessed on 29 June 2018).
- Applied Biosystems. PrepFiler™ and PrepFiler™ BTA Automated Forensic DNA Extraction Kits User Guide. Publication Number 4463349 Revision E. Thermofisher Scientific. 2021. Available online: https://www.thermofisher.com/order/catalog/product/4463349 (accessed on 29 June 2021).
- Applied Biosystems. PrepFiler™ and PrepFiler™ BTA Forensic DNA Extraction Kits. Publication Number 4463348 Revision C. Thermofisher Scientific. 2012. Available online: https://www.thermofisher.com/order/catalog/product/4463348 (accessed on 28 July 2013).
- Applied Biosystems. PrepFiler™ Automated Forensic DNA Extraction Kit. Publication Number 4463349 Revision B. Thermofisher Scientific. 2011. Available online: https://www.thermofisher.com/order/catalog/product/4463353 (accessed on 28 November 2011).
- White, T.J.; Rye, M.S.; Tay, J.W. Developmental validation of an efficient differential separation method incorporating the i-sep® DL spin column with high sperm DNA recovery for the processing of sexual assault samples. J. Forensic Sci. 2022, 67, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Hennekens, C.M.; Cooper, E.S.; Cotton, R.W.; Grgicak, C.M. The effects of differential extraction conditions on the premature lysis of spermatozoa. J. Forensic Sci. 2013, 58, 744–752. [Google Scholar] [CrossRef]
- Hedman, J.; Albinsson, L.; Noren, L.; Ansell, R. Evaluation of three new forensic DNA profiling kits on PCR-inhibitory crime scene samples. Forensic Sci. Int. Genet. Suppl. Ser. 2011, 3, e457–e458. [Google Scholar] [CrossRef]
- Hedman, J.; Nordgaard, A.; Dufva, C.; Rasmusson, B.; Ansell, R.; Radstrom, P. Synergy between DNA polymerases increases polymerase chain reaction inhibitor tolerance in forensic DNA analysis. Anal. Biochem. 2010, 405, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Takayama, T.; Hirata, K.; Yamada, S.; Nagai, A.; Nakamura, I.; Bunai, Y.; Ohya, I. DNA typing from cigarette butts. Leg. Med. 2003, 5, S177–S179. [Google Scholar] [CrossRef]
- Sidstedt, M.; Radstrom, P.; Hedman, J. PCR inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal. Bioanal. Chem. 2020, 412, 2009–2023. [Google Scholar] [CrossRef] [PubMed]
- Sidstedt, M.; Jansson, L.; Nilsson, E.; Noppa, L.; Forsman, M.; Rådström, P.; Hedman, J. Humic substances cause fluorescence inhibition in real-time polymerase chain reaction. Anal. Biochem. 2015, 487, 30–37. [Google Scholar] [CrossRef] [PubMed]
| Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).