Abstract
Bones and teeth are highly challenging sources of DNA in forensic science and human remains identification, requiring multiple laborious processing steps. In this study, we compared an organic phenol–chloroform method to the QIAamp® DNA Investigator and PrepFiler Express BTA™ methods in order to identify the most efficient automated DNA extraction method for bones and teeth. Results from individual tooth powder replicates showed that the PrepFiler Express BTA™ method extracted the highest yields of DNA per mg of tooth powder, returning a minimum of 20/21 PowerPlex® 21 loci. Samples extracted using the organic extraction or QIAamp® DNA Investigator methods produced PowerPlex® 21 profiles displaying a ski-slope morphology. The improved DNA quality and yield from the PrepFiler Express BTA™ method was verified using aged samples, where higher DNA yields per mg of powder and more informative profiles were obtained. Furthermore, the PrepFiler Express BTA™ method subsequently provided useful DNA profiles for two forensic cases involving degraded bone samples. Overall, this study showed that the PrepFiler Express BTA™ chemistry is a reliable and robust method for DNA extraction from bone and teeth samples, and will allow larger numbers of samples to be efficiently extracted in the event of a Disaster Victim Identification event.
1. Introduction
Bone and teeth samples are some of the most laborious and problematic sources of DNA in forensic casework investigations; however, these sources of DNA are often all that remains and can be reliable sources of DNA in scenarios with decomposed bodies, such as mass disasters and unidentified remains [,,]. The success of DNA extraction from bone and teeth samples is also influenced by environmental factors, i.e., type and intensity of environmental exposure, such as fire [], and length of time exposed to the weather prior to collection. Bone samples, in particular vary widely in presentation on receipt, from intact solid and dense bones to bones that are heavily decomposed due to environmental exposure, resulting in bones that are brittle, soft or flexible, or have a spongy, sludge-like texture.
Traditionally, bone and teeth samples are cleaned of debris, processed into smaller fragments, and cryogenically pulverised using liquid nitrogen and bone mills prior to DNA extraction. While these steps are time-consuming, attempts to bypass the pulverisation process through extracting DNA from bone fragments have generally resulted in lower DNA quantity [,]. Optimisation of the DNA extraction process provides the most potential to improve DNA profile success rates for bone and teeth. Once pulverised, several laboratories use a standard phenol–chloroform DNA extraction protocol [,,,], which utilises hazardous chemicals. A number of studies have compared DNA extraction methods and techniques such as full demineralisation in an attempt to improve DNA profiles, with varying rates of success [,,,,,]. Demineralisation with EDTA is used to remove calcium and mineral ions from bone and teeth samples, which are known PCR inhibitors [].
A range of automated DNA extraction methods have been designed specifically for extraction from bone and tooth samples, which can lower the potential for contamination and increase throughput as well as reduce exposure to hazardous chemicals. This study undertook an evaluation of a selection of DNA extraction chemistries and methods available for processing bones and teeth. A comparison was performed between the traditional phenol–chloroform method and the automated QIAamp® DNA Investigator and PrepFiler Express BTATM methods. This article presents the first comparison of five variations of DNA extraction methods, with an end goal of identifying an improved, automated DNA extraction method that does not require hazardous chemicals or time-consuming incubation steps to extract DNA from challenging sample sources.
2. Materials and Methods
2.1. Comparison of DNA Extraction Methods from Tooth Samples
Ten good quality aged baby tooth samples that fell out naturally and were provided with informed parental consent were cleaned to remove debris or tissue from the tooth and manually processed into small pieces. A reference buccal swab was provided by each donor, which was processed using routine laboratory procedures to provide a reference profile for comparison. A 6750 Freezer/Mill® (SPEX® SamplePrep, Metuchen, NJ, USA) was used to pulverise tooth samples immersed in liquid nitrogen into a fine powder. Vials containing ~200 mg of each powder were extracted using an organic phenol–chloroform method. Briefly, 500 µL of Extraction Buffer made from 20 µL of DTT (1 M) + 480 µL of Digest buffer (10 mM Tris-HCL pH 7.5, 10 mM EDTA. 50 mM NaCl, 2% SDS) and 10 µL of Proteinase K (10 mg/mL) was added to each sample and incubated at 56 °C overnight. After incubation, 500 µL of a phenol–chloroform–isoamyalcohol solution was added, mixed well and centrifuged for 2 min at 13,000 RPM to separate the phases. The aqueous (upper) phase containing DNA was then purified as per manufacturer’s instructions for the QIAquick® PCR Purification Kit protocol [], performed manually on a QIAvac 24 Plus vacuum manifold (QIAGEN, Hilden, Germany).
The QIAamp® DNA Investigator (QIAGEN, Hilden, Germany) automated protocol for bones and teeth was performed as per manufacturer’s instructions [], which included manual processing of 100 mg of the ground tooth powder to generate a supernatant. Briefly, 360 µL of Buffer ATL and 20 µL of Proteinase K (20 mg/mL) was added to each sample and incubated at 56 °C overnight. After incubation, 300 µL of Buffer AL was added and incubated at 70 °C for 10 min. The supernatant was then transferred to a new set of tubes and DNA was subsequently extracted on the QIAcube (QIAGEN, Hilden, Germany) using an automated protocol created by the QIAGEN Application Lab.
DNA from aliquots of 50 mg tooth powder was extracted using three variations of the PrepFiler Express BTATM (Thermo Fisher Scientific, Waltham, MA, USA) method, with automated DNA extraction performed by an AutoMateTM Express instrument (Thermo Fisher Scientific, Waltham, MA, USA) []. Two of the methods followed the manufacturer’s recommended lysis input volumes for PrepFiler Express BTATM extraction of bone and tooth samples, one with incubation at 56 °C for 2 h at 1100 RPM (standard method; PrepFiler-STD) and one with incubation at 56 °C for 18 h at 750 RPM (extended method; PrepFiler-EXT) []. The third method was adapted from Zupanic Pajnic et al. (2015) []. Briefly, 1 mL of 0.5 M EDTA was added to 50 mg of ground tooth and incubated at 37 °C overnight (~18 h), then the supernatant was discarded and the tooth pellet was washed with sterile water. After this stage, the supernatant was discarded again, and DNA was extracted from the remaining tooth powder as per the PrepFiler-STD method (full demineralisation method; PrepFiler-FD).
2.2. Compatibility of DNA Extraction Methods Using Aged Teeth and Bone Samples
Eight ground tooth samples and a ground bone sample that had been stored for an extended period of time were extracted identically using the phenol–chloroform, standard PrepFiler Express BTATM (PrepFiler-STD) and the full demineralisation PrepFiler Express BTATM (PrepFiler-FD) methods described above. All tooth samples had been in storage as ground powder for 10–11 years prior to the study, while the bone sample was from a femur that had been buried for up to nine years, then ground to a powder and stored for eight years prior to this study.
2.3. DNA Quantification and Profiling
All DNA samples were quantified using the QuantifilerTM Trio DNA Quantification Kit (Thermo Fisher Scientific, Waltham, MA, USA) on an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) as per manufacturer instructions []. Samples were amplified on a ProFlex PCR System (Thermo Fisher Scientific, Waltham, MA, USA) using the PowerPlex® 21 System (Promega, Madison, WI, USA) according to manufacturer instructions [], with an input of 0.5 ng or maximum DNA volume of 15 µL for low quantity samples. Capillary electrophoresis was performed on an Applied Biosystems 3500xL Genetic Analyser (Thermo Fisher Scientific, Waltham, MA, USA), injected at 1.2 kV for 24 s using a 36 cm capillary and POP4 polymer. Data was analysed using GeneMapper ID-X software v1.5 using an analytical threshold for all dyes of 250 RFU, a homozygote threshold of 1200 RFU and a minimum locus balance ratio of ≥50% (Thermo Fisher Scientific, Waltham, MA, USA).
3. Results
3.1. Comparison of DNA Extraction Methods from Tooth Samples
The results of the comparison of the five tested methods are shown in Figure 1 and Supplementary Figure S1. The phenol–chloroform method extracted DNA at the second lowest concentration per mg of tooth. These samples produced DNA profiles that had a ski-slope morphology where large molecular weight loci often dropped below the PowerPlex® 21 threshold. The DNA Investigator method gave the lowest concentration for nine out of ten tooth samples, and also produced PowerPlex® 21 profiles with a ski slope morphology and drop-out of large molecular weight loci. Samples extracted using the PrepFiler-STD and PrepFiler-EXT methods returned a relatively high quantity of DNA per mg of tooth powder used and all three PrepFiler Express BTATM methods produced a full, balanced PowerPlex® 21 profile, with only one sample producing 20/21 loci (PrepFiler-EXT T9; Figure 1).
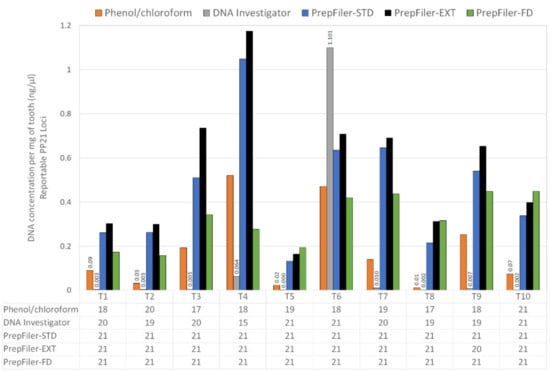
Figure 1.
Quantification and amplification results for ten tooth samples extracted using five different methods. The quantification results are shown as ng/µL per mg of tooth powder extracted, with quantification values below 0.1 ng/µL shown above the respective sample. Numbers represent how many balanced PowerPlex® 21 loci were detected (maximum of 21).
Due to both the low DNA quantity and sub-optimal DNA profiling results, such as ski-slope profiles and lower numbers of reportable loci, the DNA Investigator method was removed from further evaluation. While the phenol–chloroform method also performed poorly, it was retained for comparison as a long-standing method used for DNA extraction from bone and teeth. In addition, the comparison between PrepFiler-STD and PrepFiler-EXT showed there was no value in increasing the incubation time to 18 h. While this method resulted in a slightly higher DNA concentration compared to the PrepFiler-STD method, the DNA profiles were not improved, with Tooth 9 (T9) producing only 20/21 loci. As such, to improve routine workflow processes, only the shorter incubation time in PrepFiler-STD was included for further evaluation.
3.2. Compatibility of DNA Extraction Methods Using Aged Teeth and Bone Samples
A comparison of the phenol–chloroform method to the PrepFiler-STD and PrepFiler-FD methods on eight ground tooth powders and one ground bone powder stored frozen at −20 °C for at least 10 years is shown in Table 1. The PrepFiler-FD method was selected on the basis of being designed for aged bone and tooth samples. The DNA yield per mg of tooth powder used was higher with the PrepFiler-STD and PrepFiler-FD methods compared to the phenol–chloroform method. The degradation index, calculated comparing the short and large quantification values was variable between methods, with the lowest value generally observed for samples extracted with the PrepFiler-FD method.

Table 1.
Quantification and amplification results for DNA extracted from nine aged tooth and bone powders stored for ≥10 years extracted using the phenol–chloroform method and two variations of the PrepFiler Express BTATM method. The number of balanced PowerPlex® 21 loci that were detected are shown.
The DNA profile results showed that the PrepFiler-STD method consistently resulted in a higher number of balanced PowerPlex® 21 loci being detected compared to the phenol–chloroform method (Table 1). There was less variation between the PrepFiler-STD and PrepFiler-FD methods, with the standard method producing a slightly higher average number of detected and balanced PowerPlex® 21 loci.
3.3. Case Study of Forensic Bone Exhibits
The success of the newly implemented PrepFiler Express BTATM methodology was demonstrated in two forensic cases involving bone samples processed using either the PF-STD or PF-FD method (Table 2; Figure 2). The first case involved the remains of a heavily decomposed hand with no useable fingerprint ridges that was found in the Western Australian bushland. The age of this sample and the time of exposure to the elements was unknown. Fragments of tissue were extracted using the PrepFiler Express BTATM method as per manufacturer protocol [], but returned a low DNA quantification value of 0.0007 ng/µL and minimal PowerPlex® 21 loci, which were all below reportable analysis thresholds used in the laboratory (Figure 2). In contrast, processing 50 mg of the bone fragments using the PF-FD method obtained a DNA concentration of 0.076 ng/µL and 17/21 reportable PowerPlex® 21 loci. While DNA extraction from bone is more challenging than tissue, the full demineralisation method extracted DNA of sufficient quality to generate a useable profile for this case.

Table 2.
Quantification and PowerPlex® 21 profile results for two forensic cases with bone fragments.
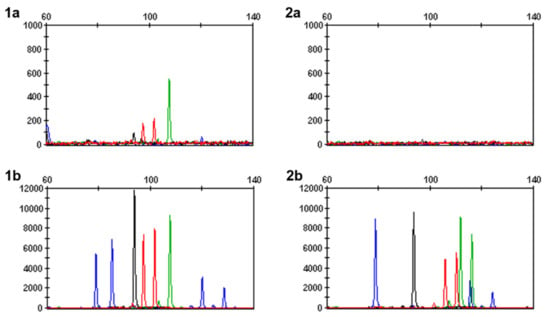
Figure 2.
Snapshot of PowerPlex® 21 loci for two forensic cases involving bone fragments. Case 1 extracted DNA from tissue (1a) with minimal loci, all below reportable analytical thresholds, while DNA extracted from bone using the PF-FD method (1b) amplified 17 reportable loci. In case 2, bone fragments extracted with PF-STD (2a) did not amplify any loci, compared to bone extracted with PF-FD (2b) which produced 16 reportable loci. Note: samples (1a,2a) have been scaled to 1000 RFU, while samples (2a,2b) are scaled to 12,000 RFU.
The second case involved a vertebral bone sample from a body in an advanced state of decomposition that was fully exposed to the Western Australian environment for seven months. The sample was initially processed using the PF-STD method with 50 mg bone powder; however, despite a DNA concentration of 0.012 ng/µL, no PowerPlex® 21 loci were amplified (Figure 2). In contrast, extraction of 50 mg bone powder using the PF-FD method produced 16 reportable PowerPlex® 21 loci. Between these methods, there was a decrease in degradation index from 6.87 with the PF-STD method to 2.24 using PF-FD. This followed the trend observed with the aged tooth and bone samples where the PF-FD method frequently produced DNA extracts with lower degradation indices compared to PF-STD.
4. Discussion
In this study, a review of several methods available for extraction of DNA from difficult sample substrates revealed that the PrepFiler Express BTATM methods produced the most optimal results. This was observed both in relation to the DNA quantification results and the quality of DNA profiles obtained using PowerPlex® 21. This outcome was expected, as the PrepFiler Express BTATM Forensic DNA Extraction kit has been specifically designed for lysis of bone, teeth and adhesive substrates [].
In the method comparison, the highest concentration was observed using the phenol–chloroform method; however, this method used 200 mg of powder compared to 50 mg using the PrepFiler Express BTATM method. When amount of tooth powder used was considered, the yield per mg of tooth powder used was higher with the three PrepFiler Express BTATM methods tested. This aligns with findings from other studies that small amounts of tooth or bone powder are more easily decalcified by EDTA [,]. Smaller amounts of powder may also decrease the amount of inhibitors purified with the sample, which increases the chance of producing a more informative profile. In addition, the poor performance of the QIAamp® DNA Investigator method for DNA extraction from bones and teeth was also observed in studies performed by Kus et al. (2016) [] and Harrel et al. (2019) []. Overall, the standard PrepFiler Express BTATM method gave the most optimal DNA yields, using an incubation time of 2 h, with the option to extend this incubation time to a maximum of 18 h. This is in agreement with the results presented by Hasap et al. (2019) [] and Zgonjanin et al. (2017) [], who obtained similar or better results for bone and tooth samples extracted using the PrepFilerTM BTA Forensic DNA extraction kit compared to the phenol–chloroform–isopropanol method. Analysis of PowerPlex® 21 profiles also showed the standard method of the PrepFiler Express BTATM chemistry consistently produced a higher number of loci, meeting threshold requirements compared to the phenol–chloroform method. Interestingly, for tooth samples extracted using the full demineralisation version of the PrepFiler Express BTATM method, the degradation index was frequently the lowest. The full demineralisation method sequesters the divalent ions, removing impurities such as calcium ions that can otherwise impact qPCR, which may explain the lower degradation index seen with this method in comparison to the standard PrepFiler Express BTATM method. There was less variation in DNA profiles between the standard and full demineralisation PrepFiler Express BTATM methods, with the standard method producing a slightly higher average number of PowerPlex® 21 loci meeting threshold requirements. Taken together, the results from this study suggest the standard PrepFiler Express BTATM method is suitable for processing tooth samples; however, the case studies show that the full demineralisation method may assist in cases where samples such as bone are extensively degraded as a second option for DNA extraction from challenging and aged samples.
One limitation in this study was the low number of bone samples available for comparison. Teeth are frequently available from baby teeth or dental extractions and, as such, teeth were more readily available for testing. In addition, there was limited potential for replication from each sample as the majority of powder was consumed by comparison across the five methods. While there was only a single bone sample available for processing using the full demineralisation method, there were two subsequent case studies that demonstrated the effectiveness of the method. For both case studies, the use of the full demineralisation method produced an informative PowerPlex® 21 profile for identification purposes in cases where either simpler sample sources or the regular bone extraction method failed to yield a useable DNA profile. The study by Harrel et al. (2019) [] also investigated increasing PCR cycles as a method for improving profile results from bone and tooth samples with limited DNA. This is a valid avenue for future studies to assess whether the number of reportable loci can be improved by adjustments to PCR and electrophoresis parameters.
5. Conclusions
The PrepFiler Express BTATM chemistry produced the most optimal results overall for DNA extraction from tooth and bone samples. The BTA lysis buffer contained within the PrepFiler Express BTATM method has been shown in this study to be more efficient for DNA lysis from challenging sources of DNA, and the option to use a full demineralisation variation of the method proved especially useful in two case studies involving degraded bone remains.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/forensicsci1030017/s1, Figure S1: Electropherograms of representative PowerPlex 21 profiles generated from a tooth sample.
Author Contributions
Conceptualization, J.Z., J.W.T., N.S.M. and M.S.R.; Formal analysis, J.Z., J.W.T. and M.S.R.; Methodology, J.Z., J.W.T., N.S.M. and M.S.R.; Supervision, J.W.T. and M.S.R.; Validation, N.S.M.; Writing–original draft, M.S.R.; Writing–review & editing, J.Z., J.W.T. and N.S.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
This case report was exempt from review by the Sir Charles Gairdner Osborne Park Health Care Group (SCGOPHCG) Human Research Ethics Committee (HREC). The institution has established a non-HREC ethical review process for case studies and case series, described in the SCGOPHCG HREC Standard Operating Procedures (2021), to ensure case reporting is undertaken appropriately and in keeping with the requirements of the National Statement on Ethical Conduct in Human Research as well as the expectations of the health service and the public. This activity has been assessed as meeting these requirements and is endorsed.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
The authors would like to acknowledge Colby Hymus for his assistance with the case study samples, and Tammara White for her review of the internal verification report.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Boer, H.H.; Roberts, J.; Delabarde, T.; Mundorff, A.Z.; Blau, S. Disaster victim identification operations with fragmented, burnt, or commingled remains: Experience-based recommendations. Forensic Sci. Res. 2020, 5, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Latham, K.E.; Miller, J.J. DNA recovery and analysis from skeletal material in modern forensic contexts. Forensic Sci. Res. 2019, 4, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prinz, M.; Carracedo, A.; Mayr, W.R.; Morling, N.; Parsons, T.J.; Sajantila, A.; Scheithauer, R.; Schmitter, H.; Schneider, P.M. DNA Commission of the International Society for Forensic Genetics (ISFG): Recommendations regarding the role of forensic genetics for disaster victim identification (DVI). Forensic Sci. Int. Genet. 2007, 1, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Emery, M.V.; Bolhofner, K.; Winingear, S.; Oldt, R.; Montes, M.; Kanthaswamy, S.; Buikstra, J.E.; Fulginiti, L.C.; Stone, A.C. Reconstructing full and partial STR profiles from severely burned human remains using comparative ancient and forensic DNA extraction techniques. Forensic Sci. Int. Genet. 2020, 46, 102272. [Google Scholar] [CrossRef] [PubMed]
- Grdina, S.; Friš, E.L.; Podovšovnik, E.; Zupanc, T.; Zupanič Pajnič, I. Storage of Second World War bone samples: Bone fragments versus bone powder. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 175–176. [Google Scholar] [CrossRef]
- Harrel, M.; Hughes-Stamm, S. A Powder-free DNA Extraction Workflow for Skeletal Samples. J. Forensic Sci. 2020, 65, 601–609. [Google Scholar] [CrossRef]
- Adler, C.J.; Haak, W.; Donlon, D.; Cooper, A.; The Genographic Consortium. Survival and recovery of DNA from ancient teeth and bones. J. Archaeol. Sci. 2011, 38, 956–964. [Google Scholar] [CrossRef]
- Jakubowska, J.; Maciejewska, A.; Pawłowski, R. Comparison of three methods of DNA extraction from human bones with different degrees of degradation. Int. J. Leg. Med. 2012, 126, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsuwan, J.; Somboonchokepisal, T.; Akaraputtiporn, T.; Srimuang, T.; Phuengsukdaeng, P.; Suwannarat, A.; Mutirangura, A.; Kitkumthorn, N. A method for extracting DNA from hard tissues for use in forensic identification. Biomed. Rep. 2018, 9, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Silva, R.C.F.; Ekert, M.H.F.; Mazanek, M.L.; Miranda, C.S.; Santos, A.L.N.; dos Santos, A.R.; do Monte, S.M.C.; de Castro, S.G.; de Souza, A.C.; Ramalho-Neto, C.E. Alternative methodology for extraction of high-quality DNA from ancient bones by demineralization without pulverization. Forensic Sci. Criminol. 2018, 3, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Harrel, M.; Gangitano, D.; Hughes-Stamm, S. The effects of extra PCR cycles when amplifying skeletal samples with the GlobalFiler((R)) PCR Amplification Kit. Int. J. Leg. Med. 2019, 133, 745–750. [Google Scholar] [CrossRef]
- Zupanic Pajnic, I.; Debska, M.; Gornjak Pogorelc, B.; Vodopivec Mohorcic, K.; Balazic, J.; Zupanc, T.; Stefanic, B.; Gersak, K. Highly efficient automated extraction of DNA from old and contemporary skeletal remains. J. Forensic Leg. Med. 2016, 37, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Kus, M.; Ossowski, A.; Zielinska, G. Comparison of three different DNA extraction methods from a highly degraded biological material. J. Forensic Leg. Med. 2016, 40, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Duijs, F.E.; Sijen, T. A rapid and efficient method for DNA extraction from bone powder. Forensic Sci. Int. Rep. 2020, 2, 100099. [Google Scholar] [CrossRef]
- Opel, K.L.; Chung, D.; McCord, B.R. A Study of PCR Inhibition Mechanisms Using Real Time PCR. J. Forensic Sci. 2010, 55, 25–33. [Google Scholar] [CrossRef] [PubMed]
- QIAGEN. QIAquick® Spin Handbook. Publication No: HB-1196-004. Revised: November 2018. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=95f10677-aa29-453d-a222-0e19f01ebe17 (accessed on 20 September 2021).
- QIAGEN. QIAamp® DNA Investigator Handbook. Publication No: 1072908. Revised: June 2012. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=26ef8f2c-7c2a-49e6-b2d2-39d4e130b3cc&lang=en (accessed on 20 September 2021).
- Applied Biosystems. PrepFiler® Express and PrepFiler® Express BTA Forensic DNA Extraction Kits–User Guide. Publication No: 4442699. Revised: March 2012. Available online: https://tools.thermofisher.com/content/sfs/manuals/cms_081933.pdf (accessed on 20 September 2021).
- Holt, A.; Wootton, S.C.; Mulero, J.J.; Brzoska, P.M.; Langit, E.; Green, R.L. Developmental validation of the Quantifiler® HP and Trio Kits for human DNA quantification in forensic samples. Forensic Sci. Int. Genet. 2016, 21, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Promega Corporation. PowerPlex® 21 System for Ussse on the Applied Biosystems® Genetic Analyzers–Technical Manual. Publication No: TMD034. Revised: April 2017. Available online: https://www.promega.de/resources/protocols/technical-manuals/101/powerplex-21-system-for-use-on-the-applied-biosystems-genetic-analyzers-protocol/ (accessed on 20 September 2021).
- Hasap, L.; Chotigeat, W.; Pradutkanchana, J.; Asawutmangkul, W.; Kitpipit, T.; Thanakiatkrai, P. Comparison of two DNA extraction methods: PrepFiler® BTA and modified PCI-silica based for DNA analysis from bone. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 669–670. [Google Scholar] [CrossRef]
- Zgonjanin, D.; Antov, M.; Alghafri, R.; Petković, S.; Vuković, R.; Stojiljković, G.; Toljić, D. DNA analysis from human skeletal remains in forensic casework. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e342–e345. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).