More Knowledge, Fewer Species: New Insights into the Systematics of Lygodactylus heeneni de Witte 1933 (Gekkota: Gekkonidae) of Central Africa
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Methods
2.3. Morphological Characters


2.4. Geographic Distribution
3. Results
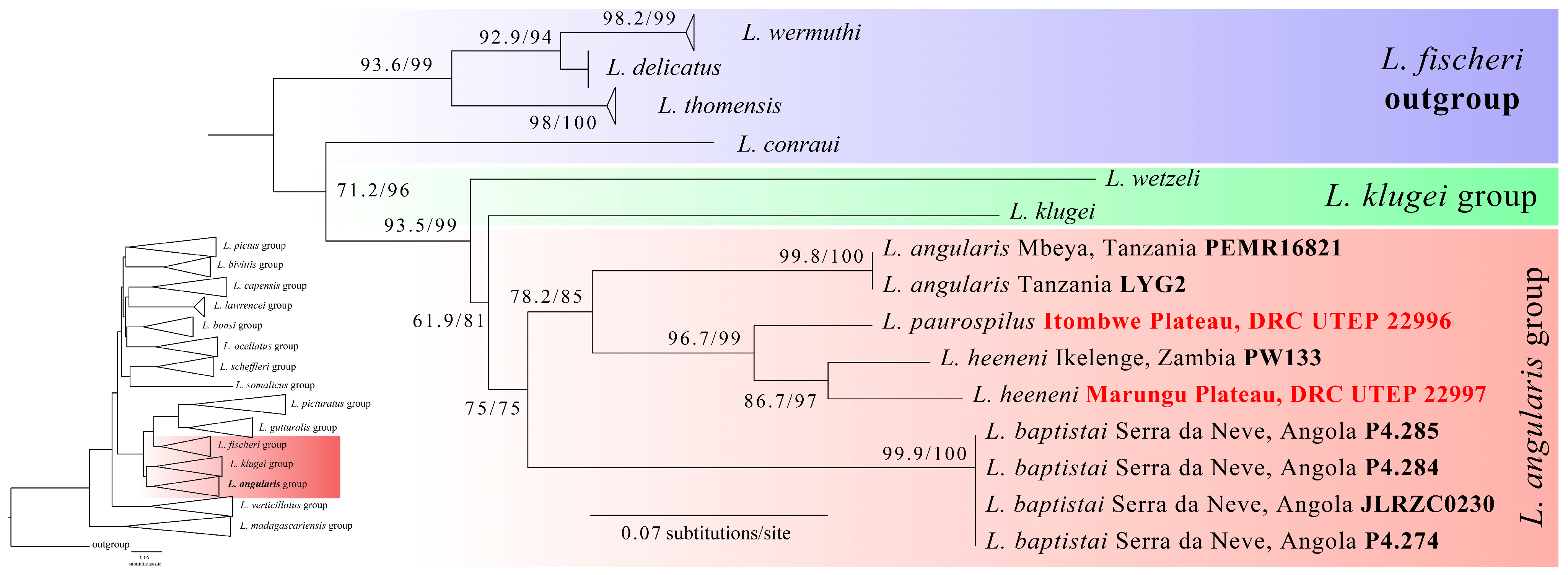
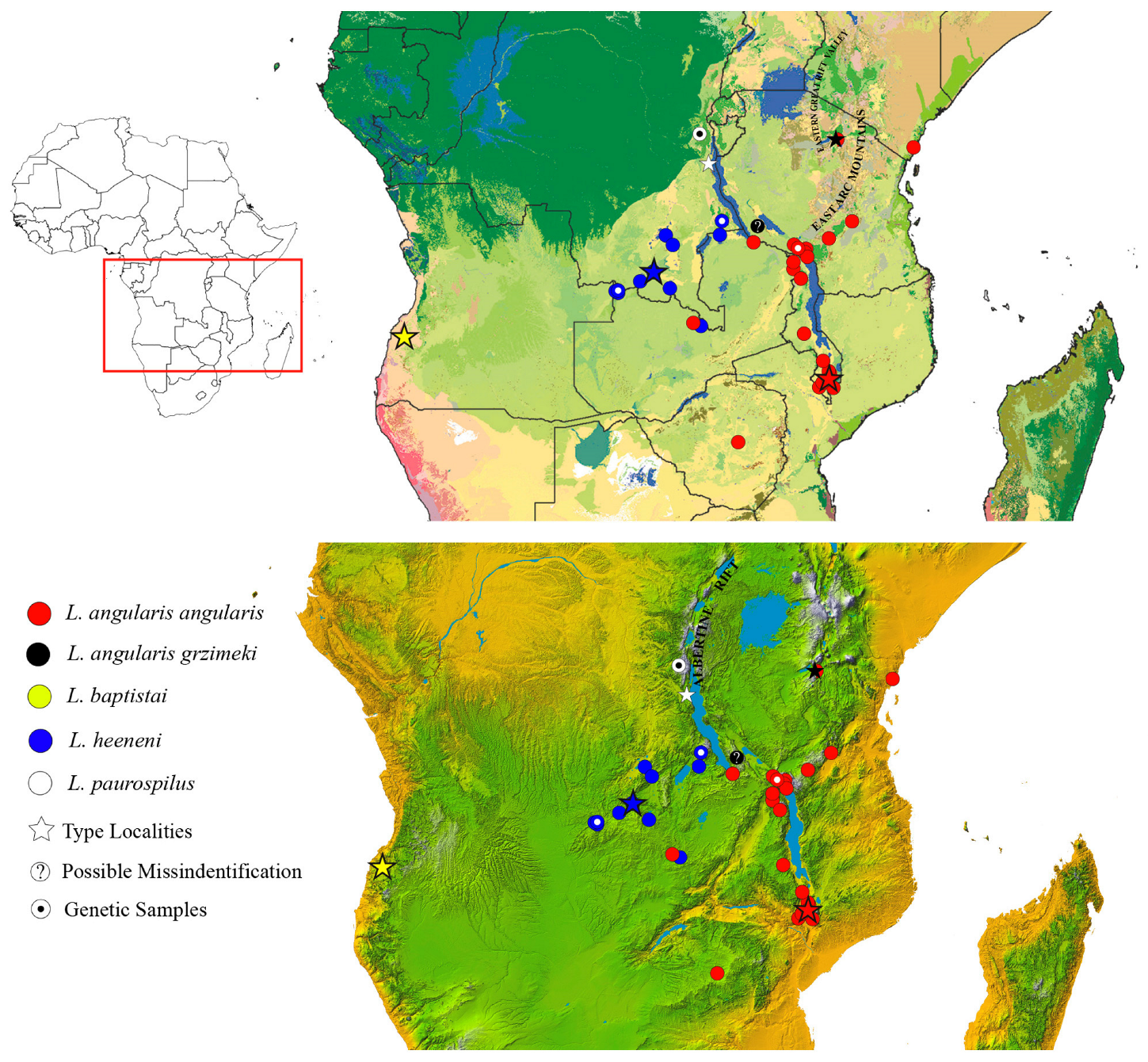
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gippner, S.; Travers, S.L.; Scherz, M.D.; Colston, T.J.; Lyra, M.L.; Mohan, A.V.; Multzsch, M.; Nielsen, S.V.; Rancilhac, L.; Glaw, F.; et al. Comprehensive phylogeny of dwarf geckos of the genus Lygodactylus, with insights into their systematics and morphological variation. Mol. Phyl. Evol. 2021, 165, 107311. [Google Scholar] [CrossRef] [PubMed]
- Lobón-Rovira, J.; Marugán-Lobón, J.; Nebreda, S.M.; Buckley, D.; Stanley, E.L.; Köhnk, S.; Glaw, F.; Conradie, W.; Bauer, A.M. Adaptive or non-adaptive? Cranial evolution in a radiation of miniaturized day geckos. BMC Ecol. Evol. 2024, 24, 150. [Google Scholar] [CrossRef]
- Malonza, P.K.; Granthon, C.; Williams, D.A. A new species of dwarf gecko in the genus Lygodactylus (Squamata: Gekkonidae) from central Kenya. Zootaxa 2016, 4061, 418–428. [Google Scholar] [CrossRef]
- Rakotoarison, A.; Gippner, S.; Multzsch, M.; Miralles, A.; Razafimanafo, D.; Hasiniaina, A.; Glaw, F.; Vences, M. Molecular systematics of the enigmatic dwarf gecko Lygodactylus ornatus and description of a new species of the L. verticillatus group from the North West of Madagascar. Zootaxa 2025, 5621, 420–436. [Google Scholar] [CrossRef] [PubMed]
- Röll, B.; Sanchez, M.; Gippner, S.; Bauer, A.M.; Travers, S.L.; Glaw, F.; Hawlitschek, O.; Vences, M. Phylogeny of dwarf geckos of the genus Lygodactylus (Gekkonidae) in the Western Indian Ocean. Zootaxa 2023, 5311, 232–250. [Google Scholar] [CrossRef]
- Travers, S.L.; Jackman, T.R.; Bauer, A.M. A molecular phylogeny of Afromontane dwarf geckos (Lygodactylus) reveals a single radiation and increased species diversity in a South African montane center of endemism. Mol. Phylogenet. Evol. 2014, 80, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Vences, M.; Multzsch, M.; Gippner, S.; Miralles, A.; Crottini, A.; Gehring, P.S.; Rakotoarison, A.; Ratsoavina, F.M.; Glaw, F.; Scherz, M.D. Integrative revision of the Lygodactylus madagascariensis group reveals an unexpected diversity of little brown geckos in Madagascar’s rainforest. Zootaxa 2022, 5179, 1–61. [Google Scholar] [CrossRef]
- Vences, M.; Multzsch, M.; Zerbe, M.; Gippner, S.; Andreone, F.; Crottini, A.; Glaw, F.; Köhler, J.; Rakotomanga, S.; Rasamison, S.; et al. Taxonomizing a truly morphologically cryptic complex of dwarf geckos from Madagascar: Molecular evidence for new species-level lineages within the Lygodactylus tolampyae complex. Zootaxa 2024, 5468, 416–448. [Google Scholar] [CrossRef]
- Vences, M.; Herrmann, C.; Multzsch, M.; Gippner, S.; Razafimanafo, D.; Rahagalala, N.A.; Rakotomanga, S.; Rakotoarison, A.; Glaw, F.; Miralles, A. Revision of the Lygodactylus tolampyae complex, with descriptions of three additional new dwarf gecko species from Madagascar’s North West. Zootaxa 2025, 5665, 301–328. [Google Scholar] [CrossRef]
- Lanna, F.M.; Werneck, F.P.; Gehara, M.; Fonseca, E.M.; Colli, G.R.; Sites, J.W., Jr.; Rodrigues, M.T.; Garda, A.A. The evolutionary history of Lygodactylus lizards in the South American open diagonal. Mol. Phylogenet. Evol. 2018, 127, 638–645. [Google Scholar] [CrossRef]
- Lobón-Rovira, J.; Bauer, A.M.; Vaz Pinto, P.; Trape, J.F.; Conradie, W.; Kusamba, C.; Julio, T.; Cael, G.; Stanley, E.L.; Hughes, D.F.; et al. Integrative revision of the Lygodactylus gutturalis (Bocage, 1873) complex unveils extensive cryptic diversity and traces its evolutionary history. Zool. J. Linn. Soc. 2024, 201, 447–492. [Google Scholar] [CrossRef]
- Greenbaum, E. Venomous River: Changing Climate, Imperiled Forests, and a Scientist’s Race to Find New Species in the Congo; High Road Books (Imprint of The University of New Mexico Press): Albuquerque, NM, USA, 2025; xvii + 298p. [Google Scholar]
- Röll, B.; Vaz Pinto, P.; Lobón-Rovira, J. A new species of African diurnal dwarf geckos (Gekkonidae: Lygodactylus) from the Lower Guinea rainforest. Zootaxa 2024, 5538, 561–574. [Google Scholar] [CrossRef]
- Röll, B.; Pröhl, H.; Hoffmann, K.P. Multigene phylogenetic analysis of Lygodactylus dwarf geckos (Squamata: Gekkonidae). Mol. Phylogenet. Evol. 2010, 56, 327–335. [Google Scholar] [CrossRef]
- Castiglia, R.; Annesi, F. The phylogenetic position of Lygodactylus angularis and the utility of using the 16S rDNA gene for delimiting species in Lygodactylus (Squamata, Gekkonidae). Acta Herpetol. 2022, 6, 35–45. [Google Scholar]
- Portik, D.M.; Travers, S.L.; Bauer, A.M.; Branch, W.R. A new species of Lygodactylus (Squamata: Gekkonidae) endemic to Mount Namuli, an isolated ‘sky island’ of northern Mozambique. Zootaxa 2013, 3710, 415–435. [Google Scholar] [CrossRef]
- Marques, M.P.; Ceríaco, L.M.; Buehler, M.D.; Bandeira, S.A.; Janota, J.M.; Bauer, A.M. A revision of the Dwarf Geckos, genus Lygodactylus (Squamata: Gekkonidae), from Angola, with the description of three new species. Zootaxa 2020, 4853, 301–352. [Google Scholar] [CrossRef] [PubMed]
- Laurent, R.F. Reptiles et batraciens nouveaux du massif du Mont Kabobo et du plateau des Marungu. Rev. Zool. Bot. Afr. 1952, 46, 18–34. [Google Scholar]
- Pasteur, G. Recherches sur l’évolution des lygodactyles, lézards Afro-Malgaches actuels. Trav. Inst. Sci. Chérifien Sér. Zool. 1965, 29, 1–132. [Google Scholar]
- de Witte, G.F. Reptiles. In Exploration du Parc National de l’Upemba, Mission G. F. de Witte en Collaboration avec W. Adam, A. Janssens, L. Van Meel et R. Verheyen (1946–1949); De Witte, G.F., Ed.; Institut des Parcs Nationaux du Congo Belge: Bruxelles, Belgium, 1953; Volume 6, 322p, pls. XLI. [Google Scholar]
- Kluge, A.G. Checklist of gekkonid lizards. Smithson. Herpetol. Inf. Serv. Ser. 1991, 85, 1–35. [Google Scholar] [CrossRef]
- Kluge, A.G. Gekkonoid Lizard Taxonomy; International Gecko Society: San Diego, CA, USA, 1993. [Google Scholar]
- Kluge, A.G. Gekkotan lizard taxonomy. Hamadryad 2001, 26, 1–209. [Google Scholar]
- Timberlake, J. Miombo Ecoregion Vision Report 2001 (rev. August 2011); Biodiversity Foundation for Africa: Bulawayo, Zimbabwe, 2011. [Google Scholar]
- Hirschfeld, M.; Blackburn, D.C.; Burger, M.; Greenbaum, E.; Zassi-Boulou, A.G.; Rödel, M.O. Two new species of long-fingered frogs of the genus Cardioglossa (Anura: Arthroleptidae) from Central African rainforests. Afr. J. Herpetol. 2015, 64, 81–102. [Google Scholar] [CrossRef]
- Broadley, D.G. Pachydactylus katanganus de Witte 1953, a species endemic to the Upemba National Park (Sauria: Gekkonidae). Afr. J. Herpetol. 2003, 52, 69–70. [Google Scholar] [CrossRef]
- Owiunji, I.; Plumptre, A.J. The importance of cloud forest sites in the conservation of endemic and threatened species of the Albertine Rift. In Tropical Montane Cloud Forests: Science for Conservation and Management; Bruijnzeel, L.A., Scatena, F.N., Hamilton, L.S., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 164–171. [Google Scholar]
- Greenbaum, E.; Kusamba, C. Conservation implications following the rediscovery of four frog species from the Itombwe Natural Reserve, eastern Democratic Republic of the Congo. Herpetol. Rev. 2012, 43, 253–259. [Google Scholar]
- Bruford, M.W.; Hanotte, O.; Brookfield, J.F.Y.; Burke, T. Single-locus and multilocus DNA fingerprint. In Molecular Genetic Analysis of Populations: A Practical Approach; Hoelzel, A.R., Ed.; Irl Press: Oxford, UK, 1992; pp. 225–270. [Google Scholar]
- Palumbi, S.R. The polymerase chain reaction. In Molecular Systematics, 2nd ed.; Hillis, D., Moritz, C., Mable, B., Eds.; Sinauer Associates: Sunderland, UK, 1996; pp. 205–247. [Google Scholar]
- Pramuk, J.B.; Robertson, T.; Sites, J.W., Jr.; Noonan, B.P. Around the world in 10 million years: Biogeography of the nearly cosmopolitan true toads (Anura: Bufonidae). Glob. Ecol. Biogeogr. 2008, 17, 72–83. [Google Scholar] [CrossRef]
- Wong, T.K.F.; Ly-Trong, N.; Ren, H.; Banos, H.; Roger, A.J.; Susko, E.; Bielow, C.; De Maio, N.; Goldman, N.; Hahn, M.W.; et al. IQ-TREE 3: Phylogenomic Inference Software Using Complex Evolutionary Models. 2025. Available online: https://ecoevorxiv.org/repository/view/8916/ (accessed on 15 January 2025).
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2025, 35, 518–522. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- VertNet. Available online: https://www.vertnet.org (accessed on 2 October 2025).
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.nhe7cq (accessed on 17 October 2025).
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation. 2021. Available online: http://qgis.org (accessed on 15 January 2025).
- Sayre, R.G.; Comer, P.; Hak, J.; Josse, C.; Bow, J. A New Map of Standardized Terrestrial Ecosystems of Africa; Association of American Geographers: Washington, DC, USA, 2013; 22p. [Google Scholar]
- Pasteur, G. Notes préliminaires sur les lygodactyles (gekkonidés). II. Diagnose de quelques Lygodactylus d’Afrique. Bull. Inst. Franç. Afrique Noire 1962, 24, 606–614. [Google Scholar]
- Jesus, J.; Brehm, A.; Harris, D.J. Phylogenetic relationships of Lygodactylus geckos from the Gulf of Guinea islands: Rapid rates of mitochondrial DNA sequence evolution? Herpetol. J. 2006, 16, 291–295. [Google Scholar]
- Broadley, D.G.; Cotterill, F.P.D. The reptiles of southeast Katanga, an overlooked ‘hot spot’. Afr. J. Herpetol. 2004, 53, 35–61. [Google Scholar] [CrossRef]
- Bocage, J.V.B. Mammiferos, aves e reptis da Hanha, no sertào de Benguella. Jorn. Sci. Math. Phys. Nat. 1896, 14, 105–114. [Google Scholar]
- Rebelo, A.D.; Bates, M.F.; Burger, M.; Branch, W.R.; Conradie, W. Range expansion of the Common Dwarf Gecko, Lygodactylus capensis: South Africa’s most successful reptile invader. Herpetol. Notes 2019, 12, 643–650. [Google Scholar]
- Huntley, B.J.; Ferrand, N. Biodiversity of Angola; Science & Conservation: A Modern Synthesis; Springer: Cham, Switzerland, 2019; 549p. [Google Scholar]
- Branch, W.R.; Baptista, N.L.; Vaz Pinto, P.; Conradie, W. The reptiles of Angola—History, diversity, endemism and hotspots. In Biodiversity of Angola; Huntley, B.J., Ferrand, N., Eds.; Science & Conservation: A Modern Synthesis; Springer: Cham, Switzerland, 2019; pp. 283–326. [Google Scholar]


| Groups | Species | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|---|
| L. angularis group | 1. L. baptistai | 0.0 | ||||||||
| 2. L. angularis | 13.56 | 0.7 | ||||||||
| 3. L. paurospilus Itombwe | 13.44 | 12.62 | – | |||||||
| 4. L. heeneni Ikelenge | 13.21 | 11.79 | 6.37 | – | ||||||
| 5. L. heeneni Pepa | 12.50 | 11.32 | 4.95 | 4.01 | – | |||||
| L. fischeri group | 6. L. wermuthi | 14.03 | 13.56 | 12.15 | 12.85 | 12.15 | 0.1 | |||
| 7. L. thomensis | 14.81 | 13.63 | 12.50 | 13.98 | 13.27 | 7.29 | 2.5 | |||
| 8. L. delicatus | 14.62 | 14.15 | 12.50 | 12.97 | 12.26 | 4.13 | 5.81 | 0.0 | ||
| 9. L. conraui | 14.86 | 15.45 | 12.97 | 14.39 | 14.39 | 12.38 | 11.68 | 12.03 | – |
| Species | Catalog Number | Tissue Sample | Locality | Sex | Dorsal Pattern | Precloacal Pores | Supralabials (R/L) |
|---|---|---|---|---|---|---|---|
| L. heeneni (Holotype) | RMCA_Vert_R.8477 | – | Kapiri, Katanga Region | M | 1 | 10 | 9/8 |
| L. heeneni (Paratype) | RMCA_Vert_R.8478 | – | Kapiri, Katanga Region | M | 1 | 10 | 8/8 |
| L. heeneni (Paratype) | MCZ R-42859 | – | Kapiri, Katanga Region | F | 1 | – | –/7 |
| L. heeneni * | UTEP 22997 | EBG 2982 | Marungu Plateau, Pepa | M | 2 | 10 | 7/7 |
| L. heeneni | UTEP 22998 | EBG 2983 | Marungu Plateau, Pepa | F | 0 | – | 7/7 |
| L. heeneni | UTEP 22999 | EBG 2984 | Marungu Plateau, Pepa | F | 2 | – | 8/8 |
| L. heeneni | UTEP 23000 | EBG 2985 | Marungu Plateau, Pepa | F | 0 | – | 7/8 |
| L. heeneni | UTEP 23001 | EBG 2986 | Marungu Plateau, Pepa | M | 0 | 10 | 7/7 |
| L. heeneni | UTEP 23002 | EBG 2987 | Marungu Plateau, Pepa | M | 2 | 9 | 8/7 |
| L. heeneni | UTEP 23003 | EBG 2988 | Marungu Plateau, Pepa | F | 2 | – | 7/7 |
| L. heeneni | UTEP 23004 | EBG 2989 | Marungu Plateau, Pepa | F | 2 | – | 6/6 |
| L. heeneni | UTEP 23005 | EBG 2990 | Marungu Plateau, Pepa | M | 0 | 7 | 7/7 |
| L. heeneni | UTEP 23006 | EBG 2991 | Marungu Plateau, Pepa | M | 1 | 10 | 8/9 |
| L. heeneni | PEM R20761 | – | Fungurume, Lualaba | F | 2 | – | 8/8 |
| L. heeneni | RBINS 6103 | – | Upemba National Park | F | 1 | – | 8/– |
| L. heeneni | RBINS 6104 | – | Upemba National Park | F | 2 | – | 8/– |
| L. heeneni | RBINS 6106 | – | Upemba National Park | M | 1 | 7 | 7/8 |
| L. heeneni | RBINS 6107 | – | Upemba National Park | M | 1 | 7 | 8/8 |
| L. heeneni | RBINS 6110 | – | Upemba National Park | M | 2 | 8 | 7/8 |
| L. heeneni | RBINS 6113 | – | Upemba National Park | F | 1 | – | 9/9 |
| L. paurospilus (Holotype) | RMCA_Vert_R.27408 | – | Kabobo Plateau, Lubitshako | F | 1 | – | 7/7 |
| L. paurospilus (Paratype) | RMCA_Vert_R.27409 | – | Kabobo Plateau, Lubitshako | M | 0 | 7 | 7/8 |
| L. paurospilus * | UTEP 22996 | DFH 4790 | Itombwe Plateau, Kilumbi | M | 0 | 7 | 7/7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Lobón-Rovira, J.; Devaney, C.; Kusamba, C.; Greenbaum, E. More Knowledge, Fewer Species: New Insights into the Systematics of Lygodactylus heeneni de Witte 1933 (Gekkota: Gekkonidae) of Central Africa. Taxonomy 2026, 6, 9. https://doi.org/10.3390/taxonomy6010009
Lobón-Rovira J, Devaney C, Kusamba C, Greenbaum E. More Knowledge, Fewer Species: New Insights into the Systematics of Lygodactylus heeneni de Witte 1933 (Gekkota: Gekkonidae) of Central Africa. Taxonomy. 2026; 6(1):9. https://doi.org/10.3390/taxonomy6010009
Chicago/Turabian StyleLobón-Rovira, Javier, Calum Devaney, Chifundera Kusamba, and Eli Greenbaum. 2026. "More Knowledge, Fewer Species: New Insights into the Systematics of Lygodactylus heeneni de Witte 1933 (Gekkota: Gekkonidae) of Central Africa" Taxonomy 6, no. 1: 9. https://doi.org/10.3390/taxonomy6010009
APA StyleLobón-Rovira, J., Devaney, C., Kusamba, C., & Greenbaum, E. (2026). More Knowledge, Fewer Species: New Insights into the Systematics of Lygodactylus heeneni de Witte 1933 (Gekkota: Gekkonidae) of Central Africa. Taxonomy, 6(1), 9. https://doi.org/10.3390/taxonomy6010009






