Abstract
Benchmarking in taxonomy is viewed both as establishing a specimen as a standard of reference and as a process for optimizing that process. Here, it is founded on vision theory that recognition of specimens, as for all objects, is personal to the observer and is based on stored exemplars (benchmark images) in their memory. A special feature of a holotype as a scientific benchmark is that it has been published with a Linnaean name permanently attached. This concept is generalized to include all specimens published by subsequent taxonomists with that name attached (a labeled specimen knowledge base). As a record of usage, it integrates all published images with a Linnaean name. It promotes an inquiry into processes for the selection of such specimens. In the conventional model of practice, taxonomists categorize specimens using their stored representations of already identified individuals; the process is immediate, acute, and autonomous, but is largely concealed; a specimen may be selected as a benchmark, but its typicality is not revealed. As a remedy, a population model of practice is advocated wherein the basic autonomous visual process is supplemented by objective data about a specimen and the probability of its position within a potential source population.
1. Introduction
In its original usage 200 years ago, a benchmark was a surveying term that referred to a mark in the ground which enabled instruments to be re-established in their original location: it was a reliable point of reference, a ground truth location. Although this usage continues, it has evolved in the digital age to include processes seeking to optimize performance [1,2] and is widely used in science and technology. A principal application in the biological sciences has been in computational genomics, where it refers to the optimization of processes for decoding molecular data.
Considered as a point of reference, as in its original use, ‘benchmark’ is seldom used in Linnaean taxonomy either by practicing taxonomists or by philosophers of science and semantics, yet, as identified by [3], the holotype specimen appears to be a prime example. Examined here is its use by [4] in their review of the taxonomy of living planktonic foraminifera. They aim to benchmark species concepts, noting that the taxonomy should be robust and operational. Their usage of ‘benchmark’ is simple: for species, they confirm that a holotype specimen exists and that the name is not a subjective synonym [3] (art 61). Otherwise expressed, they assess whether the species name has been validly typified. While this is a strict interpretation of a benchmark specimen in Linnaean taxonomy, it overlooks its role in the recognition of such taxa, either as new species or as specimens of already named species. Several aspects of these roles are considered here.
2. Morphospecies: Their Recognition
In cognitive neuroscience, ‘categorization’ (more generally ‘classification’) denotes a process whereby a group of objects is picked out by an individual’s mental representation (concept) of the group. It allows for the determination of objects belonging to the group [5]; visual similarity is a primary determinant of membership. Current research indicates that the mental representation may be based on exemplars (members) of the group already stored in visual memory [6,7], or on prototypes formed from summed similarities of exemplars. These processes are located separately in the brain but may operate in tandem [8]. Category learning is aided by the extent of similarity among exemplars and may be assisted by textual data [9]. Importantly, all of these processes are personal to each observer.
In practice, building knowledge of a morphospecies begins with a taxonomist recognizing a new (= unnamed, i.e., no imagery recall) group within their collections or in the literature, depositing a specimen (holotype) in a recognized repository, and publishing a Linnaean name and description for it. This makes the species available in the public domain as a named taxon, as [4] showed. The holotype is the name bearer and, as such, qualifies as a benchmark, or equivalently, as a voucher specimen in a machine learning application. That the species name is bound to that specimen creates a widely discussed problem in its use [10] and is a focus of this commentary.
How the author’s concept is to be promulgated is not addressed by [3] and seldom is in the wider literature, yet it is vital for its use as a referent. The following representation seeks to mirror conventional practice for referencing the name of a newly recognized taxon as a visual entity. In it, ‘labeled’ is used for specimens, including images thereof, to which the Linnaean name is attached, say as a label on the specimen or on its mountant, or in captions for illustrations of the specimen. In this sense, the holotype is the primary labeled specimen (PLS). Knowledge of the species is incremented and disseminated as other taxonomists view the deposited museum specimen, or its published image, and recognize similar specimens in their collections. They might isolate and label such specimens, subjectively determined (SLS), with that name for their reference or publish illustrations, thereby extending the set of labeled specimens. Some may have inspected the holotype (Figure 1), but commonly this is infeasible. Increasingly, available sources of visual knowledge of species are SLS specimens shown in print or digital publications. They form an essential guide for today’s taxonomists to develop their personal concept of a species and also serve as an important adjunct to the limited capacity of our short-term memory [11].
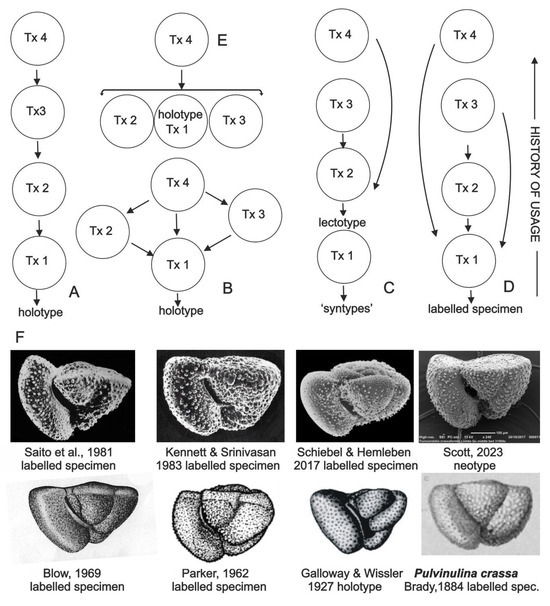
Figure 1.
(A–E) Hypothetical LSKB viewed as flow maps of visual information about species. Arrows indicate a previously labeled specimen used as a guide for a taxonomist (Tx n) to identify and label a specimen in his collection. (A) compares with the chain-like structure of the historical theory of causal reference in semantics [12] and might map a preference for viewing the last-published image because of improvements in image quality over time. (B) Tx 4 views all available imagery. (C) models some planktonic foraminiferal taxa named in the early 19th century [13] for which authors isolated several specimens (‘syntypes’) but did not label a holotype; published images were of models. (D) shows the significance of scientific atlases [14] in taxonomy; in this example, as in several real examples, the atlas is not rooted in the holotype. (E) shows the role of online image aggregator sites, which are becoming the default sites for viewing labeled specimens. (F) Labeled specimens of Truncorotalia crassaformis Galloway & Wissler, 1927 [15] from selected references [15,16,17,18,19,20,21,22].
For a given species, labeled specimens form an incremental record of its identification that expands out from its original publication. Ostensibly, the expansion is a historical record of specimens identified by later taxonomists labeled using the name of the holotype specimen: it is termed a labeled specimen knowledge base (LSKB). For a recently described species, individual entries in the LSKB are readily traceable online, and the history of usage is viewable. However, as taxonomists seldom record which prior labeled specimens they inspected when forming their concept of a species, for many taxa, their LSKB resembles a virtual entity.
3. Discussion
3.1. Conventional Model
The conventional model (CM) relies on the operations of our vision system for the construction of an LSKB and inherits its defects for the recognition of groups based on a primary labeled specimen. Nominally, an LSKB is an archive of images whose name is a homonym of the holotype: it uses the name of the holotype but references different specimens. This referencing problem is generally ignored by taxonomists (e.g., [23,24]) but is widely discussed in the philosophy of the natural sciences [10,25]. Of relevance here is [26] p.12195, who regards the holotype as a sample and argues that “One needs a particular explanatory purpose to pick out a particular population by a species baptism. If that is left open, the name may not latch onto any determinate population”. Although this is not an operational solution, it recognizes the need to relate a holotype to its source population and is consistent with the following model.
3.2. Population Model
The population model (PM) accepts that ‘species’ is a group concept for biological specimens recognized as similar tangible objects, a view that compares with [27] p.159 “To sum up, I believe that species come to be tolerably well-defined objects …”. However, to remedy the problems of CM, PM follows [28] p.65 that “… Populations, not individuals, are the units of systematics …” and Beckner’s [29] view that species are polytypic populations in space/time; a realist view of ‘species’. The PM problem then for a species proposal is to identify a specimen to serve as the primary benchmark (PLS) for that population, or as a benchmark for a local named population. It leads to a quite different, probabilistic search strategy wherein the opaque operations of the personal perception process of CM are supplemented by strategies that create probabilistic, contestable data about the position of the holotype or other labeled specimen in its source population. The schema of the population model used here follows these steps. 1. The taxonomist uses their visual perception to select specimens they provisionally identify as species A or similar thereto. 2. Morphometric data on a functional trait are gathered. 3. Statistical analyses provide probabilities of the selected specimens belonging to one population. 4. The taxonomist evaluates this statistical grouping against their knowledge of the functional significance of the trait. 5. A benchmark or other voucher specimens are selected, informed by the probability that they belong to Species A.
The trait in the examples of this study is the axial outline of planktonic foraminifera shells. Experimental neuroscience shows that, subliminally, the outline is an important guide to object recognition [30,31]. Here, it captures most of a specimen’s ontogeny and is regarded as serving hydrodynamic and trophic functions. The normal model is preferred for analysis because it provides a suitable measure of central tendency, which might be associated with adaptation towards an optimum form in the shape trait.
The primary focus of PM is to demonstrate the probability that specimens in a sample are drawn from one population (Figure 2). Its value for accessing the relation of benchmark specimens to other populations is shown in Figure 3. Theoretically, PM might be viewed as an optimizing strategy for the selection of representative specimens such as benchmarks. Nevertheless, the statistical data remain open to interpretation, biologically: the taxonomist retains discretionary judgment when, for example (Figure 2 #30), a specimen is flagged as unlikely to belong to the population when it is interpreted as a juvenile [32].
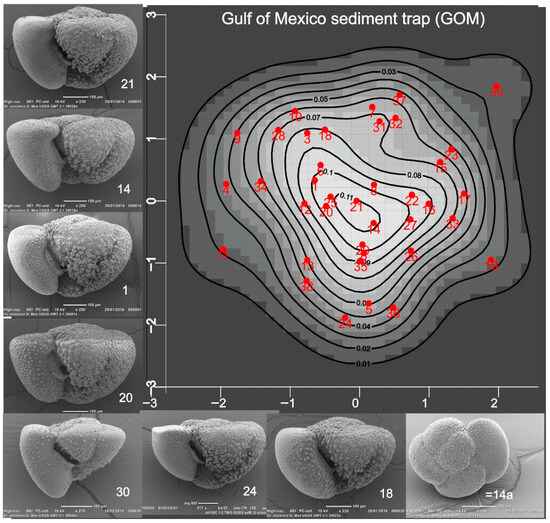
Figure 2.
Density map showing probabilities that specimens in the Gulf of Mexico GMT21 sediment trap, a time series (2008–2012) of foraminiferal and particulate flux at 700 m on the northern Gulf of Mexico continental shelf (27.5° N; 90.3° W), belong to a normal (Gaussian) shape population. A benchmark specimen might be selected from those with the highest probabilities. Refer to [32] for details.
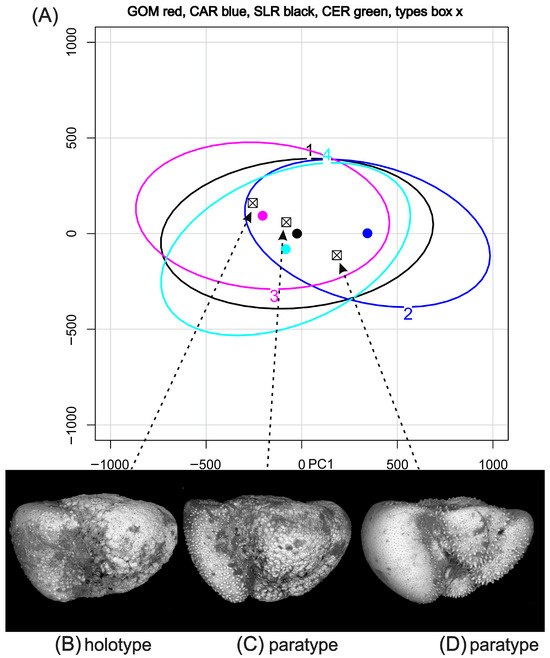
Figure 3.
(A) Data ellipses (95% confidence around mean values shown as colored dots) for a procrustes analysis of pooled samples from the Gulf of Mexico (GOM), Cariaco Basin (CAR), and Ceara Rise (CER). Sierra Leone Rise (SLR) axial samples, with the addition of the three named specimens of Truncorotalia oceanica. These specimens lie within the confidence limit of each sample. All of the material is from the source region of Truncorotalia oceanica Cushman & Bermudez, 1949. (B–D) Holotype and paratypes of Truncorotalia oceanica. Refer to [32] for details.
Operationally, PM requires that the population from which the holotype was sourced is analyzed and that data from that analysis are publicly available to allow them to be plotted in an analysis of a local population that a taxonomist considers to represent the holotype population (e.g., Figure 3). This is a project that organizations like those represented by [4] might consider.
4. Conclusions
Brummer & Kucera [4] offer an account of the application of benchmarking modern planktonic taxa that is limited to verifying the existence of a formally named specimen, its subjective synonyms, and specification of diagnostic characters, but without imagery thereof. They do not address how an operational taxonomy is to be developed from these data.
ICZN [3] rules relate only to the formation of a valid name for a species and the curation of one specimen, the holotype or benchmark specimen. They do not provide guidance on the usage of the name of that specimen: its role as a referent. In conventional practice, taxonomists identify individual specimens using their personal concept of a species, which is informed by their stored mental representations of previously viewed specimens, or their images, to which the name has been applied. Collectively, the named images form a database of referents, which, excepting the holotype, are subjective interpretations of the name and personal to each taxonomist’s species concept.
In practice, morphospecies comprise tangible specimens primarily grouped by their common visual attributes. Although conventional practice (CM), reliant on the autonomous processes of visual memory, is highly efficient for specimen recognition and grouping of such objects, its mechanisms are cryptic, and outcomes are personal to the viewer. They do not provide objective data about the relation of a labeled specimen to the name-bearing specimen (benchmark) and are unsuitable for species when viewed as polytypic populations. Contrarily, for the realist model (PM), the taxonomic problem is to identify their populations and generate contestable, objective data about specimen relationships to facilitate the selection of representative specimens.
Expectedly, because of its history of usage, efficiency, and general acceptance, CM will remain normative into the future, and accusations of “stamp collecting” [33] will continue. Acceptance of PM, or similar models, will be contingent on a wider appreciation of the constraints imposed on recognition of a natural group when its name is bound to one specimen. Impeding wide usage of PM for many cohorts is the present limited availability and cost of laboratory equipment for rapid specimen positioning and capture of 2D-3D image data.
Funding
This research received no external funding.
Data Availability Statement
No new data were created or analyzed in this study.
Acknowledgments
I am grateful to GNS Science for access to facilities and to Polly Winsor for perceptive advice.
Conflicts of Interest
The author declares no conflict of interest.
References
- Bartz-Beielstein, T.; Doerr, C.; van den Berg, D.; Bossek, J.; Chandrasekaran, S.; Eftimov, T.; Fischbach, A.; Kerschke, P.; Cava, W.L.; Lopez-Ibanez, M.; et al. Benchmarking in Optimization: Best Practice and Open Issues. arXiv 2020. [Google Scholar] [CrossRef]
- Moriarty, J.P.; Smallman, C. En Route to a Theory of Benchmarking. Benchmarking Int. J. 2009, 16, 484–503. [Google Scholar] [CrossRef]
- Ride, W.D.L. International Code of Zoological Nomenclature: Code International de Nomenclature Zoologique, 4th ed.; International Commission on Zoological Nomenclature, Ride, W.D.L., International Trust for Zoological Nomenclature, Natural History Museum (London, England), International Union of Biological Sciences, Eds.; International Trust for Zoological Nomenclature, c/o Natural History Museum: London, UK, 1999. [Google Scholar]
- Brummer, G.-J.A.; Kučera, M. Taxonomic Review of Living Planktonic Foraminifera. J. Micropalaeontology 2022, 41, 29–74. [Google Scholar] [CrossRef]
- Medin, D.L.; Colely, J.D. Concepts and Categorization. In Perception and Cognition at Century’s End; Academic Press: San Diego, CA, USA, 1998; pp. 403–440. [Google Scholar]
- Smith, J.D. Prototypes, Exemplars, and the Natural History of Categorization. Psychon. Bull. Rev. 2014, 21, 312–331. [Google Scholar] [CrossRef] [PubMed]
- Ashby, F.G.; Rosedahl, L.A. Neural Interpretation of Exemplar Theory. Psychol. Rev. 2017, 124, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Blank, H.; Bayer, J. Functional Imaging Analyses Reveal Prototype and Exemplar Representations in a Perceptual Single-Category Task. Commun. Biol. 2022, 5, 896. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.I.; Thomas, A.K. Visual Category Learning: Navigating the Intersection of Rules and Similarity. Psychon. Bull. Rev. 2021, 28, 711–731. [Google Scholar] [CrossRef] [PubMed]
- Michaelson, E. The Stanford Encyclopaedia of Philosophy; Stanford University: Stanford, CA, USA, 2024. [Google Scholar]
- Brady, T.F.; Konkle, T.; Alvarez, G.A. A Review of Visual Memory Capacity: Beyond Individual Items and Toward Structured Representations. Available online: http://www.journalofvision.org/content/11/5/4 (accessed on 29 June 2025).
- Michaelson, E. The Vagaries of Reference. Ergo 2022, 9, 1433–1448. [Google Scholar] [CrossRef]
- Banner, F.T.; Blow, W.H. Some Primary Types of Species Belonging to the Superfamily Globigerinaceae. Contrib. Cushman Found. Foraminifer. Res. 1960, 11, 1–40. [Google Scholar]
- Daston, L.; Galison, P. The Image of Objectivity. Representations 1992, 40, 81–128. [Google Scholar] [CrossRef]
- Galloway, J.J.; Wissler, S.G. Pleistocene Foraminifera from the Lomita Quarry, Palos Verde Hills, California. J. Paleontol. 1927, 1, 35–87. [Google Scholar]
- Saito, T.; Thompson, P.R.; Breger, D. Systematic Index of Recent and Pleistocene Planktonic Foraminifera. 190 Pp., 56 Plates; Price £15.00.; University of Toyko: Toyko, Japan, 1981; ISBN 0 86008 280 6. [Google Scholar]
- Kennett, J.P.; Srinivasan, M.S. Neogene Planktonic Foraminifera: A Phylogenetic Atlas; Hutchinson Ross: Stroudsburg, PA, USA, 1983. [Google Scholar]
- Schiebel, R.; Hemleben, C. Planktic Foraminifers in the Modern Ocean; Springer: Berlin, Germany, 2017. [Google Scholar]
- Scott, G.H. A Replacement Neotype for Globigerina crassaformis Galloway & Wissler, 1927. J. Foraminifer. Res. 2023, 53, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Blow, W.H. Late Middle Eocene to Recent Planktonic Foraminiferal Biostratigraphy. In Proceedings of the First International Conference on Planktonic Microfossils; Brill: Leiden, The Netherlands, 1969; Volume 1, pp. 199–422. [Google Scholar]
- Parker, F.L. Planktonic Foraminiferal Species in Pacific Sediments. Micropaleontology 1962, 8, 219–254. [Google Scholar] [CrossRef]
- Brady, H.B. Report on the Foraminifera Dredged by HMS Challenger during the Years 1873-1876. In Report of Scientific Results of the Exploration Voyage of HMS Challenger; Zoology; Challenger Office: Edinburgh, UK, 1884; Volume 9, pp. 1–814. [Google Scholar]
- Braby, M.F.; Hsu, Y.-F.; Lamas, G. How to Describe a New Species in Zoology and Avoid Mistakes. Zool. J. Linn. Soc. 2024, 202, zlae043. [Google Scholar] [CrossRef]
- Ruedas, L.A.; Norris, R.W.; Timm, R.M. Best Practices for the Naming of Species. J. Mammology 2025, 106, 523–531. Available online: https://pdxscholar.library.pdx.edu/bio_fac (accessed on 29 June 2025).
- Ereshevsfy, M.; Reydon, T.A. Scientific Kinds. Philos. Stud. 2015, 172, 969–986. [Google Scholar] [CrossRef]
- Crane, J.K. Two Approaches to Natural Kinds. Synthese 2021, 1999, 12177–12198. [Google Scholar] [CrossRef]
- Darwin, C. Origin of Species; D. Appleton: New York, NY, USA, 1861. [Google Scholar]
- Simpson, G.G. Principles of Animal Taxonomy; Columbia University Press: New York, NY, USA, 1961; Available online: https://www.degruyter.com/document/doi/10.7312/simp92414/html (accessed on 29 June 2025).
- Beckner, M. The Biological Way of Thought; Columbia University Press: New York, NY, USA, 1959. [Google Scholar] [CrossRef]
- Spröte, P.; Schmidt, F.; Fleming, R.W. Visual Perception of Shape Altered by Inferred Causal History. Sci. Rep. 2016, 6, 36245. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.H. Shape from Contour: Computation and Representation. Annu. Rev. Vis. Sci. 2018, 4, 423–450. [Google Scholar] [CrossRef] [PubMed]
- Scott, G.H. Testing Expert Opinion in Synonymies: An Example from the Zooplankton. GNS Science, Lower Hutt, New Zealand (manuscript in preparation).
- Johnson, K. Natural History as Stamp Collecting: A Brief History. Arch. Nat. Hist. 2007, 34, 244–258. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).