Description of Silvibacterium acidisoli sp. nov. and Edaphobacter albus sp. nov. and a Proposal for Taxonomic Rearrangements Within the Family Acidobacteriaceae Based on Comparative Genome Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains Collection
2.2. Phenotypic Characterisation
2.3. Genome Selection and Quality Assessment
2.4. Phylogenetic Analyses Based on 16S rRNA Gene and Genome Sequences
2.5. Genome Relatedness Indices Calculation
2.6. Species and Genus Assignments Based on Genotypic and Phenotypic Data
3. Results and Discussion
3.1. Phenotypic Characterisation
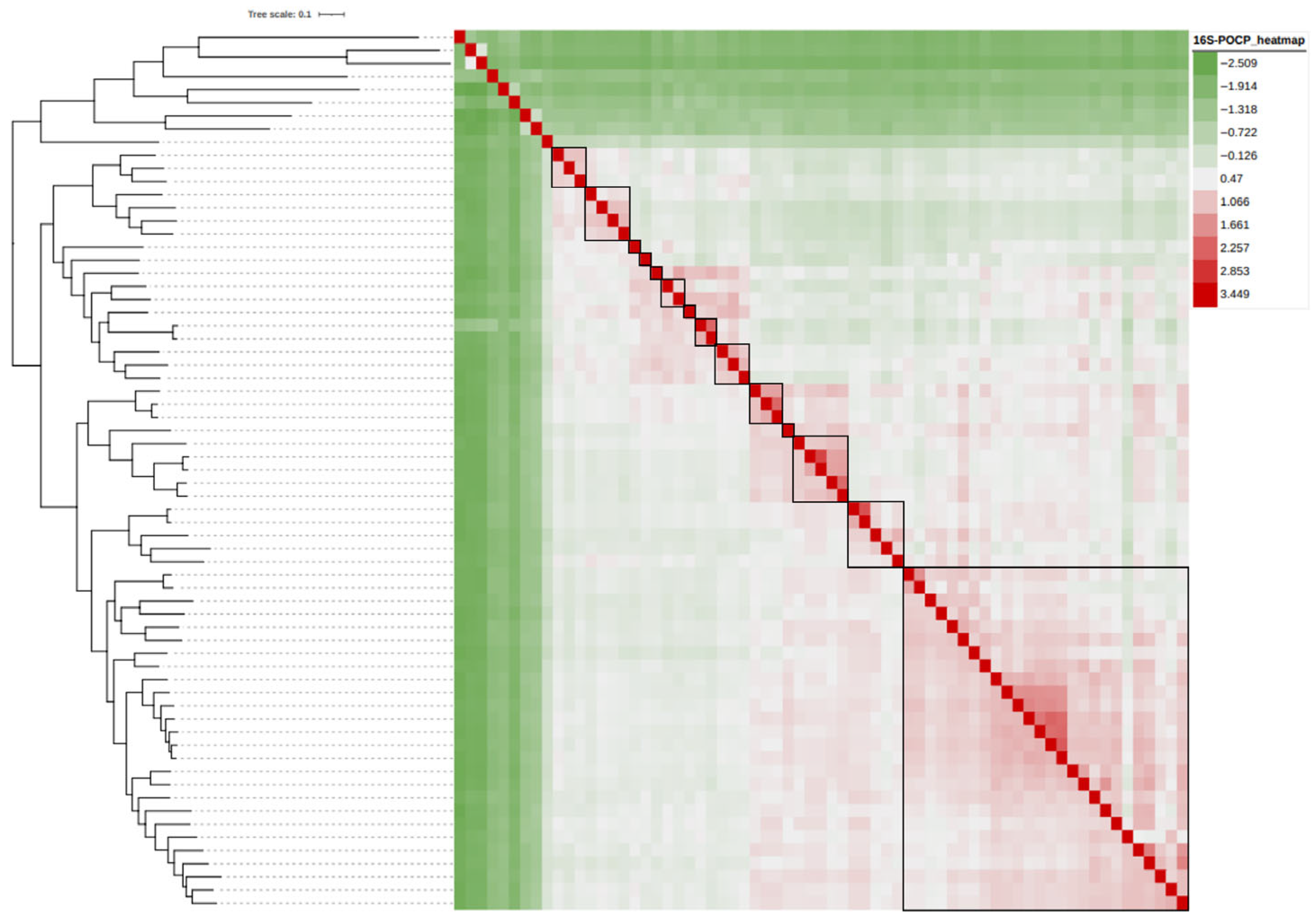
3.2. The 16S rRNA Gene-Based Analysis
3.3. Genome-Based Phylogenetic Analysis
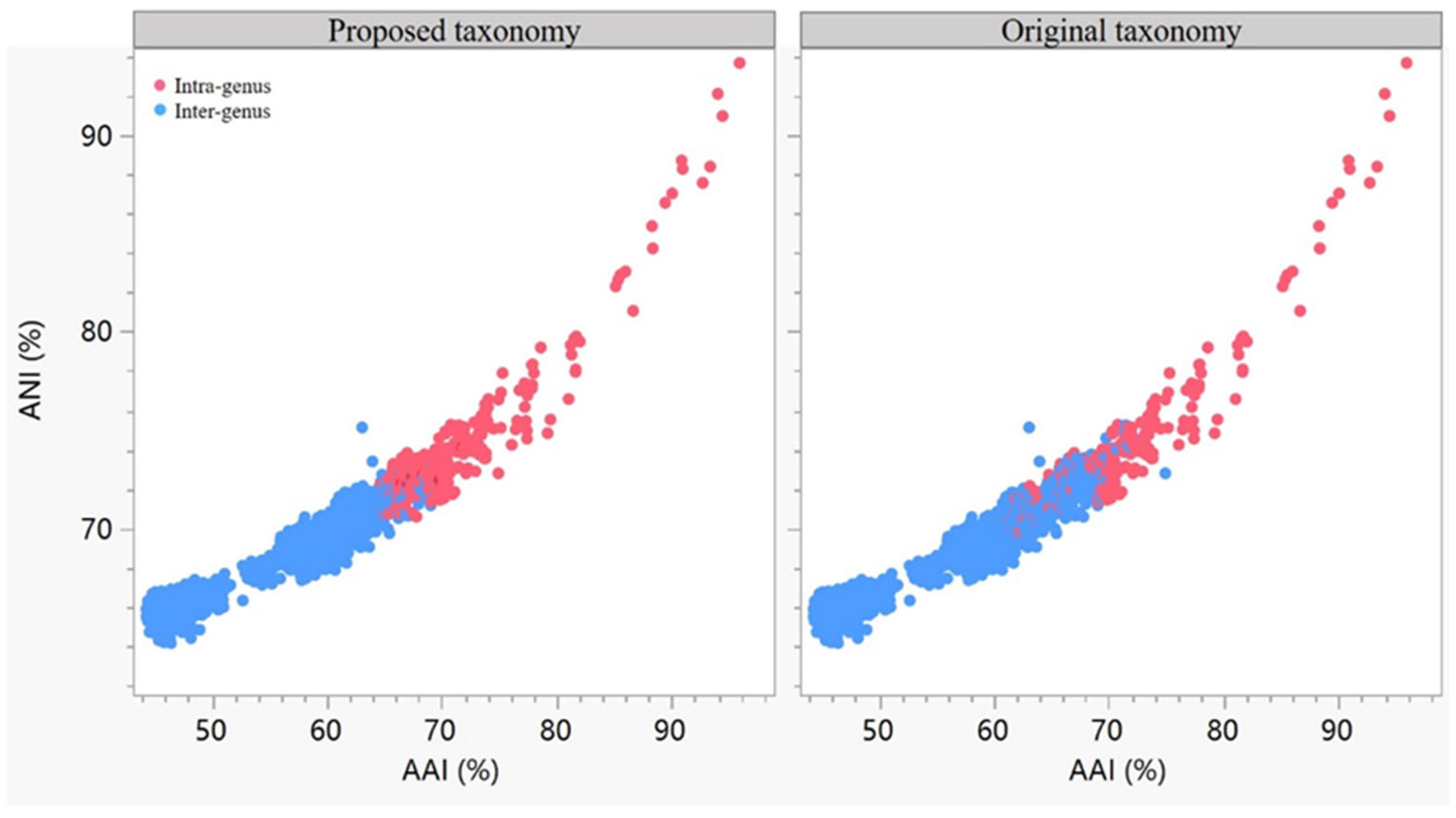
3.4. Reclassification Supported by AAI, ANI and POCP Values
3.5. Reclassification Supported by Phenotypic Characteristics and Fatty Acid Profiles
4. Conclusions
5. Description of Alloterriglobus gen. nov.
6. Description of Rhizacidiphilus gen. nov.
7. Description of Edaphobacter albus sp. nov.
8. Description of Silvibacterium acidisoli sp. nov.
9. Description of Alloterriglobus saanensis comb. nov.
10. Description of Edaphobacter arcticus comb. nov.
11. Description of Edaphobacter pectinivorans comb. nov.
12. Description of Edaphobacter roseus comb. nov.
13. Description of Edaphobacter sapmiensis comb. nov.
14. Description of Edaphobacter sibiricus comb. nov.
15. Description of Edaphobacter tundricola comb. nov.
16. Description of Edaphobacter xylanilyticus comb. nov.
17. Description of Granulicella elongata comb. nov.
18. Description of Occallatibacter gabretensis comb. nov.
19. Description of Pseudacidobacterium dinghuense comb. nov.
20. Description of Rhizacidiphilus albidus comb. nov.
21. Description of Rhizacidiphilus tenax comb. nov.
22. Emended Description of Edaphobacter Koch et al. 2008
23. Emended Description of Occallatibacter Foesel et al. 2016
24. Emended Description of Pseudacidobacterium Myers et al. 2016, Zhang et al. 2022
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent Understanding of Soil Acidobacteria and Their Ecological Significance: A Critical Review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef] [PubMed]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed]
- Aszalos, J.M.; Krett, G.; Anda, D.; Marialigeti, K.; Nagy, B.; Borsodi, A.K. Diversity of extremophilic bacteria in the sediment of high-altitude lakes located in the mountain desert of Ojos del Salado volcano, Dry-Andes. Extremophiles 2016, 20, 603–620. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, N.; Kosako, Y.; Tano, T. Acidobacterium capsulatum gen. nov., sp. nov.: An acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Current Microbiol 1991, 22, 1–7. [Google Scholar] [CrossRef]
- Liesack, W.; Bak, F.; Kreft, J.U.; Stackebrandt, E. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 1994, 162, 85–90. [Google Scholar] [CrossRef]
- Lonergan, D.J.; Jenter, H.L.; Coates, J.D.; Phillips, E.J.; Schmidt, T.M.; Lovley, D.R. Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 1996, 178, 2402–2408. [Google Scholar] [CrossRef]
- Ludwig, W.; Bauer, S.H.; Bauer, M.; Held, I.; Kirchhof, G.; Schulze, R.; Huber, I.; Spring, S.; Hartmann, A.; Schleifer, K.H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 1997, 153, 181–190. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Zimmermann, J.; Gonzalez, J.M.; Saiz-Jimenez, C.; Ludwig, W. Detection and phylogenetic relationships of highly diverse uncultured acidobacterial communities in altamira cave using 23S rRNA sequence analyses. Geomicrobiol. J. 2005, 22, 379–388. [Google Scholar] [CrossRef]
- Barns, S.M.; Cain, E.C.; Sommerville, L.; Kuske, C.R. Acidobacteria phylum sequences in uranium-contaminated subsurface sediments greatly expand the known diversity within the phylum. Appl. Environ. Microbiol. 2007, 73, 3113–3116. [Google Scholar] [CrossRef]
- Fukunaga, Y.; Kurahashi, M.; Yanagi, K.; Yokota, A.; Harayama, S. Acanthopleuribacter pedis gen. nov., sp. nov., a marine bacterium isolated from a chiton, and description of Acanthopleuribacteraceae fam. nov., Acanthopleuribacterales ord. nov., Holophagaceae fam. nov., Holophagales ord. nov. and Holophagae classis nov. in the phylum ‘Acidobacteria’. Int. J. Syst. Evol. Microbiol. 2008, 58, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Wust, P.K.; Geppert, A.; Foesel, B.U.; Huber, K.J.; Overmann, J. Novel isolates double the number of chemotrophic species and allow the first description of higher taxa in Acidobacteria subdivision 4. Syst. Appl. Microbiol. 2015, 38, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; da Costa, M.S.; Garrity, G.M.; Rainey, F.A.; Rossello-Mora, R.; Schink, B.; Sutcliffe, I.; Trujillo, M.E.; Whitman, W.B. Proposal to include the rank of phylum in the International Code of Nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2015, 65, 4284–4287. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Yilmaz, P. Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 3796–3806. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Okamura, K.; Kawai, A.; Yamada, T.; Hiraishi, A. Acidipila rosea gen. nov., sp. nov., an acidophilic chemoorganotrophic bacterium belonging to the phylum Acidobacteria. FEMS Microbiol. Lett. 2011, 317, 138–142. [Google Scholar] [CrossRef]
- Koch, I.H.; Gich, F.; Dunfield, P.F.; Overmann, J. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int. J. Syst. Evol. Microbiol. 2008, 58, 1114–1122. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Kirsanova, L.A.; Kaparullina, E.N.; Kevbrin, V.V.; Dedysh, S.N. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria, and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int. J. Syst. Evol. Microbiol. 2012, 62, 430–437. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef]
- Rawat, S.R.; Mannisto, M.K.; Bromberg, Y.; Haggblom, M.M. Comparative genomic and physiological analysis provides insights into the role of Acidobacteria in organic carbon utilization in Arctic tundra soils. FEMS Microbiol. Ecol. 2012, 82, 341–355. [Google Scholar] [CrossRef]
- Belova, S.E.; Ravin, N.V.; Pankratov, T.A.; Rakitin, A.L.; Ivanova, A.A.; Beletsky, A.V.; Mardanov, A.V.; Sinninghe Damsté, J.S.; Dedysh, S.N. Hydrolytic Capabilities as a Key to Environmental Success: Chitinolytic and Cellulolytic Acidobacteria From Acidic Sub-arctic Soils and Boreal Peatlands. Front. Microbiol. 2018, 9, 2775. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Trojan, D.; Roux, S.; Herbold, C.; Rattei, T.; Woebken, D. Genomic insights into the Acidobacteria reveal strategies for their success in terrestrial environments. Environ. Microbiol. 2018, 20, 1041–1063. [Google Scholar] [CrossRef] [PubMed]
- Llado, S.; Benada, O.; Cajthaml, T.; Baldrian, P.; Garcia-Fraile, P. Silvibacterium bohemicum gen. nov. sp. nov., an acidobacterium isolated from coniferous soil in the Bohemian Forest National Park. Syst. Appl. Microbiol. 2016, 39, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Fraile, P.; Benada, O.; Cajthaml, T.; Baldrian, P.; Llado, S. Terracidiphilus gabretensis gen. nov., sp. nov., an Abundant and Active Forest Soil Acidobacterium Important in Organic Matter Transformation. Appl. Environ. Microbiol. 2016, 82, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Foesel, B.U.; Mayer, S.; Luckner, M.; Wanner, G.; Rohde, M.; Overmann, J. Occallatibacter riparius gen. nov., sp. nov. and Occallatibacter savannae sp. nov., acidobacteria isolated from Namibian soils, and emended description of the family Acidobacteriaceae. Int. J. Syst. Evol. Microbiol. 2016, 66, 219–229. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Shi, C.; Huang, Z.; Zhang, Y.; Li, S.; Li, Y.; Ye, J.; Yu, C.; Li, Z.; et al. SOAPnuke: A MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience 2018, 7, gix120. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward Defining the Course of Evolution: Minimum Change for a Specific Tree Topology. Syst. Biol. 1971, 20, 406–416. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Na, S.I.; Kim, Y.O.; Yoon, S.H.; Ha, S.M.; Baek, I.; Chun, J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018, 56, 280–285. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U.; et al. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 2500. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Ouk Kim, Y.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Qin, Q.L.; Xie, B.B.; Zhang, X.Y.; Chen, X.L.; Zhou, B.C.; Zhou, J.; Oren, A.; Zhang, Y.Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef]
- Liu, Y.; Du, J.; Pei, T.; Du, H.; Feng, G.-D.; Zhu, H. Genome-based taxonomic classification of the closest-to-Comamonadaceae group supports a new family Sphaerotilaceae fam. nov. and taxonomic revisions. Syst. Appl. Microbiol. 2022, 45, 126352. [Google Scholar] [CrossRef]
- Falagan, C.; Foesel, B.; Johnson, B. Acidicapsa ferrireducens sp. nov., Acidicapsa acidiphila sp. nov., and Granulicella acidiphila sp. nov.: Novel acidobacteria isolated from metal-rich acidic waters. Extremophiles 2017, 21, 459–469. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Kostina, L.A.; Valaskova, V.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; de Boer, W.; Dedysh, S.N. Acidicapsa borealis gen. nov., sp. nov. and Acidicapsa ligni sp. nov., subdivision 1 Acidobacteria from Sphagnum peat and decaying wood. Int. J. Syst. Evol. Microbiol. 2012, 62, 1512–1520. [Google Scholar] [CrossRef]
- Matsuo, H.; Kudo, C.; Li, J.; Tonouchi, A. Acidicapsa acidisoli sp. nov., from the acidic soil of a deciduous forest. Int. J. Syst. Evol. Microbiol. 2017, 67, 862–867. [Google Scholar] [CrossRef]
- Ou-Yang, T.N.; Xia, F.; Qiu, L.H. Acidicapsa dinghuensis sp. nov., a novel acidobacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 2364–2369. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Breznak, J.A.; Schmidt, T.M. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 2007, 73, 2708–2717. [Google Scholar] [CrossRef]
- Myers, M.R.; King, G.M. Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat. Int. J. Syst. Evol. Microbiol. 2016, 66, 5328–5335. [Google Scholar] [CrossRef]
- Zhang, Q.-M.; Fu, J.-C.; Chen, Z.-Q.; Qiu, L.-H. Paracidobacterium acidisoli gen. nov., sp. nov. and Alloacidobacterium dinghuense gen. nov., sp. nov., two acidobacteria isolated from forest soil, and reclassification of Acidobacterium ailaaui and Acidipila dinghuensis as Pseudacidobacterium ailaaui gen. nov., comb. nov. and Silvibacterium dinghuense comb. nov. Int. J. Syst. Evol. Microbiol. 2022, 72, 005415. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.W.; Wang, J.; Chen, M.H.; Lv, Y.Y.; Qiu, L.H. Acidipila dinghuensis sp. nov., an acidobacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Wust, P.K.; Geppert, A.; Foesel, B.U.; Huber, K.J.; Overmann, J. Terriglobus albidus sp. nov., a member of the family Acidobacteriaceae isolated from Namibian semiarid savannah soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3297–3304. [Google Scholar] [CrossRef] [PubMed]
- Whang, K.S.; Lee, J.C.; Lee, H.R.; Han, S.I.; Chung, S.H. Terriglobus tenax sp. nov., an exopolysaccharide-producing acidobacterium isolated from rhizosphere soil of a medicinal plant. Int. J. Syst. Evol. Microbiol. 2014, 64, 431–437. [Google Scholar] [CrossRef]
- Mannisto, M.K.; Rawat, S.; Starovoytov, V.; Haggblom, M.M. Terriglobus saanensis sp. nov., an acidobacterium isolated from tundra soil. Int. J. Syst. Evol. Microbiol. 2011, 61, 1823–1828. [Google Scholar] [CrossRef]
- Baik, K.S.; Choi, J.S.; Kwon, J.; Park, S.C.; Hwang, Y.M.; Kim, M.S.; Kim, E.M.; Seo, D.C.; Cho, J.S.; Seong, C.N. Terriglobus aquaticus sp. nov., isolated from an artificial reservoir. Int. J. Syst. Evol. Microbiol. 2013, 63, 4744–4749. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Kulichevskaya, I.S.; Serkebaeva, Y.M.; Mityaeva, M.A.; Sorokin, V.V.; Suzina, N.E.; Rijpstra, W.I.C.; Sinninghe Damste, J.S. Bryocella elongata gen. nov., sp. nov., a member of subdivision 1 of the Acidobacteria isolated from a methanotrophic enrichment culture, and emended description of Edaphobacter aggregans Koch et al. 2008. Int. J. Syst. Evol. Microbiol. 2012, 62, 654–664. [Google Scholar] [CrossRef]
- Pankratov, T.A.; Dedysh, S.N. Granulicella paludicola gen. nov., sp. nov., Granulicella pectinivorans sp. nov., Granulicella aggregans sp. nov. and Granulicella rosea sp. nov., acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 2010, 60, 2951–2959. [Google Scholar] [CrossRef]
- Mannisto, M.K.; Rawat, S.; Starovoytov, V.; Haggblom, M.M. Granulicella arctica sp. nov., Granulicella mallensis sp. nov., Granulicella tundricola sp. nov. and Granulicella sapmiensis sp. nov., novel acidobacteria from tundra soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Okuno, Y.; Meng, X.Y.; Tamaki, H.; Kamagata, Y.; Hanada, S. Granulicella cerasi sp. nov., an acidophilic bacterium isolated from cherry bark. Int. J. Syst. Evol. Microbiol. 2014, 64, 2781–2785. [Google Scholar] [CrossRef] [PubMed]
- Oshkin, I.Y.; Kulichevskaya, I.S.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Rakitin, A.L.; Ravin, N.V.; Dedysh, S.N. Granulicella sibirica sp. nov., a psychrotolerant acidobacterium isolated from an organic soil layer in forested tundra, West Siberia. Int. J. Syst. Evol. Microbiol. 2019, 69, 1195–1201. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Cai, Y.M.; Chen, D.X.; Qiu, L.H. Edaphobacter acidisoli sp. nov., an acidobacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4260–4265. [Google Scholar] [CrossRef]
- Xia, F.; Ou-Yang, T.N.; Gao, Z.H.; Qiu, L.H. Edaphobacter flagellatus sp. nov. and Edaphobacter bradus sp. nov., two acidobacteria isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 2530–2537. [Google Scholar] [CrossRef]
- Wang, J.; Chen, M.H.; Lv, Y.Y.; Jiang, Y.W.; Qiu, L.H. Edaphobacter dinghuensis sp. nov., an acidobacterium isolated from lower subtropical forest soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 276–282. [Google Scholar] [CrossRef]
- Belova, S.E.; Suzina, N.E.; Rijpstra, W.I.C.; Sinninghe Damste, J.S.; Dedysh, S.N. Edaphobacter lichenicola sp. nov., a member of the family Acidobacteriaceae from lichen-dominated forested tundra. Int. J. Syst. Evol. Microbiol. 2018, 68, 1265–1270. [Google Scholar] [CrossRef]
- Damste, J.S.; Rijpstra, W.I.; Hopmans, E.C.; Weijers, J.W.; Foesel, B.U.; Overmann, J.; Dedysh, S.N. 13,16-Dimethyl octacosanedioic acid (iso-diabolic acid), a common membrane-spanning lipid of Acidobacteria subdivisions 1 and 3. Appl. Environ. Microbiol. 2011, 77, 4147–4154. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1,000+ Type-Strain Genomes Substantially Improves Taxonomic Classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Barco, R.A.; Garrity, G.M.; Scott, J.J.; Amend, J.P.; Nealson, K.H.; Emerson, D. A Genus Definition for Bacteria and Archaea Based on a Standard Genome Relatedness Index. mBio 2020, 11, e02475-19. [Google Scholar] [CrossRef]
- Luo, C.; Rodriguez, R.L.; Konstantinidis, K.T. MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014, 42, e73. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
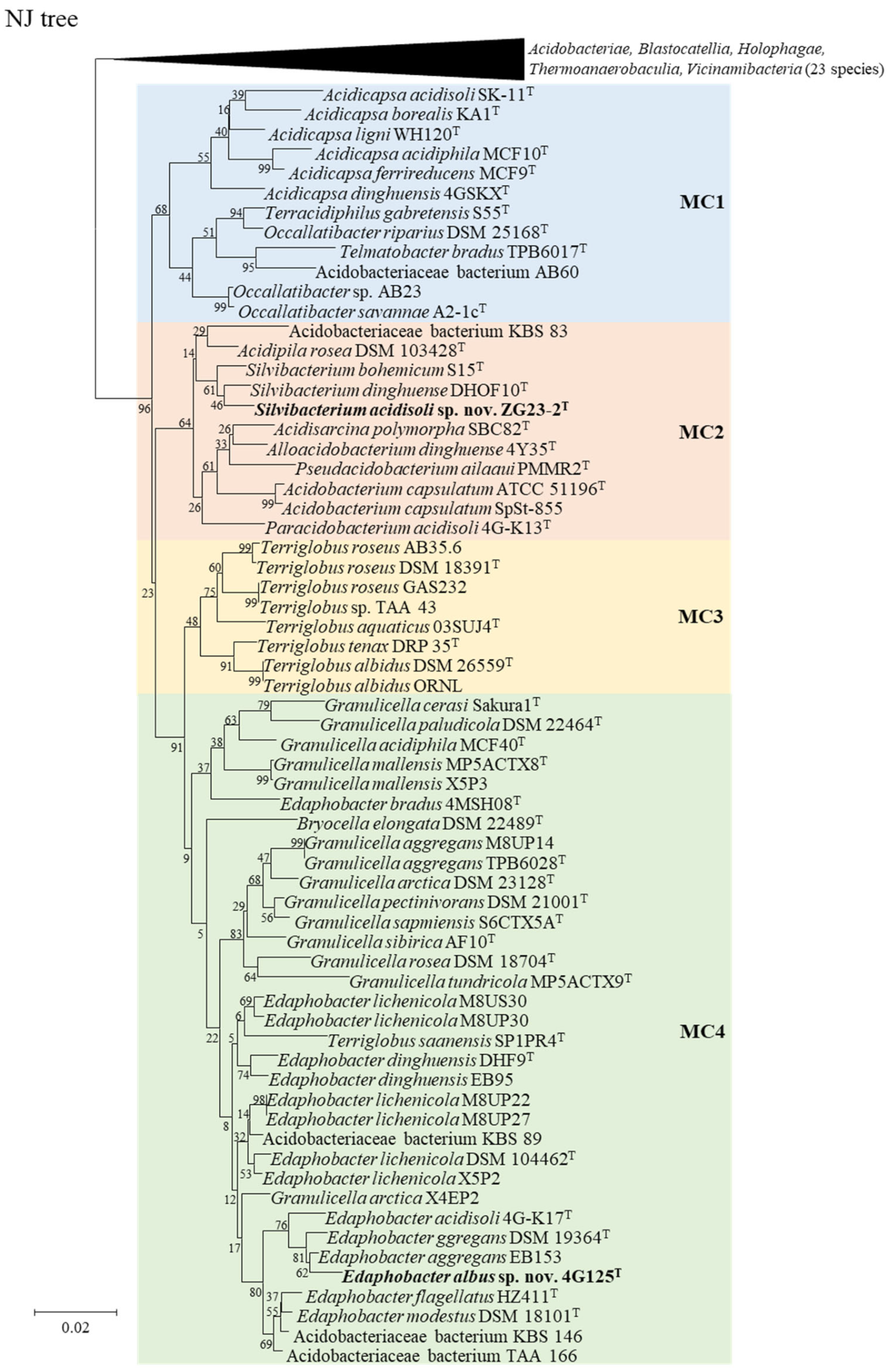
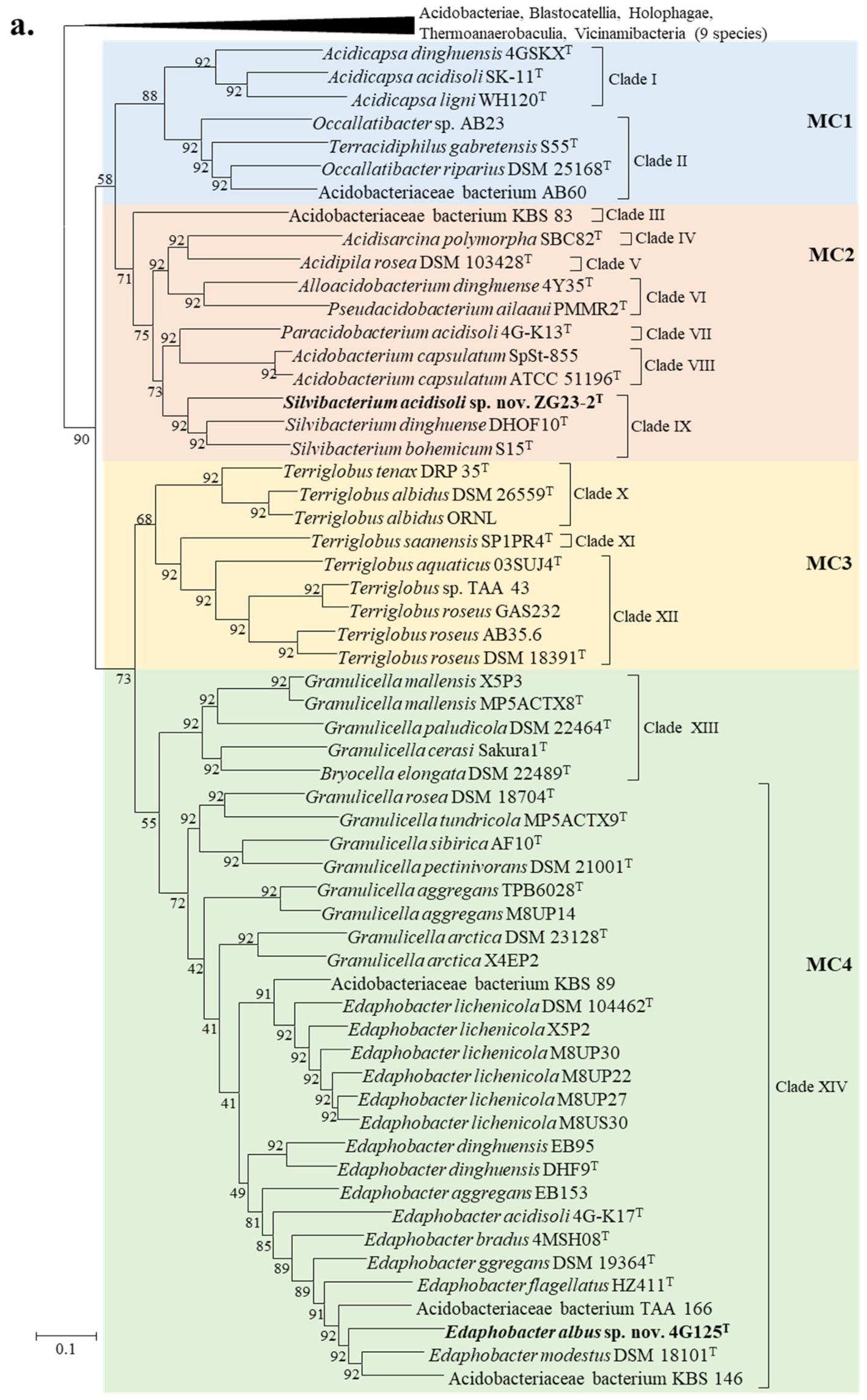
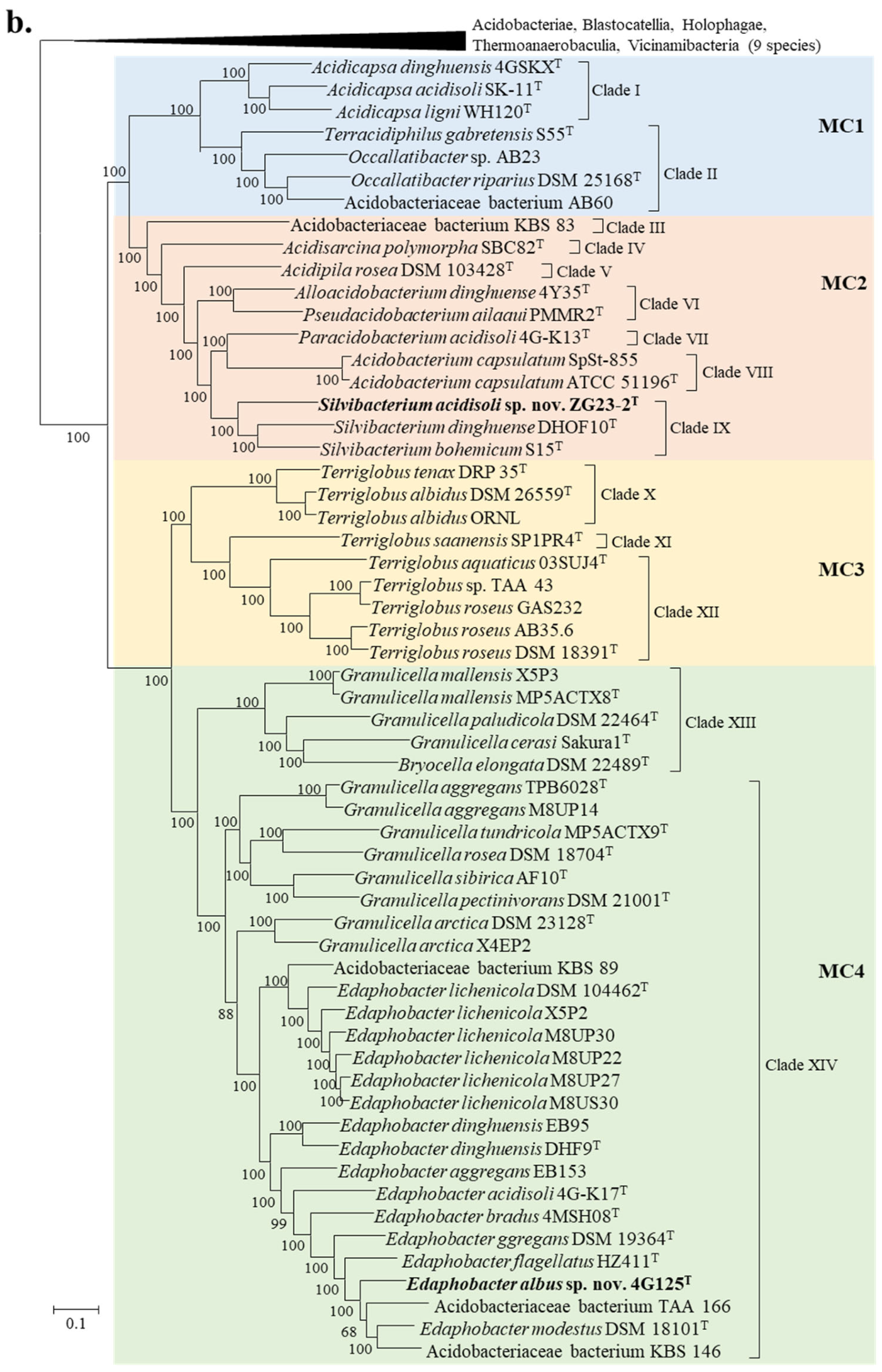
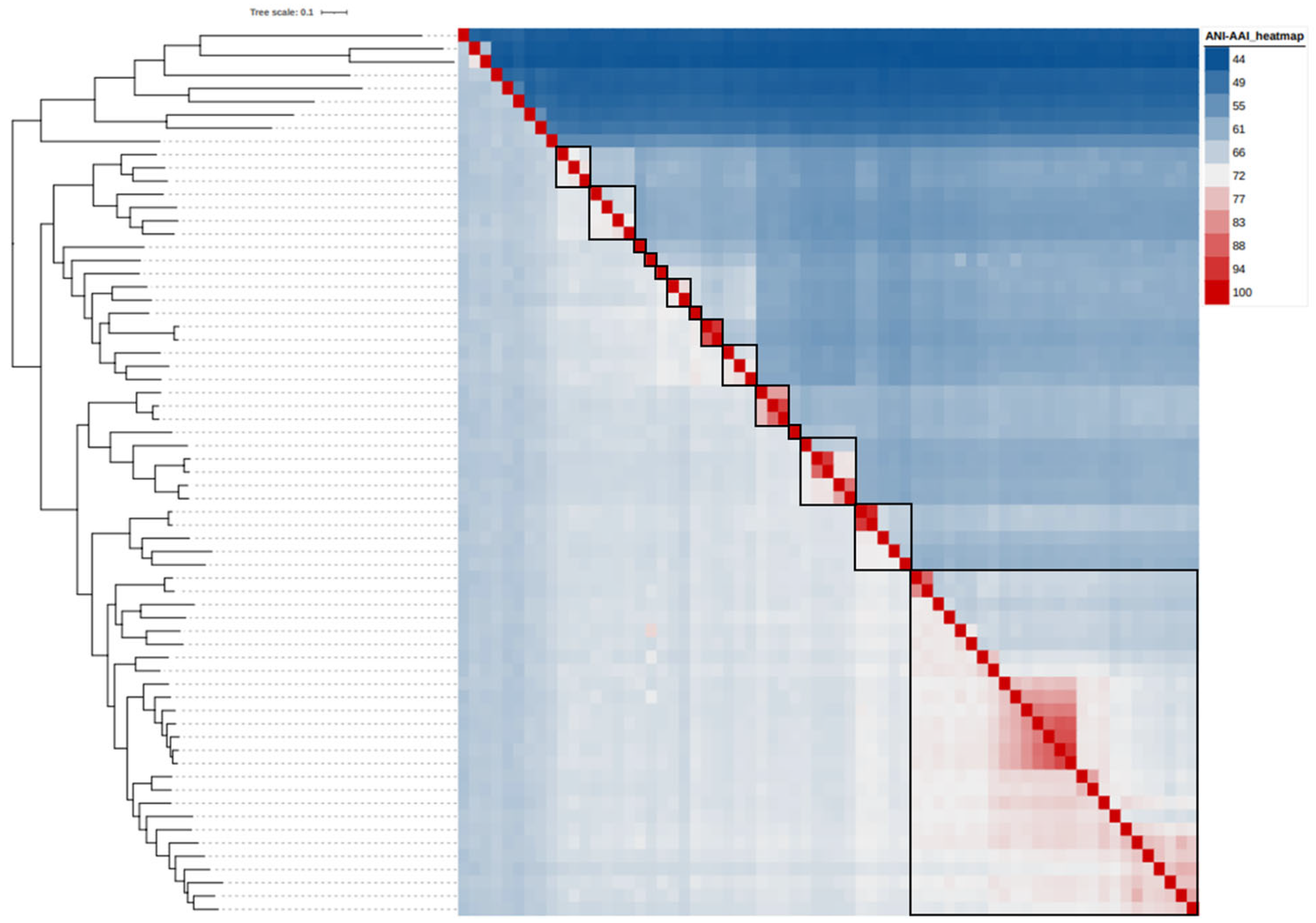
| Strain | Genome Accession | Genome Size (Mb) | Scaffold count | N50 Scaffold (bp) | (G+C)% | Gene Number |
|---|---|---|---|---|---|---|
| Acidicapsa dinghuensis 4GSKXT | JAGSYH000000000 | 6.17 | 17 | 816,156 | 56.47 | 5125 |
| Acidicapsaligni WH120T | JAGSYG000000000 | 5.68 | 17 | 961,249 | 55.41 | 4697 |
| Acidicapsa acidisoli SK-11T | JAGSYI000000000 | 7.11 | 18 | 1,906,827 | 57.13 | 5905 |
| Terracidiphilus gabretensis S55T | GCA_001449115.1 | 5.35 | 17 | 729,622 | 57.30 | 4426 |
| Occallatibacter riparius DSM 25168T | CP093313 | 6.79 | 1 | 6,794,547 | 60.23 | 5637 |
| Acidobacteriaceae bacterium AB60 | GCA_008682415.1 | 6.65 | 33 | 466,258 | 61.86 | 5574 |
| Occallatibacter sp. AB23 | GCA_003131205.1 | 6.28 | 18 | 795,662 | 59.10 | 5356 |
| Acidobacteriaceae bacterium KBS 83 | GCA_000381585.1 | 6.25 | 21 | 826,801 | 59.14 | 5419 |
| Acidipilarosea DSM 103428T | GCA_004339725.1 | 4.21 | 15 | 1,147,137 | 58.81 | 3513 |
| Silvibacterium bohemicum S15T | GCA_001006305.1 | 6.46 | 25 | 883,337 | 58.22 | 5345 |
| Acidipila dinghuensis DHOF10T | GCA_004123295.1 | 5.12 | 9 | 1,191,396 | 60.50 | 3998 |
| Silvibacterium acidisolisp. nov. ZG23-2T | CP085323 | 5.36 | 1 | 5,359,900 | 58.40 | 4344 |
| Acidisarcina polymorpha SBC82T | GCA_003330725.1 | 7.60 | 5 | 7,112,011 | 56.87 | 6702 |
| Pseudacidobacterium dinghuensesp. nov. 4Y35T | GCA_014274465.1 | 6.27 | 1 | 6,271,114 | 55.79 | 5475 |
| Acidobacterium ailaaui PMMR2T | GCA_000688455.1 | 3.69 | 1 | 3,686,523 | 56.49 | 3163 |
| Acidobacterium capsulatum ATCC 51196T | GCA_000022565.1 | 4.13 | 1 | 4,127,356 | 60.50 | 3365 |
| Acidobacterium capsulatum SpSt-855 | GCA_011333575.1 | 3.16 | 81 | 66,937 | 60.99 | 2719 |
| Paracidobacterium acidisoligen. nov., sp. nov. 4G-Kl3T | GCA_003428625.2 | 5.00 | 16 | 846,090 | 60.46 | 4022 |
| Terriglobus roseus AB35.6 | GCA_900105625.1 | 4.81 | 2 | 4,742,757 | 59.89 | 4003 |
| Terriglobus roseus DSM 18391T | GCA_000265425.1 | 5.23 | 1 | 5,227,858 | 60.28 | 4347 |
| Terriglobus roseus GAS232 | GCA_900102185.1 | 4.85 | 1 | 4,852,066 | 56.45 | 4097 |
| Terriglobus sp. TAA 43 | GCA_000800015.1 | 4.95 | 7 | 3,477,257 | 56.69 | 4213 |
| Terriglobus aquaticus 03SUJ4T | JAGSYB000000000 | 4.16 | 2 | 4,126,416 | 62.71 | 3452 |
| Terriglobus saanensis SP1PR4T | GCA_000179915.2 | 5.10 | 1 | 5,095,226 | 57.34 | 4267 |
| Terriglobus tenax DRP35T | JAGSYA000000000 | 4.36 | 4 | 3,021,852 | 59.76 | 3697 |
| Teriglobus albidus DSM26559T | JAGTAS000000000 | 6.10 | 58 | 220,284 | 57.56 | 4963 |
| Terriglobus albidus ORNL | GCA_008000815.1 | 6.41 | 1 | 6,405,582 | 57.90 | 5217 |
| Granulicella cerasi SakuralT | JAGSYD000000000 | 4.08 | 7 | 978,916 | 60.82 | 3418 |
| Granulicella paludicola DSM22464T | JAGSYC000000000 | 4.96 | 26 | 630,162 | 58.99 | 4031 |
| Granulicella mallensis MP5ACTX8T | GCA_000178955.2 | 6.24 | 1 | 6,237,577 | 57.91 | 4899 |
| Granulicella mallensis X5P3 | GCA_014203225.1 | 6.47 | 52 | 253,602 | 57.80 | 5186 |
| Bryocella elongata DSM 22489T | GCA_900108185.1 | 5.67 | 16 | 661,247 | 62.00 | 4535 |
| Granulicella aggregans M8UP14 | GCA_014203275.1 | 7.30 | 83 | 844,690 | 58.59 | 6188 |
| Granulicella aggregans TPB6028T | JAGSYE000000000 | 5.74 | 14 | 1,416,590 | 59.83 | 4653 |
| Granulicella pectinivorans DSM 21001T | GCA_900114625.1 | 5.28 | 3 | 4,439,413 | 61.28 | 4282 |
| Grandicella sibirica AF10T | GCA_004115155.1 | 6.14 | 16 | 1,421,513 | 59.82 | 5140 |
| Grandicella rosea DSM 18704T | GCA_900188085.1 | 5.29 | 24 | 586,566 | 62.91 | 4296 |
| Granulicella arctica DSM 23128T | JAGTUT000000000 | 5.85 | 9 | 4,736,692 | 57.57 | 5138 |
| Granulicella arctica X4EP2 | GCA_013410065.1 | 4.31 | 3 | 2,159,447 | 58.26 | 3645 |
| Granulicella tundricola MP5ACTX9T | GCA_000178975.2 | 5.50 | 6 | 4,309,153 | 59.98 | 4715 |
| Acidobacteriaceae bacterium KBS 89 | GCA_000381605.1 | 6.01 | 13 | 943,631 | 57.55 | 5055 |
| Edaphobacter lichenicola DSM 104462T | CP073696 | 5.66 | 1 | 5,662,239 | 56.48 | 4891 |
| Edaphobacter lichenicola X5P2 | GCA_014201335.1 | 6.18 | 23 | 507,847 | 56.41 | 5415 |
| Edaphobacter lichenicola M8UP30 | GCA_013410115.1 | 5.29 | 4 | 2,888,932 | 57.56 | 4466 |
| Edaphobacter lichenicola M8UP22 | GCA_013410875.1 | 5.07 | 5 | 2,909,133 | 57.73 | 4322 |
| Edaphobacter lichenicola M8UP27 | GCA_014201315.1 | 4.88 | 15 | 1,190,613 | 58.40 | 4146 |
| Edaphobacter lichenicola M8US30 | GCA_014201375.1 | 4.83 | 15 | 711,311 | 58.18 | 4171 |
| Edaphobacter dinghuensisDHF9T | JAGSYJ000000000 | 4.54 | 11 | 1,438,063 | 56.93 | 3732 |
| Edaphobacter dinghuensis EB95 | GCA_003633965.1 | 4.46 | 2 | 3,541,529 | 55.56 | 3854 |
| Edaphobacter acidisoli 4G-K17T | JAGSYK000000000 | 4.20 | 9 | 1,158,799 | 58.44 | 3493 |
| Edaphobacter aggregans DSM 19364T | GCA_000745965.1 | 8.18 | 345 | 76,811 | 58.70 | 7425 |
| Edaphobacter aggregans EB153 | GCA_003945235.1 | 6.15 | 1 | 6,152,256 | 57.25 | 5109 |
| Edaphobacter bradus 4MSH08T | JAGSYF000000000 | 4.75 | 13 | 1,458,508 | 59.44 | 4076 |
| Edaphobacter albussp.nov.4G125T | GCA_014274685.1 | 4.25 | 1 | 4,254,614 | 55.03 | 3605 |
| Edaphobacter flagellatus HZ411T | CP073697 | 4.46 | 1 | 4,445,627 | 57.41 | 3727 |
| Edaphobacter modestus DSM 18101T | GCA_004217555.1 | 7.45 | 8 | 6,121,180 | 57.10 | 6642 |
| Acidobacteriaceae bacterium KBS 146 | GCA_000688615.1 | 5.00 | 2 | 4,996,384 | 56.74 | 4161 |
| Acidobacteriaceae bacterium TAA 166 | GCA_000421065.1 | 6.14 | 3 | 4,712,210 | 58.83 | 5269 |
| Candidatus Koribacter versatilis Ellin345 | GCA_000014005.1 | 5.65 | 1 | 5,650,368 | 58.38 | 4995 |
| Candidatus Solibacter usitatus Ellin6076 | GCA_000014905.1 | 9.97 | 1 | 9,965,640 | 61.90 | 8193 |
| Bryobacter aggregatus MPL3T | GCA_000702445.1 | 5.75 | 4 | 4,257,809 | 57.95 | 5060 |
| Chloracidobacterium thermophilum BT | GCA_000226295.1 | 3.70 | 2 | 2,683,362 | 61.34 | 3046 |
| Pyrinomonas methylaliphatogenes K22T | GCA_000820845.2 | 3.79 | 16 | 443,598 | 59.37 | 3165 |
| Luteitalea pratensis DSM 100886T | GCA_001618865.1 | 7.48 | 1 | 7,480,314 | 67.22 | 6260 |
| Holophagafoetida DSM 6591T | GCA_000242615.3 | 4.13 | 3 | 3,443,192 | 62.90 | 3586 |
| Geothrix fermentans DSM 14018T | GCA_000428885.1 | 3.29 | 36 | 164,722 | 68.85 | 2903 |
| Thermoanaerobaculum aquaticum MP-01T | GCA_000687145.1 | 2.66 | 68 | 115,892 | 63.01 | 2324 |
| Strain | Color | Motility | Relation to Oxygen | Capsule | Temperature Range | pH Range | NaCl (w/v,%) |
|---|---|---|---|---|---|---|---|
| 1 | Pink | + | Aerobic | ND | 10–32 | 3.2–5.2 | >300 mM |
| 2 | White | + | Aerobic | ND | 10–32 | 2.3–5.1 | >200 mM |
| 3 | Pink | − | Aerobic | + | 10–33 | 2.5–7.3 | 0–2.0 |
| 4 | White | − | Aerobic | + | 10–33 | 3.5–6.4 | 0–2.0 |
| 5 | White | − | Aerobic | + | 10–35 | 4.0–5.5 | 0–0.4 |
| 6 | Cream | − | Aerobic | − | 12–37 | 4.0–6.5 | 0–1.0 |
| 7 | Opaque | + | Facultatively anaerobic | − | 4–35 | 3.0–6.0 | 0–0.06 |
| 8 | White | + | Aerobic | + | 11–40 | 3.5–8.5 | 0–1.0 |
| 9 | Pink | + | Aerobic | + | 11–40 | 3.5–6.5 | 0–0.5 |
| 10 | White | + | Aerobic | + | 12–30 | 3.0–6.0 | 0–0.5 |
| 11 | White | − | Microaerophilic | + | 12–37 | 4.5–6.0 | ND |
| 12 | White | − | Facultatively anaerobic | − | 5–36 | 4.0–7.7 | 0–1.5 |
| 13 | Pink | − | Aerobic | + | 22–37 | 3.0–6.0 | 0–1.0 |
| 14 | Cream | − | Facultatively anaerobic | + | 15–55 | 4.5–7.0 | 0–1.0 |
| 15 | White | − | Aerobic | − | 12–37 | 3.5–6.5 | 0–2.0 |
| 16 | Pale yellow | − | Aerobic | − | 12–37 | 3.5–6.5 | 0–1.0 |
| 17 | Orange | + | Aerobic | + | 25–37 | 3.0–6.0 | 0–3.5 |
| 18 | Transparent | − | Aerobic | + | 12–33 | 3.5–6.0 | 0–2.5 |
| 19 | White | + | Aerobic | − | 20–30 | 3.0–6.0 | 0–0.1 |
| 20 | White | − | Aerobic | + | 10–37 | 3.5–8.0 | 0–1.0 |
| 21 | White | − | Aerobic | − | 10–42 | 3.9–9.8 | ND |
| 22 | Cream | − | Aerobic | ND | 15–45 | 3.5–7.0 | 0–1.0 |
| 23 | Cream | − | Aerobic | − | 4–30 | 4.5–7.5 | ND |
| 24 | Pink | − | Aerobic | ND | 15–30 | 6.0–7.0 | 0–1.0 |
| 25 | Pink | − | Facultatively anaerobic | + | 12–23 | 5.0–7.0 | ND |
| 26 | White | − | Facultatively anaerobic | + | 12–23 | 5.0–6.5 | ND |
| 27 | Pink | − | Aerobic | + | 6–32 | 3.2–6.6 | 0–3.0 |
| 28 | Red | − | Aerobic | − | 2–33 | 3.0–7.5 | 0–3.5 |
| 29 | White | − | Aerobic | ND | 10–32 | 2.3–5.3 | >300 mM |
| 30 | White | − | Aerobic | − | 4–28 | 3.5–6.5 | 0–1.5 |
| 31 | Pink | − | Aerobic | − | 10–30 | 4.5–8.5 | 0–1.0 |
| 32 | Red | − | Aerobic | − | 2–33 | 3.0–7.5 | 0–3.5 |
| 33 | Red | − | Aerobic | − | 2–33 | 3.0–7.5 | 0–3.5 |
| 34 | Pink | − | Aerobic | − | 2–33 | 3.0–7.5 | 0–3.5 |
| 35 | Pink | − | Aerobic | − | 4–28 | 3.5–6.5 | 0–1.0 |
| 36 | Pale–pink | − | Aerobic | − | 20–25 | 4.5–5.0 | 0–1.5 |
| 37 | White | − | Aerobic | − | 4–26 | 3.5–7.0 | 0–1.0 |
| 38 | Pink | − | Aerobic | − | 4–28 | 3.5–6.5 | 0–1.0 |
| 39 | Cream | − | Aerobic | − | 10–42 | 3.0–7.0 | 0–2.5 |
| 40 | Cream | + | Aerobic | − | 15–30 | 4.5–7.0 | ND |
| 41 | Cream | − | Aerobic | − | 15–37 | 4.0–7.0 | ND |
| 42 | Faint yellow | + | Aerobic | − | 10–37 | 3.5–6.5 | 0–2.5 |
| 43 | White | + | Aerobic | + | 12–37 | 4.0–7.0 | 0–3.0 |
| 44 | Faint yellow | − | Aerobic | − | 10–37 | 3.0–6.5 | 0–3.0 |
| 45 | Cream | − | Aerobic | − | 10–33 | 3.5–5.5 | 0–2.0 |
| 46 | Pink | − | Aerobic | − | 7–37 | 3.4–7.0 | 0–1.0 |
| 47 | White | − | Facultatively anaerobic | + | 12–23 | ND | ND |
| 48 | White | − | Facultatively anaerobic | + | 12–23 | 5.0–7.0 | ND |
| Analyses Method | Strain | C14:0 | C15:0 | C16:0 | C17:0 | C18:0 | iso-C13:0 | iso-C15:0 | C14:0 anteiso | C14:1ω5c | iso-C15:0 3-OH | C16:1ω7c | C17:1ω8c | C18:1ω9c | iso-C17:0 | C17:0 cyclo | iso-C17:1ω7c | 13,16-dimethyl-octacosanedioic acid | Summed Features:1 | Summed Features:8 | Summed Features:9 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIDI | 1 | 0.3 | 0.1 | 0.8 | 0.8 | 0.2 | 69.3 | 0.1 | 2.9 | 0.5 | 0.1 | 5.9 | 17.2 | 0.7 | |||||||
| MIDI | 2 | 0.5 | 80.7 | 0.7 | 0.9 | 1.9 | 13.0 | 1.8 | |||||||||||||
| GC-MS | 3 | 0.9 | 13 | 45.9 | 3.4 | 21.9 | 21.9 | ||||||||||||||
| GC-MS | 4 | 0.4 | 1.4 | 0.7 | 40.7 | 3.9 | 13 | 32.8 | |||||||||||||
| MIDI | 5 | 2.9 | 0.5 | 0.5 | 55.4 | 2.2B | 0.5 | 16.7 | 0.6 | 17.7 | |||||||||||
| MIDI | 6 | 7.3 | 48.8 | 2.1B | 5.2 | 8.1 | 3.5 | 14.7 | |||||||||||||
| MIDI | 7 | 0.1 | 1.5 | 0.2 | 0.5 | 69.8 | 0.4A | 5.4 | 14.8C | 1.9 | |||||||||||
| MIDI | 8 | 0.5 | 61.9 | 0.4B | 5.3 | 29 | |||||||||||||||
| MIDI | 9 | 0.6 | 54.8 | 0.9B | 9.5 | 30.7 | |||||||||||||||
| MIDI | 10 | 3.94 | 0.76 | 45.78 | 4.0B | 12.17 | 22.37 | ||||||||||||||
| GC-MS | 11 | 0.4 | 8.1 | 0.9 | 29.4 | 20.4 | 1.5 | 30.7 | |||||||||||||
| GC-MS | 12 | 0.5–1.6 | 8.7–13.3 | 1.5–2.1 | 1.3–1.9 | 49.9–53.1 | 25.3–26.5 | 1.5–2.4 | |||||||||||||
| MIDI | 13 | 0.8 | 2.5 | 0.1 | 3.8 | 60.6 | 1.1 | 0.2 | 0.1 | 4.2B | 0.1 | 0.4 | 11.4 | 1 | 8.8 | ||||||
| MIDI | 14 | 1.14 | 6.49 | 0.97 | 1.43 | 0.09 | 52.3 | 2.41 | 0.15 | 10.47B | 1.77 | 5.39 | 4.74 | 3.12 | 5.77 | ||||||
| MIDI | 15 | 0.4 | 3 | 2.69 | 3.8 | 56.61 | 0.08 | 6.24B | 3.1 | 3.25 | 6.09 | 1.47 | 8.08 | ||||||||
| MIDI | 16 | 1.36 | 4.21 | 0.86 | 2.05 | 0.12 | 50.91 | 0.18 | 5.09B | 16.25 | 1.76 | 0.1 | 0.57 | 1.59 | |||||||
| MIDI | 17 | 1.37 | 5.86 | 1.69 | 4 | 37.66 | 0.37 | 15.23B | 3.19 | 8.54 | 1.64 | 1.74 | 2.96 | 3.51 | |||||||
| MIDI | 18 | 0.96 | 5.28 | 0.2 | 1.99 | 54.56 | 0.34 | 0.07 | 13.31B | 0.92 | 7.28 | 4.3 | 0.99 | 1.5 | 4.58 | ||||||
| MIDI | 19 | 1.7 | 8.6 | 2.7 | 3.7 | 48.6 | 11.5B | 17 | |||||||||||||
| MIDI | 20 | 0.6 | 3.52 | 0.07 | 0.6 | 60.19 | 1.88 | 0.32 | 0.14 | 23.6B | 0.08 | 0.76 | 0.72 | 1.64 | 1.59 | ||||||
| MIDI | 21 | 0.94 | 6.1 | 0.08 | 0.72 | 0.05 | 53.34 | 0.73 | 0.45 | 0.07 | 32.79B | 0.03 | 0.65 | 0.42 | 1.24 | 0.65 | |||||
| GC-MS | 22 | 2.1 | 0.4 | 8.4 | 0.5 | 0.4 | 9.8 | 39.9 | 28.4 | 1.7 | |||||||||||
| MIDI | 23 | 6.8 | 9.1 | 2.8 | 44.5 | 2.7 | 22.5B | 0.8 | |||||||||||||
| MIDI | 24 | 3.4 | 7.77 | 43.88 | 27.08B | ||||||||||||||||
| MIDI | 25 | 4.25 | 0.74 | 9.14 | 0.68 | 36.33 | 0.67 | 30.24A | 0.79 | ||||||||||||
| MIDI | 26 | 0.96 | 5.02 | 0.97 | 0.27 | 47.58 | 0.46 | 30.15B | 1.47 | 0.71 | 0.88 | 1.83 | |||||||||
| MIDI | 27 | 1.8 | 13.2 | 0.9 | 0.5 | 13.1 | 43.3 | 1.1 | 0.3 | 0.1 | 18.3B | 0.1 | 0.1 | 1.9 | 0.1 | 0.2 | 0.4 | ||||
| MIDI | 28 | 0.6 | 3.2 | 68.5 | 21B | 0.8 | 3.7 | 0.9 | |||||||||||||
| MIDI | 29 | 1.39 | 11.15 | 0.11 | 0.47 | 10.64 | 45.26 | 0.37 | 0.14 | 22.54B | 1.49 | 0.17 | 0.76 | 0.67 | |||||||
| MIDI | 30 | 4.45 | 6.29 | 0.22 | 6.83 | 49.24 | 2.45 | 24.6B | 0.1 | 0.5 | 0.36 | 0.29 | |||||||||
| MIDI | 31 | 2.5 | 0.4 | 6 | 0.5 | 0.3 | 0.4 | 52.8 | 27.7B | 1.5 | |||||||||||
| MIDI | 32 | 4.04 | 14.63 | 0.66 | 0.85 | 0.08 | 43.76 | 0.89 | 25.64B | 0.15 | 1.43 | 1.31 | 0.13 | 0.5 | |||||||
| MIDI | 33 | 1.07 | 7.44 | 0.3 | 1.16 | 0.84 | 62.28 | 0.25 | 0.15 | 18.81B | 0.08 | 3.16 | 0.02 | 0.34 | 0.83 | 1.04 | |||||
| MIDI | 34 | 1.09 | 5.99 | 0.5 | 0.84 | 31.03 | 1.13 | 6.95 | 35.63B | 0.22 | 0.19 | 0.87 | 0.18 | 0.92 | |||||||
| GC-MS | 35 | 3 | 6.6 | 0.6 | 0.4 | 46.4 | 35 | 3.3 | |||||||||||||
| GC-MS | 36 | 1.9 | 7.5 | 0.2 | 0.7 | 18.7 | 27 | 1.6 | 38.9 | ||||||||||||
| GC-MS | 37 | 1.6 | 4.6 | 0.2 | 51.5 | 35.3 | 1.8 | ||||||||||||||
| MIDI | 38 | 2.3 | 17.3 | 6.3 | 44.9 | 20.2B | 1.8 | ||||||||||||||
| MIDI | 39 | 2.25 | 11.29 | 0.53 | 0.49 | 38.93 | 0.83 | 35.83 | 1.67 | 3.05 | 1.84C | ||||||||||
| MIDI | 40 | 1.98 | 7.13 | 0.1 | 0.49 | 44.09 | 1.52 | 0.25 | 34.3 | 0.16 | 2 | 0.37 | 0.1 | 2.11 | |||||||
| MIDI | 41 | 2 | 9.6 | 2.2 | 39.8 | 38.1 | |||||||||||||||
| MIDI | 42 | 0.99 | 8.95 | 0.67 | 1.52 | 40.44 | 4.7 | 0.21 | 29.37B | 0.18 | 0.39 | 2.18 | 2.17 | ||||||||
| MIDI | 43 | 1.9 | 8 | 1.2 | 46.1 | 29.5 | |||||||||||||||
| MIDI | 44 | 1.6 | 10.2 | 2.2 | 4.3 | 36.7 | 31.6B | ||||||||||||||
| MIDI | 45 | 1.58 | 13.58 | 0.17 | 0.7 | 2.94 | 52.05 | 0.5 | 20.56B | 0.24 | 2.54 | 0.7 | 0.38 | 0.76 | |||||||
| MIDI | 46 | 1.16 | 7.33 | 0.75 | 4.12 | 47.09 | 1 | 26.18A | 1.25C | ||||||||||||
| MIDI | 47 | 3.68 | 8.71 | 0.59 | 37.06 | 0.57 | 26.99A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, L.; Cao, L. Description of Silvibacterium acidisoli sp. nov. and Edaphobacter albus sp. nov. and a Proposal for Taxonomic Rearrangements Within the Family Acidobacteriaceae Based on Comparative Genome Analysis. Taxonomy 2025, 5, 40. https://doi.org/10.3390/taxonomy5030040
Qiu L, Cao L. Description of Silvibacterium acidisoli sp. nov. and Edaphobacter albus sp. nov. and a Proposal for Taxonomic Rearrangements Within the Family Acidobacteriaceae Based on Comparative Genome Analysis. Taxonomy. 2025; 5(3):40. https://doi.org/10.3390/taxonomy5030040
Chicago/Turabian StyleQiu, Lihong, and Lixiang Cao. 2025. "Description of Silvibacterium acidisoli sp. nov. and Edaphobacter albus sp. nov. and a Proposal for Taxonomic Rearrangements Within the Family Acidobacteriaceae Based on Comparative Genome Analysis" Taxonomy 5, no. 3: 40. https://doi.org/10.3390/taxonomy5030040
APA StyleQiu, L., & Cao, L. (2025). Description of Silvibacterium acidisoli sp. nov. and Edaphobacter albus sp. nov. and a Proposal for Taxonomic Rearrangements Within the Family Acidobacteriaceae Based on Comparative Genome Analysis. Taxonomy, 5(3), 40. https://doi.org/10.3390/taxonomy5030040





