Abstract
The genus Xanthomonas primarily comprises phytopathogenic species. By carrying out deep phylo-taxonogenomics, we recently reported that the genera Xylella, Stenotrophomonas, and Pseudoxanthomonas are misclassified and belong to the genus Xanthomonas. Considering the importance of Xanthomonas/Xylella as plant pathogens and to further determine the taxonomic and phylogenetic breadth of this genus, we extended our earlier study by including all the reported genera and families in the order. This investigation revealed that at least four more genera belong to the genus Xanthomonas, with a notable case being Lysobacter, after which the family and order are named. Similarly, our investigation also allowed us to reveal the expanded taxonomic breadth of the related genus Rhodanobacter. This finding of a major related genus that lacks plant pathogenic species will allow for taxonomy-based comparative studies. The phylo-taxonogenomic revelations were further supported by complete 16S rRNA-based sequence boundaries proposed for genus delineation. Accordingly, we propose a taxonomic revision of these major and closely related genera along with their constituent families within the order Lysobacteraceae (Xanthomonadaceae). The identification of a major related genus lacking plant pathogenic species will be important in investigating the origin and success of pathogenic species/lineages in the genus Xanthomonas.
1. Introduction
According to the current standing in nomenclature, the order Lysobacterales (also known as Xanthomonadales) constitutes two families, namely, Lysobacteraceae and Rhodanobacteraceae. Both of these complex families consist of more than 30 genera and around 300 species [1]. These include diverse species of phytopathogens, environmental bacteria, and opportunistic human pathogens [2]. The Lysobacteraceae (Xanthomonadaceae) family contains Xanthomonas and Xylella, the two major phytopathogenic genera with significant economic and agricultural impacts [3,4,5,6]. The member species of the Xanthomonas and Xylella genera infect a wide range of crops and plants across the globe. On the other hand, the family also includes the metabolically versatile genus Stenotrophomonas, with one of its species, S. maltophilia, being a multidrug-resistant opportunistic pathogen responsible for hospital-acquired infections in immunocompromised patients [7]. Interestingly, Stenotrophomonas was earlier classified as Xanthomonas, and there are an increasing number of reports of Stenotrophomonas spp. as plant pathogens [8,9,10]. At the same time, there are several reports of novel species of Xanthomonas that are non-pathogenic in nature [8,9], suggesting plant associated and environmental lifestyle of Xanthomonas. Other environmental genera in this family include Pseudomarimonas, Lysobacter, and Vulcaniibacterium, which consist of species of industrial importance causing the leaching of heavy metals and bioremediation [10,11,12]. The synthesis of extracellular enzymes by members of the Lysobacter group has attracted considerable attention. The group is also considered as a prolific source for the creation of novel antibiotics, including β-lactams [13], macrocyclic lactams, and antibiotics that contain macrocyclic peptides or depsipeptides, such as katanosins [14]. Recent research has shown that Lysobacter species, including L. enzymogenes C3, have the potential to act as biological pest controllers for plant diseases [15]. On the other hand, the family Rhodanobacteraceae [1] consists of genera encoding functions required for the bioremediation of hydrocarbons, etc. [16,17]. Members of the genus Dyella act as potential biocontrol agents against grapevine yellows [18]. Genera like Fulvimonas and Aerosticca are isolated from soil after enrichment with acetylated starch plastic and from crude oil-contaminated soil [17]. The species of the genus Oleiagrimonas, like Oleiagrimonas soli, play a special role in oil contamination bioremediation [19]. The member species of Rhodanobacter like R. denitrificans and R. thiooxydans are acid-tolerant denitrifiers that are well-suited to acidic, nitrate-rich subsurface conditions. pH is proven to be the primary driver of bacterial community structure in this contaminated subterranean environment [20]. In acidic tundra soils that were experimentally modified with long-term nutrient fertilization, Campbell and co-workers reported that bacteria belonging to the Dyella and Rhodanobacter lineages were present in significant abundance [21].
The integration of classical taxonomy with genomic evidence has revolutionized the field of microbial systematics [22,23]. Apart from species-level reclassifications, the implementation of genome-based methods has enabled reconciliations on the order, family, and genus levels [24,25]. Average nucleotide identity (ANI) and digital DNA-DNA hybridization (dDDH) are widely used for species delineation; however, these indices are not ideal for genus-level taxonomy because of their reliance on nucleotide identity [26,27]. Average amino acid identity (AAI) [28] and the percentage of conserved proteins (POCP) [29] have been proposed for genus-level classification. AAI is based on the similarity calculation of protein sequences common to the two bacterial isolates and allows for the comparison of diversely related isolates. Unlike nucleotide information, the conservation reflected in the amino acid sequence similarities enables the delineation of high taxa, especially genera [24,28]. However, the POCP only accounts for the presence or absence of proteins and is not suitable when the relatives have a reduced genome size, as in the case of Xylella, which was reported as a variant Xanthomonas lineage with a highly reduced genome size and G+C content [30]. Hence, AAI is more robust for investigating taxonomy on the level of the genus, irrespective of a reduced genome size and G+C content [8,26,28]. Though the genus level threshold for AAI is assigned as 60–80% [26,31,32], Meehan and co-workers used 65% and Zheng et al. used 68% as cut-offs for the genus delineation of the genera Mycobacterium and Lactobacillus, respectively [24,25]. Furthermore, whereas AAI is typically calculated from all the genes shared between a pair of organisms [28,33], the AAI values for the core genes (cAAI) are employed as a robust parameter for genus delineation. In addition to these genome similarity criteria, whole genome-based phylogeny provides us with a robust phylogenomic framework with which to verify appropriate AAI criteria based on the specific thresholds or boundaries of identified lineage(s) that do not violate the monophyly rule [34,35,36]. Hence, there is an excellent opportunity and need to understand the taxonomic boundaries of the genus Xanthomonas within the family Lysobacteraceae (Xanthomonadaceae) and the order Lysobacterales (Xanthomonadales).
The current taxonomy of the order Lysobacterales (Xanthomonadales) is based on 16S rRNA analysis, a limited number of molecular markers, conserved sequence indels (CSI), and a phylogenetic tree based on no more than 30 conserved proteins [1]. In an earlier study, by carrying out an in-depth phylo-taxonogenomic investigation of the order Lysobacterales, we reported major reshufflings on the family level, in addition to revealing the boundary of the order and its outliers [10]. In a follow-up study, by carrying out an in-depth phylo-taxonogenomic investigation of the genus Xanthomonas and its close relatives, we also reported that the member species of the genera Pseudoxanthomonas, Stenotrophomonas, and Xylella belong to a single genus [30]. As an extension of this study, herein, we re-evaluate the genus boundary of Xanthomonas by including all the species of all the reported genera within the family and the order Lysobacterales (Xanthomonadales). In addition to the genus Xanthomonas and its family, this study also allowed us to revise the taxonomic and phylogenetic breadth of the genus Rhodanobacter and its family within the order Lysobacterales.
2. Materials and Methods
2.1. Genome Access and Quality Assessment
All the information about the members within the order Lysobacterales (Xanthomonadales) was obtained from their LPSN (List of Prokaryotic Names with Standing in Nomenclature) (https://lpsn.dsmz.de/order/lysobacterales, accessed on 14 October 2021). The genomes of the strains used in the study were obtained from NCBI microbes (https://www.ncbi.nlm.nih.gov/genome/microbes/, accessed on 14 October 2021). The quality check of the genomes was carried out using CheckM v1.1.3 [37]. The genomes that passed the QC (<5% contamination and >95% completeness) were considered and annotated using Prokka v1.14.6 [38].
2.2. Phylogenomic Investigation of the Members of the Order Lysobacterales (Xanthomonadales)
The core genome phylogeny was generated using PhyloPhlAn 3.0, which is based on nearly 400 conserved genes. The PhyloPhlAn tool employs a phylogenetic pipeline that is both modular and parallel and can be customized to suit the user’s needs. The pipeline commences with the identification of phylogenetic markers from the input sequences, culminating in the inference of the final tree [39]. Here, USEARCH v5.2.32 [40] was implemented for ortholog searching, MUSCLE v3.8.3 [41] was used for multiple sequence alignment, and FastTree v2.1 [42] was used for phylogenetic construction. In order to obtain a more robust phylogeny, we fetched the core gene using PIRATE (GNU GPL v3.0) [43]. PIRATE is suitable for the identification of ortholog groups of divergent genomes using amino acid identity thresholds of 50%, 60%, 70%, 80%, 90%, and 95%. PIRATE executes a pangenome pipeline to fetch the core genome with a high level of robustness (https://figshare.com/s/bcc8e94bca580fbb57f8).
2.3. Taxonogenomic Assessment of the Members of the Order Lysobacterales (Xanthomonadales)
The genome relatedness amongst the genomes was assessed using the average amino acid identity (AAI) with CompareM v0.0.23 (https://github.com/dparks1134/CompareM), which uses the mean amino acid identity of orthologous genes between a given pair of genomes. Furthermore, for the core average amino acid identity (cAAI), the core genes amongst the type/representative species genomes were fetched from the PIRATES pangenome analysis (https://figshare.com/s/bcc8e94bca580fbb57f8). These core genes were then used to evaluate the cAAI values. The 16S rRNA phylogeny and similarity were generated using BLASTn. All the complete 16S rRNA sequences used in this study were obtained from LPSN (https://lpsn.dsmz.de/).
3. Results and Discussion
3.1. Phylogenomic Evaluation of the Families and Their Boundaries within the Order Lysobacterales (Xanthomonadales)
A total of 213 genomes belonging to type species and type strains of the order and families within Lysobacterales (Xanthomonadales) are summarized in the table (Supplementary Table S3). We also evaluated all the conserved genes of 213 species using fastANI, validating that there are no synonyms on the species level (https://figshare.com/s/bcc8e94bca580fbb57f8). Species of the genus Xanthomonas have a genome size of 5 Mb, whereas Pseudoxanthomonas and Stenotrophomonas have 3–5 Mb genomes, and Xylella’s genome is 2.5 Mb in size. Other members of the order, such as Lysobacter, Luteimonas, Thermomonas, and Vulcaniibacterium, have genomes ranging in size from 2 to 4 Mb. The average GC content of species of the genera Xanthomonas and Pseudoxanthomonas and Stenotrophomonas genomes is 60–70%, whereas species of the genus Xylella have a reduced GC of around 51%. Other species of this genus have a GC content of 65–71%. In contrast, other species of the remaining genera, such as Thermomonas, Rhodanobacter, Frateuria, Dyella, Luteibacter, etc., have 2.5–5 Mb genomes with a 60–65% GC.
A whole-genome-based phylogeny obtained using PhyloPhlAn, including the type species of all the genera reported in the order Lysobacterales (Xanthomonadales) (n = 33), revealed two major clades corresponding to the reported families of Lysobacteraceae (Xanthomonadaceae) and Rhodanobacteraceae (Figure 1). Pseudofulvimonas gallinarii and Ahniella affigens formed outgroups for these families, respectively. A whole-genome-based phylogeny, including all the type species and type strains (n = 213) of all the reported genera of the order Lysobacterales (Xanthomonadales), also revealed two major clades corresponding to the families of Lysobacteraceae (Xanthomonadaceae) and Rhodanobacteraceae (Figure 2). PhyloPhlAn uses more than 400 conserved genes from across the bacterial world, thereby providing a robust phylogenomic tree. The phylogenomic positioning of the constituent species was further confirmed using a core pangenome-based tree parameter. The implementation of PIRATE (pangenome investigation) resulted in core genome extraction from all the strains under study (n = 213). A phylogenomic tree obtained using FastTree resulted in a phylogenomic tree similar to that obtained using PhyloPhlAn and PIRATE (Figure 2 and Figure 3). In all three phylogenomic trees’ construction, we used Ignatzshineria larvae DSM 13226T and Pseudomonas aeruginosa DSM50071T as the outgroups. In these trees, Pseudofulvimonas gallinarii and Ahniella affigens also formed outgroups for these families, as mentioned earlier. In the current study, we are not proposing them as distinct families, and at the same time, we are excluding them from the known families based on the phylogenomic trees (Figure 1, Figure 2 and Figure 3). However, we are including them in the order.
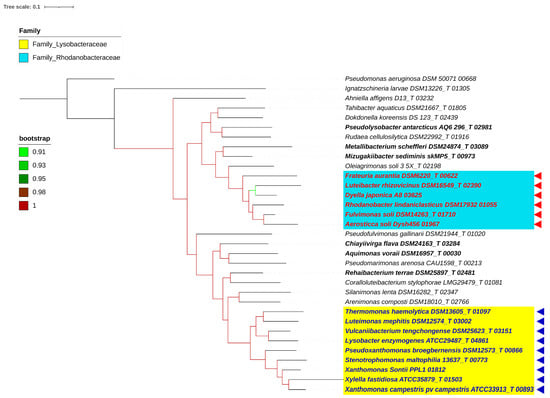
Figure 1.
PhyloPhlAn of type species. PhyloPhlAn tree comprising 33 type species and two outgroups, Pseudomonas aeruginosa DSM 50071 00668Tand Ignatzschineria larvae DSM13226T 01305. Yellow represents the Lysobacteraceae family, with blue triangles representing proposed species in the genus Xanthomonas. The sky-blue tint represents the Rhodanobacteracea family, while the red triangles represent the proposed species of the Rhodanobacter genus. The bootstrap values are displayed with color branches. Reshuffled genera are shown in bold.
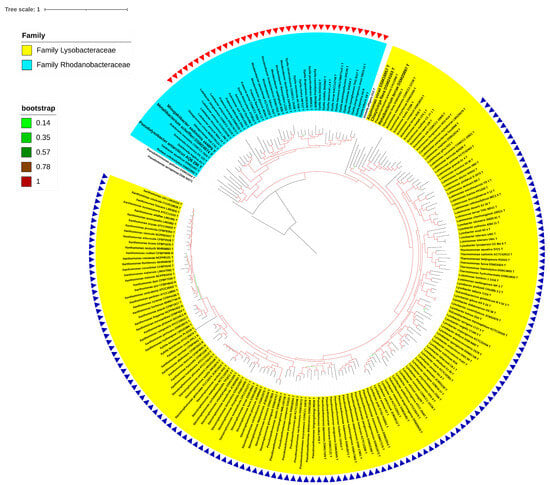
Figure 2.
PhyloPhlAn of type strains. PhyloPhlAn tree depicting 213 Type strains, with Pseudomonas aeruginosa DSM 50071 00668T serving as an outgroup. Yellow shade represents the Lysobacteraceae family, with blue triangles representing the proposed species belonging to the genus Xanthomonas. The Rhodanobacteraceae family is represented by the sky-blue hue, while the proposed species belonging to the genus Rhodanobacter are represented by the red triangles. The bootstrap values are displayed as numbers with color branches.
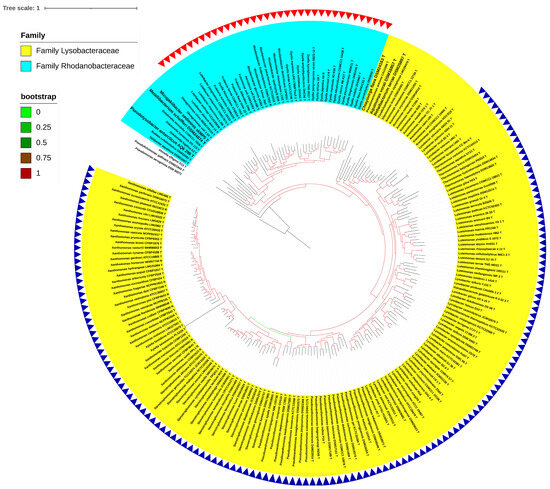
Figure 3.
PIRATE of type strains. A core-genome-based PIRATE tree is shown with 213 type strains with Pseudomonas aeruginosa DSM 50071 00668T as an outgroup. The yellow hue represents the Lysobacteraceae family, with the blue-colored triangles representing the proposed species belonging to the genus Xanthomonas. The Rhodanobacteracea family is represented by the sky-blue hue, while the proposed species belonging to the genus Rhodanobacter are represented by the red triangles. The bootstrap values are indicated as numbers with color branches.
3.2. Reshuffling of Six Genera within the Families Lysobacteraceae and Rhodanobacteraceae
Based on the previous results, we propose moving the genera Aquimonas Saha et al. 2005 [44], Chiayiivirga Hsu et al. 2013 [45], and Rehaibacterium Yu et al. 2013 [46] from the family Rhodanobacteraceae to the family Lysobacteraceae and reshuffling Mizugakiibacter Kojima et al. 2014 [47], Metallibacterium Ziegler et al. 2013 [48], and Pseudolysobacter Wei et al. 2020 [49] from the family Lysobacteraceae to the family Rhodanobacteraceae. These member genera of this family were identified through the deep order-level phylo-taxonogenomic analysis in this study.
3.3. Phylo-Taxonogenomic Assessment of the Genera of Xanthomonas and Rhodanobacter within Their Respective Families
All three phylogenomic-based trees revealed one major sub-lineage or sub-clade in each of the major clades corresponding to families, i.e., Lysobacteraceae (Xanthomonadaceae) and Rhodanobacteraceae (Frateuriaceae). The sub-lineage within Lysobacteraceae (Xanthomonadaceae) consisted of the genera Xanthomonas, Xylella [50], Pseudoxanthomonas [51], Stenotrophomonas [52], Lysobacter [53], Vulcaniibacterium [12], Luteimonas [51], and Thermomonas, while the sub-lineage corresponding to Rhodanobacteraceae (Frateuriaceae) consisted of Aerosticca [54], Fulvimonas [17], Rhodanobacter [16], Dyella [18] Luteibacter [55], and Frateuria [56].
Previously, using deep phylo-taxonogenomics, we reported that Xanthomonas, Xylella, Stenotrophomonas, and Pseudoxanthomonas belong to one genus [30]. These four genera are members of a sub-lineage identified in the family Lysobacteraceae (Xanthomonadaceae). It is possible that these four genera, along with the other four genera, are all misclassified and in fact belong to one genus. Similarly, six-member genera, i.e., Aerosticca, Fulvimonas, Rhodanobacter, Dyella, Luteibacter, and Frateuria, of the sub-lineage identified in Rhodanobacteraceae (Frateuriaceae) may also be misclassified and belong to one genus. Hence, it was necessary to confirm the phylogenomic grouping using genome-based taxonogenomic indices.
Accordingly, we used an overall genomic relatedness index (OGRI) for all the type/representative species representing different genera and families of the order. Here, according to the average amino acid identity (AAI) and core average amino acid identity (cAAI) cut-off values of 65% used for delineating a novel genus, the genus boundary for Xanthomonas is extended to include eight genera in total (Figure 4 and Figure 5). As indicated earlier, four of these genera were already reported to be synonyms in an earlier in-depth phylo-taxonogenomic study [30]. Here, according to the genus cut-off, all eight of these genera are synonyms of the genus Xanthomonas. The 65% AAI threshold also correlates with the whole-genome-based phylogenies, as these genera, with their constituent member species, form a distinct phylogroup within the family (constituent members are indicated by the bold font in the phylogenomic tree) (Figure 1, Figure 2 and Figure 3). When selecting a cut-off, it is important that the resulting genera be monophyletic [33,34]. Since the evolutionary history of these organisms is unobservable, we relied on sequence similarity and other computational metrics as proxies for evolutionary relationships. Accordingly, the various phylogenies that we present (Figure 1, Figure 2 and Figure 3) contain clade structures that are consistent with our proposed newly reorganized genera [36,57]. Similarly, according to the genus level cut-off for AAI and cAAI, the genus Rhodanobacter includes five more genera, Aerosticca, Fulvimonas, Dyella, Luteibacter, and Frateuria, that correspond to the sub-lineage identified within the family Rhodanobacteracea (Figure 4 and Figure 5). In this case, too, the 65% AAI threshold also correlates with the whole-genome-based phylogenies, as these genera, with their constituent member species, form a distinct phylogroup within the family (constituent members are indicated by the bold font in the phylogenomic tree) (Figure 1, Figure 2 and Figure 3). Even though it is reported that the vast majority of bacterial intra-genus AAI values are higher than 68%, for a particular or proposed genus under study, the threshold should pass the test of monophyly. Hence, by first establishing the monophyletic nature of the clade or group through deep genome-based phylogenetic trees using two different software (Figure 1, Figure 2 and Figure 3) and inspecting the AAI of the values (Figure 4 and Figure 5), in the case of Xanthomonas and Frateuria, we found that the cAAI and AAI cut-off 68% is also applicable to a large number of member species. Hence, considering monophyly test, we propose a cAAI and AAI cut-off of 65% as a threshold in the case of the genera Xanthomonas and Frateuria.
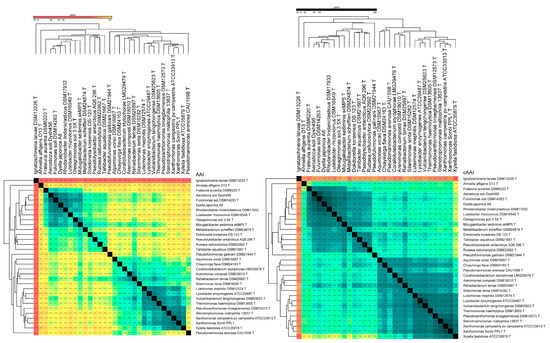
Figure 4.
AAI and cAAI of type species. Heatmap showing the average amino acid identity (AAI) and core average amino acid identity (cAAI) of 33 type species. The sky-blue color boxes symbolize the Lysobacteraceae and the Rhodanobacteraceae family.
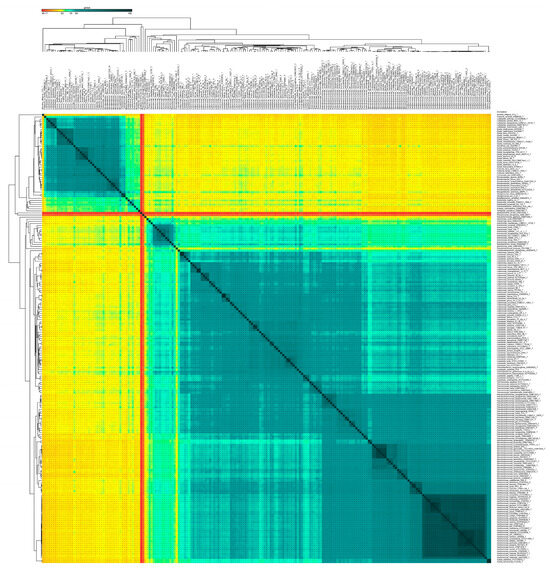
Figure 5.
AAI of type strains. Heatmap showing average amino acid identity (AAI) amongst 213 type strains. The Lysobacteraceae and the Rhodanobacteraceae families are represented by the sky-blue boxes.
3.4. Taxonomic Revision of the Families Lysobacteraceae and Rhodanobacteraceae within the Order Lysobacterales in Light of the Phylo-Taxonogenomic Findings
Since the genus Lysobacter was found to be a synonym of Xanthomonas, the genus Xanthomonas [58] has precedence over other genera, including Lysobacter [53]. Hence, there is a need to explore the validity of the family name, i.e., Lysobacteraceae, and the synonym Xanthomonadaceae, which is considered illegitimate [1]. Similarly, there is also a need to explore the validity of the order Lysobacterales [53] and its synonym, Xanthomonadales, which is considered illegitimate [2]. If the International Code of Nomenclature of Prokaryotes (2008 Revision) permits it, then there is a need to reverse the status of, or provide legitimate status to, Xanthomonadaceae and Xanthomonadales, as proposed in this study. Similarly, there is a need to check the validity of the family names, i.e., Rhodanobacteraceae and the genus Rhodanobacter, as the genus Frateuria was found to be a synonym of Rhodanobacter [16], and this genus was proposed earlier than Rhodanobacter or any other genera of that family [16].
3.5. Validating the Deep-Phylo-Taxonogenomic-Based Taxonomic Revelations at the Level of Genus with a Complete 16S rRNA Gene Sequence-Based Threshold
With the availability of a complete 16S rRNA gene sequence for all the type strains, cut-offs have been proposed for delineating higher taxa. These are 94.5% for a genus, 86.5% for a family, 82.0% for an order, 78.5% for a class, and 75.0% for a phylum [57]. Before this study, a 16S rRNA sequence identity threshold of 95%, along with a 16S rRNA-based tree, had been used for genus-level delineation [59]. Hence, in the present report, we only propose an emended or expanded description of the genera Xanthomonas and Rhodanobacter and do not propose any family or emended description of the higher taxa. Hence, we only checked the status of the genera identified in our study. The phylo-taxonogenomic findings (both the AAI threshold of 65% employed and the sub-lineages corresponding to the genera Xanthomonas and Frateuria), with the 16S rRNA boundaries proposed by Yarza and co-workers for genus delineation, support our revision of the genera Xanthomonas and Rhodanobacter (Supplementary Table S4). We do not include Oleiagrimonas soli in the emended genus Frateuria as it does not meet the strict requirement of the monophyly rule [50] and forms an outgroup in the phylogenomic tree(s) (Figure 1, Figure 2 and Figure 3). However, Xylella fastidiosa was included in the emended genus as it meets the strict requirement of the monophyly rule in the phylogenomic tree(s) (Figure 1, Figure 2 and Figure 3) meeting the 16S rRNA similarity threshold (Figure 6). In fact, Xylella fastidiosa clusters with Xanthomonas deep within the clade in the phylogenetic trees.
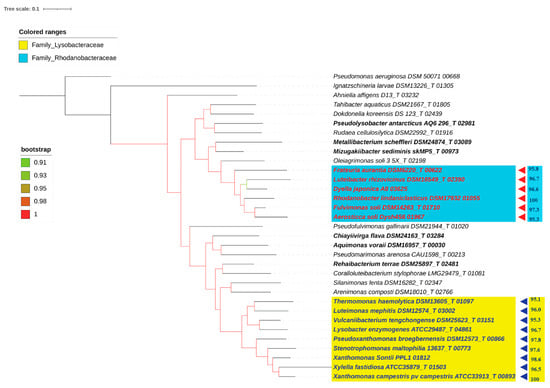
Figure 6.
PhyloPhlAn tree comprising 33 type species and two outgroups, Pseudomonas aeruginosa DSM 50071 00668T and Ignatzschineria larvae DSM13226T 01305. The 16S rRNA similarity values are shown in front of each genus. Yellow represents the Lysobacteraceae family, with blue triangles representing the proposed species in the genus Xanthomonas. The sky-blue tint represents the Rhodanobacteraceae family, while the red triangles represent the proposed species belonging to the Rhodanobacter genus. The bootstrap values are displayed with color branches. Reshuffled genera are shown in bold.
Furthermore, with our proposal for the reclassification of Lysobacter as a synonym of Xanthomonas, we propose the re-usage of the family name Xanthomonadaceae and order name Xanthomonadales that are now considered illegitimate (Figure 6).
3.6. Emended Description of the Order Xanthomonadales (Saddler and Bradbury 2005a,b) [2]
- Synonym: Lysobacterales Christensen and Cook (1978) (Approved Lists 1980)
The order consists of two families, Xanthomonadaceae and Frateuriaceae. The characteristics of the organisms in the order are as described by Naushad et al. 2014 [1] and Saddler and Bradbury (2005a,b) [60] and based on the phylo-taxonogenomic analysis of the present study. The type genus is Xanthomonas (Xanthomonas campestris (Pammel 1895) Dowson 1939 (Approved Lists 1980)), as Lysobacter (Lysobacter Christensen and Cook 1978 (Approved Lists 1980)) was re-classified as a synonym of Xanthomonas in the present study.
3.7. Emended Description of the Family Xanthomonadaceae Saddler et al. 2005
- N.L. fem. n. Xanthomonas, type genus of the family; L. fem. pl. n. suff. -aceae, ending to denote a family; N.L. fem. pl. n. Xanthomonadaceae, the Xanthomonas family.
The description of the family Xanthomonadaceae is as given by Saddler et al. 2005 and Christensen and Cook 1978 (Approved Lists 1980), with the following amendments:
- Synonym: Lysobacteraceae Christensen and Cook 1978 (Approved Lists 1980)
The family is within the order Xanthomonadales and includes the genera Xanthomonas Dowson 1939 [58], Coralloluteibacterium Chen et al. 2018 [61], Arenimonas Kwon et al. 2007 [62], Silanimonas Lee et al. 2005 [63], and Rudaea Weon et al. 2009 [64], with inclusion in Aquimonas Saha et al. 2005 [44], Chiayiivirga Hsu et al. 2013 [45], Rehaibacterium Yu et al. 2013 [46] and exclusion from Mizugakiibacter Kojima et al. 2014 [47], Metallibacterium Ziegler et al. 2013 [48], and Pseudolysobacter Wei et al. 2020 [49]. The member genera of the family were established through the latest deep phylo-taxonogenomic analysis conducted in the present study.
3.8. Description of the Family Frateuriaceae fam. nov.
Frateuriaceae (Frat.eur’i.a.a,ce’ae N.L. fem. n. Frateuria is the type genus of the family; -aceae represents the family; N.L. fem. pl. n. Frateuriaceae, the family whose nomenclature type is the genus Frateuria).
- Synonym: Rhodanobacteraceae Naushad et al. 2015 [1].
The family is within the order Xanthomonadales. The proposed family Frateuriaceae includes the genera Frateuria Swings et al. 1980 [56], Dokdonella Yoon et al. 2006 [65], Pseudolysobacter Wei et al. 2020 [49], Rudaea Weon et al. 2009 [64], and Tahibacter Makk et al. 2014 [66]. Aquimonas Saha et al. 2005 [44], Chiayiivirga Hsu et al. 2013 [45], and Rehaibacterium Yu et al. 2013 [46] are excluded from the previously described family Rhodanobacteraceae, while Mizugakiibacter Kojima et al. 2014 [47], Metallibacterium Ziegler et al. 2013 [48] and Pseudolysobacter Wei et al. 2020 are included [49]. The member genera of this family were identified through the latest deep phylo-taxonogenomic analysis conducted in the present study.
3.9. Emended Description of the Genus Xanthomonas Dowson 1939 (Approved Lists 1980)
- Xan.tho.mo.nas. Gr. masc. adj. xanthos, yellow; L. fem. n. monas, unit, monad; N.L. fem. n. Xanthomonas, yellow monad.
Synonyms: Xylella, Pseudoxanthomonas, Stenotrophomonas, Lysobacter, Vulcaniibacterium, Luteimonas and Thermomonas.
The type species is Xanthomonas campestris (Pammel 1895) Dowson 1939 (Approved Lists 1980)).
The description is as provided in Naushad et al. and Saddler et al. [1,2] and based on the deep phylo-taxonogenomic analysis conducted in the present study. The genus now includes the previously described genera and their member species, i.e., Xylella [50], Pseudoxanthomonas [51], Stenotrophomonas [52], Lysobacter [53], Vulcaniibacterium [12], Luteimonas [51], and Thermomonas [67] (Supplementary Table S1). Xanthomonas is a synonym of the previously described genera Xylella, Pseudoxanthomonas, Stenotrophomonas, Lysobacter, Vulcaniibacterium, Luteimonas, and Thermomonas (Supplementary Table S1).
Emended descriptions of the key member species of the genus Xanthomonas are provided below and in Supplementary Table S1.
- Emended description of Xanthomonas maltophilia = Stenotrophomonas maltophilia ((Hugh 1981) Swings et al. 1983).
- Description as provided in (Hugh 1981) Swings et al. 1983 [56] and the genomic analyses conducted in the present study and a previous study by Bansal et al. 2021 [30].
- Emended description of Xanthomonas fastidiosa = Xylella fastidiosa Wells et al. 1987.
- Description as provided in Wells et al. 1987 [50] and the genomic analyses conducted in the present study and a previous study by Bansal et al., 2021 [30].
- Emended description of Xanthomonas broegbernensis = Pseudoxanthomonas broegbernensis Finkmann et al. 2000.
- Description as provided for Pseudoxanthomonas broegbernensis in Finkmann et al. 2000 [51] and based on the genomic analyses in the present study and a previous study by Bansal et al., 2021 [30].
- Emended description of the genus Frateuria Swing et al. 1980.
- Frat.eur’i.a. N.L. fem. n. Frateuria, named after Joseph Frateur (1903–1974), the eminent Belgian microbiologist. The type species is Frateuria aurantia (ex Kondô and Ameyama 1958) Swings et al. 1980 [56].
- Synonyms: Aerosticca, Fulvimonas, Rhodanobacter, Dyella, and Luteibacter.
The type species is Frateuria aurantia (ex Kondô and Ameyama 1958) Swings et al. 1980.
The description is as provided in Naushad et al. 2015 [1] and based on the deep phylo-taxonogenomic analysis conducted in the present study. The genus now includes the previously described genera and their member species, i.e., the genus now includes genus Aerosticca, Fulvimonas, Rhodanobacter, Dyella, Luteibacter, and Frateuria (Supplementary Table S2).
The emended descriptions of the member species of the genus Frateuria are provided in Supplementary Table S2.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/taxonomy3040026/s1, Table S1: List of proposed species of the genus Xanthomonas; Table S2: List of proposed species of the genus Rhodanobacter; Table S3: Metadata showing the list of all of the genomes (213) and their accessions number used in this study belonging to the type species and type strains of the order Lysobacterales (Xanthomonadales); Table S4: Metadata showing 16S rRNA % similarity values using the Phylogeny server (DSMZ).
Author Contributions
K.B., S.K. and A.S.: data analysis and drafting of the manuscript. A.C.: metadata preparation. P.B.P. conceived and participated in the planning, design, and drafting of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by an institutional project grant provided to MTCC and CSIR MPL065 provided to PBP from CSIR.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Naushad, S.; Adeolu, M.; Wong, S.; Sohail, M.; Schellhorn, H.E.; Gupta, R.S. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: Proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales, the family Rhodanobacteraceae fam. nov., containing the genus Rhodanobacter and its closest relatives. Antonie Van Leeuwenhoek 2015, 107, 467–485. [Google Scholar]
- Saddler, G.; Bradbury, J. Xanthomonadales ord. nov. In Bergey’s Manual of Systematic Bacteriology; validation of the publication of new names and new combinations previously effectively published outside the IJSEM, List; Springer: Boston, MA, USA, 2005; pp. 2235–2238. [Google Scholar]
- Midha, S.; Bansal, K.; Kumar, S.; Girija, A.M.; Mishra, D.; Brahma, K.; Laha, G.S.; Sundaram, R.M.; Sonti, R.V.; Patil, P.B. Pop-ulation genomic insights into variation and evolution of Xanthomonas oryzae pv. oryzae. Sci. Rep. 2017, 7, 40694. [Google Scholar] [CrossRef] [PubMed]
- Bansal, K.; Midha, S.; Kumar, S.; Patil, P.B. Ecological and evolutionary insights into Xanthomonas citri pathovar diversity. Appl. Environ. Microbiol. 2017, 83, e02993-16. [Google Scholar] [CrossRef] [PubMed]
- Midha, S.; Patil, P.B. Genomic insights into the evolutionary origin of Xanthomonas axonopodis pv. citri and its ecological rela-tives. Appl. Environ. Microbiol. 2014, 80, 6266–6279. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chatterjee, S.; Almeida, R.P.P.; Lindow, S. Living in two worlds: The plant and insect lifestyles of Xylella fastidiosa. Annu. Rev. Phytopathol. 2008, 46, 243–271. [Google Scholar] [CrossRef]
- Brooke, J.S. Stenotrophomonas maltophilia: An emerging global opportunistic pathogen. Clin. Microbiol. Rev. 2012, 25, 2–41. [Google Scholar] [CrossRef]
- Vauterin, L.; Yang, P.; Alvarez, A.; Takikawa, Y.; Roth, D.A.; Vidaver, A.K.; Stall, R.E.; Kersters, K.; Swings, J. Identification of non-pathogenic Xanthomonas strains associated with plants. Syst. Appl. Microbiol. 1996, 19, 96–105. [Google Scholar] [CrossRef]
- Singh, A.; Kumar, S.; Bansal, K.; Patil, P.B. Taxonomic and phylogenomic assessment identifies phyto-pathogenicity potential of Stenotrophomonas maltophilia complex. Phytopathology 2023. [Google Scholar] [CrossRef]
- Kumar, S.; Bansal, K.; Patil, P.P.; Patil, P.B. Phylogenomics insights into order and families of Lysobacterales. Access Microbiol. 2019, 1, e000015. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Patil, P.P.; Sharma, S.; Patil, P.B. Genomic data resource of type strains of genus Pseudoxanthomonas. Data Brief 2022, 42, 108145. [Google Scholar] [CrossRef]
- Yu, T.-T.; Zhou, E.-M.; Yin, Y.-R.; Yao, J.-C.; Ming, H.; Dong, L.; Li, S.; Nie, G.-X.; Li, W.-J. Vulcaniibacterium tengchongense gen. nov., sp. nov. isolated from a geothermally heated soil sample, and reclassification of Lysobacter thermophilus Wei et al. 2012 as Vulcaniibacterium thermophilum comb. nov. Antonie Van Leeuwenhoek 2013, 104, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, K.; Chohnan, S.; Ohashi, H.; Hirata, T.; Masaki, T.; Sakiyama, F. Purification, bacteriolytic activity, and specificity of β-lytic protease from Lysobacter sp. IB-9374. J. Biosci. Bioeng. 2003, 95, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Panthee, S.; Hamamoto, H.; Paudel, A.; Sekimizu, K. Lysobacter species: A potential source of novel antibiotics. Arch. Microbiol. 2016, 198, 839–845. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuen, G.Y. Biological control of Bipolaris sorokiniana on tall fescue by Stenotrophomonas maltophilia strain C3. Phytopathology 1999, 89, 817–822. [Google Scholar] [CrossRef]
- Nalin, R.; Simonet, P.; Vogel, T.M.; Normand, P. Rhodanobacter lindaniclasticus gen. nov., sp. nov., a lindane-degrading bacte-rium. Int. J. Syst. Evol. Microbiol. 1999, 49, 19–23. [Google Scholar] [CrossRef]
- Mergaert, J.; Cnockaert, M.C.; Swings, J. Fulvimonas soli gen. nov., sp. nov., a gamma-proteobacterium isolated from soil after enrichment on acetylated starch plastic. Int. J. Syst. Evol. Microbiol. 2002, 52, 1285–1289. [Google Scholar]
- Iasur-Kruh, L.; Zahavi, T.; Barkai, R.; Freilich, S.; Zchori-Fein, E.; Naor, V. Dyella-like bacterium isolated from an insect as a potential biocontrol agent against grapevine yellows. Phytopathology 2018, 108, 336–341. [Google Scholar] [CrossRef]
- Fang, T.; Wang, H.; Huang, Y.; Zhou, H.; Dong, P. Oleiagrimonas soli gen. nov., sp. nov., a genome-sequenced gammaproteo-bacterium isolated from an oilfield. Int. J. Syst. Evol. Microbiol. 2015, 65, 1666–1671. [Google Scholar] [CrossRef]
- Green, S.J.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 2012, 78, 1039–1047. [Google Scholar] [CrossRef]
- Campbell, B.J.; Polson, S.W.; Hanson, T.E.; Mack, M.C.; Schuur, E.A. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbiol. 2010, 12, 1842–1854. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.; Harris, H.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Bei-jerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.J.; Barco, R.A.; Loh, Y.-H.E.; Cogneau, S.; Rigouts, L. Reconstituting the genus Mycobacterium. Int. J. Syst. Evol. Mi-crobiol. 2021, 71, 004922. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Prokaryotic taxonomy and phylogeny in the genomic era: Advancements and challenges ahead. Curr. Opin. Microbiol. 2007, 10, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005, 187, 6258–6264. [Google Scholar] [CrossRef]
- Qin, Q.-L.; Xie, B.-B.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhou, J.; Oren, A.; Zhang, Y.-Z. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Kaur, A.; Singh, A.; Patil, P.B. Deep phylo-taxono genomics reveals Xylella as a variant lineage of plant associated Xanthomonas and supports their taxonomic reunification along with Stenotrophomonas and Pseudoxanthomonas. Ge-nomics 2021, 113, 3989–4003. [Google Scholar] [CrossRef]
- Luo, C.; Rodriguez-r, L.M.; Konstantinidis, K.T. MyTaxa: An advanced taxonomic classifier for genomic and metagenomic sequences. Nucleic Acids Res. 2014, 42, e73. [Google Scholar] [CrossRef]
- Park, M.-J.; Kim, Y.J.; Park, M.; Yu, J.; Namirimu, T.; Roh, Y.-R.; Kwon, K.K. Establishment of genome based criteria for classi-fication of the family Desulfovibrionaceae and proposal of two novel genera, Alkalidesulfovibrio gen. nov. and Salidesulfovibrio gen. nov. Front. Microbiol. 2022, 13, 738205. [Google Scholar] [CrossRef] [PubMed]
- Wirth, J.S.; Whitman, W.B. Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropici-bacter, and the proposal of six novel genera. Int. J. Syst. Evol. Microbiol. 2018, 68, 2393–2411. [Google Scholar] [PubMed]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic analysis of the gammaproteobacterial methanotrophs (order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef]
- Rosselló-Mora, R.; Amann, R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001, 25, 39–67. [Google Scholar] [CrossRef] [PubMed]
- Orata, F.D.; Kits, K.D.; Stein, L.Y. Complete genome sequence of methylomonas denitrificans strain FJG1, an obligate aerobic methanotroph that can couple methane oxidation with denitrification. Genome Announc. 2018, 6, e00276-18. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Asnicar, F.; Thomas, A.M.; Beghini, F.; Mengoni, C.; Manara, S.; Manghi, P.; Zhu, Q.; Bolzan, M.; Cumbo, F.; May, U. Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Bayliss, S.C.; Thorpe, H.A.; Coyle, N.M.; Sheppard, S.K.; Feil, E.J. PIRATE: A fast and scalable pangenomics toolbox for clus-tering diverged orthologues in bacteria. Gigascience 2019, 8, giz119. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Krishnamurthi, S.; Mayilraj, S.; Prasad, G.; Bora, T.C.; Chakrabarti, T. Aquimonas voraii gen. nov., sp. nov., a novel gammaproteobacterium isolated from a warm spring of Assam, India. Int. J. Syst. Evol. Microbiol. 2005, 55, 1491–1495. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-H.; Lai, W.-A.; Lin, S.-Y.; Hameed, A.; Shahina, M.; Shen, F.-T.; Zhu, Z.-L.; Young, L.-S.; Young, C.-C. Chiayiivirga flava gen. nov., sp. nov., a novel bacterium of the family Xanthomonadaceae isolated from an agricultural soil, and emended description of the genus Dokdonella. Int. J. Syst. Evol. Microbiol. 2013, 63, 3293–3300. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.-T.; Yao, J.-C.; Yin, Y.-R.; Dong, L.; Liu, R.-F.; Ming, H.; Zhou, E.-M.; Li, W.-J. Rehaibacterium terrae gen. nov., sp. nov. isolated from a geothermally heated soil sample. Int. J. Syst. Evol. Microbiol. 2013, 63, 4058–4063. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Tokizawa, R.; Fukui, M.J. Mizugakiibacter sediminis gen. nov., sp. nov., isolated from a freshwater lake. Int. J. Syst. Evol. Microbiol. 2014, 64, 3983–3987. [Google Scholar] [CrossRef]
- Ziegler, S.; Waidner, B.; Itoh, T.; Schumann, P.; Spring, S.; Gescher, J.J. Metallibacterium scheffleri gen. nov., sp. nov., an alka-linizing gammaproteobacterium isolated from an acidic biofilm. Int. J. Syst. Evol. Microbiol. 2013, 63, 1499–1504. [Google Scholar] [CrossRef]
- Wei, Z.; Wu, T.; Huang, Y.; Zhu, G.; Zhang, Y.; Geng, Y.; Peng, F.J. Pseudolysobacter antarcticus gen. nov., sp. nov., isolated from soil in Fildes Peninsula, Antarctica. Int. J. Syst. Evol. Microbiol. 2020, 70, 1861–1867. [Google Scholar] [CrossRef]
- Wells, J.M.; Raju, B.C.; Hung, H.-Y.; Weisburg, W.G.; Mandelco-Paul, L.; Brenner, D.J. Xylella fastidiosa gen. nov., sp. nov: Gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Evol. Microbiol. 1987, 37, 136–143. [Google Scholar] [CrossRef]
- Finkmann, W.; Altendorf, K.; Stackebrandt, E.; Lipski, A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broeg-bernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 273–282. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Bradbury, J.F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Evol. Microbiol. 1993, 43, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Christensen, P.; Cook, F. Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Evol. Mi-crobiol. 1978, 28, 367–393. [Google Scholar] [CrossRef]
- Watanabe, M.; Kojima, H.; Fukui, M. Aerosticca soli gen. nov., sp. nov., an aerobic gammaproteobacterium isolated from crude oil-contaminated soil. Arch. Microbiol. 2020, 202, 1069–1076. [Google Scholar] [CrossRef] [PubMed]
- Johansen, J.E.; Binnerup, S.J.; Kroer, N.; Mølbak, L. Luteibacter rhizovicinus gen. nov., sp. nov., a yellow-pigmented gammapro-teobacterium isolated from the rhizosphere of barley (Hordeum vulgare L.). Int. J. Syst. Evol. Microbiol. 2005, 55, 2285–2291. [Google Scholar] [CrossRef] [PubMed]
- Swings, J.; Gillis, M.; Kersters, K.; De Vos, P.; Gosselé, F.; De Ley, J. Frateuria, a new genus for “Acetobacter aurantius”. Int. J. Syst. Evol. Microbiol. 1980, 30, 547–556. [Google Scholar] [CrossRef]
- Yarza, P.; Yilmaz, P.; Pruesse, E.; Glöckner, F.O.; Ludwig, W.; Schleifer, K.-H.; Whitman, W.B.; Euzéby, J.; Amann, R.; Rossel-ló-Móra, R. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 2014, 12, 635–645. [Google Scholar] [CrossRef]
- Dowson, W.J. On the systematic position and generic names of the Gram-negative bacterial plant pathogens. Zentralblatt Bakte-riol. Parasitenkd. Infekt. Hyg. 1939, 100, 177–193. [Google Scholar]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.-J.; Ludwig, W.; Kämpfer, P.J. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef]
- Saddler, G.; Bradbury, J. Family I. Xanthomonadaceae. In Bergey’s Manual of Systematic Bacteriology, Volume 2 (The Proteobacteria), Part B (The Gammaproteobacteria); Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Chen, W.-M.; Xie, P.-B.; Tang, S.-L.; Sheu, S.-Y. Coralloluteibacterium stylophorae gen. nov., sp. nov., a new member of the family Lysobacteraceae isolated from the reef-building coral Stylophora sp. Arch. Microbiol. 2018, 200, 473–481. [Google Scholar] [CrossRef]
- Kwon, S.-W.; Kim, B.-Y.; Weon, H.-Y.; Baek, Y.-K.; Go, S.-J. Arenimonas donghaensis gen. nov., sp. nov., isolated from seashore sand. Int. J. Syst. Evol. Microbiol. 2007, 57, 954–958. [Google Scholar] [CrossRef]
- Lee, E.M.; Jeon, C.O.; Choi, I.; Chang, K.-S.; Kim, C.-J. Silanimonas lenta gen. nov., sp. nov., a slightly thermophilic and alkaliphilic gammaproteobacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 2005, 55, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Weon, H.-Y.; Yoo, S.-H.; Kim, Y.-J.; Lee, C.-M.; Kim, B.-Y.; Jeon, Y.-A.; Hong, S.-B.; Anandham, R.; Kwon, S.-W. Rudaea cellulosilytica gen. nov., sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 2308–2312. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Kang, S.-J.; Oh, T.-K. Dokdonella koreensis gen. nov., sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 145–150. [Google Scholar] [CrossRef]
- Makk, J.; Homonnay, Z.G.; Kéki, Z.; Lejtovicz, Z.; Márialigeti, K.; Spröer, C.; Schumann, P.; Tóth, E.M. Tahibacter aquaticus gen. nov., sp. nov., a new gammaproteobacterium isolated from the drinking water supply system of Budapest (Hungary). Syst. Appl. Microbiol. 2011, 34, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Busse, H.-J.; Kämpfer, P.; Moore, E.; Nuutinen, J.; Tsitko, I.; Denner, E.; Vauterin, L.; Valens, M.; Rosselló-Mora, R.; Salki-noja-Salonen, M.S. Thermomonas haemolytica gen. nov., sp. nov., a gamma-proteobacterium from kaolin slurry. Int. J. Syst. Evol. Microbiol. 2002, 52, 473–483. [Google Scholar] [CrossRef] [PubMed][Green Version]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).