Morphological Description and Molecular Analysis of Newly Recorded Asteroid, Henricia djakonovi Chichvarkhin, 2017 (Asteroidea: Spinulosida: Echinasteridae), from Dokdo Island, Korea
Abstract
1. Introduction
2. Materials and Methods
3. Systematic Accounts
- 1. Abactinal skeleton generally imbricated in close meshwork --------------------------------- 2
- – Abactinal skeleton generally reticulated in open meshwork ---------------------------------- 9
- 2. Abactinal and actinal plate extension of ossicles present ----------------------- H. epiphysialis
- – Abactinal and actinal plate extension of ossicles absent ---------------------------------------- 3
- 3. Abactinal paxillae with scattered spinelets, not in distinct group ---------------- H. anomala
- – Abactinal paxillae crowded with spinelets in groups ------------------------------------------- 4
- 4. Shape of abactinal spines pillar ---------------------------------------------------------------------- 5
- – Shape of abactinal spines slender -------------------------------------------------------------------- 6
- 5. Shape of tip of abactinal spine granular ---------------------------------------------- H. leviuscula
- – Shape of tip of abactinal spine droplet-like --------------------------------------------H. djakonovi
- 6. Arms long (R/r: >5.5) ---------------------------------------------------------------------- H. reniossa
- – Arms short (R/r: <4.0) ------------------------------------------------------------------------------------ 7
- 7. Shape of abactinal plates quasi-quadrate ---------------------------------------------- H. regularis
- – Shape of abactinal plates rounded cross ------------------------------------------------------------ 8
- 8. Number of adambulacral spines more than 10 ----------------------------------------- H. elachys
- – Number of adambulacral spines less than 10 ----------------------------------------- H. nipponica
- 9. Slender arms; tightly meshed abactinal skeleton ----------------------------------------------- 10
- – Broad arms; loosely meshed abactinal skeleton ------------------------------------------------- 12
- 10. Adambulacral armature bearing 5 or 6 spines -------------------------------------- H. ohshimai
- – Adambulacral armature bearing 10 to 18 spines ------------------------------------------------ 11
- 11. Abactinal skeleton strong structured; marginal plates conspicuous ----------- H. hayashii
- – Abactinal skeleton weak structured; marginal plates inconspicuous ------------- H. pacifica
- 12. Number of adambulacral spines more than 10 ------------------------------- H. sanguinolenta
- – Number of adambulacral spines less than 10 ---------------------------------------------------- 13
- 13. Shape of abactinal spines with broad basal, rapidly taper to tip ------------ H. pachyderma
- – Shape of abactinal spines with narrow basal, slowly taper to tip --------------------------- 14
- 14. Shape of inferomarginal plates reniform ----------------------------------------------- H. oculata
- – Shape of inferomarginal plates rounded cross ---------------------------------------- H. perforata
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blake, D.B. A reassessment of the sea-star orders Valvatida and Spinulosida. J. Nat. Hist. 1981, 15, 375–394. [Google Scholar] [CrossRef]
- Lambert, P. Sea Stars of British Columbia, Southeast Alaska, and Puget Sound; University of British Columbia Press: Vancouver, BC, Canada, 2000; 186p. [Google Scholar]
- Byrne, M.; O’Hara, T.D. Australian Echinoderms: Biology, Ecology and Evolution; Csiro Publishing: Clayton, Australia, 2017; 612p. [Google Scholar]
- Clark, R.N.; Jewett, S.C. A new genus and thirteen new species of sea stars (Asteroidea: Echinasteridae) from the Aleutian Island Archipelago. Zootaxa 2010, 2571, 1–36. [Google Scholar] [CrossRef]
- Shield, C.J.; Witman, J.D. The impact of Henricia sanguinolenta (O.F. Muller) (Echinodermata: Asteroidea) predation on the finger sponges, Isodictya sp. J. Exp. Mar. Biol. Ecol. 1993, 166, 107–133. [Google Scholar] [CrossRef]
- Mah, C.; Blake, B.D. Global Diversity and Phylogeny of the Asteroidea (Echinodermata). PLoS ONE 2012, 7, e35644. [Google Scholar] [CrossRef]
- Shin, S. Sea Stars: Invertebrate Fauna of Korea Vol. 32; National Institute of Biological Resources: Incheon, Republic of Korea, 2010; 150p. [Google Scholar]
- Ubagan, M.D.; Lee, T.; Kim, P.; Shin, S. A new species of the genus Henricia (Asteroidea, Spinulosida, Echinasteridae) from South Korea. ZooKeys 2020, 997, 1–15. [Google Scholar] [CrossRef]
- Shin, S.; Ubagan, M.D. A Newly Recorded Sea Star of Genus Henricia (Asteroidea: Spinulosida: Echinasteridae) from Jeju Island, Korea. Korean J. Environ. Biol. 2015, 33, 390–393. [Google Scholar] [CrossRef]
- Ubagan, M.D.; Shin, S. A new record of sea star genus Henricia (Asteroidea: Spinulosida: Echinasteridae) from Jeju Island, Korea. J. Species Res. 2016, 5, 351–354. [Google Scholar] [CrossRef]
- Ubagan, M.D.; Shin, S. New record of a sea star of genus Henricia (Asteroidea: Spinulosida: Echinasteridae) from Jeju Island, Korea. Korean J. Environ. Biol. 2019, 37, 68–71. [Google Scholar] [CrossRef]
- Ubagan, M.D.; Shin, S. New record of a sea star, Henricia perforata (Asteroidea: Spinulosida: Echinasteridae), in the East Sea, Korea. Korean J. Environ. Biol. 2020, 38, 388–391. [Google Scholar] [CrossRef]
- Chichvarkhin, A. Henricia djakonovi sp. nov. (Echinodermata, Echinasteridae): A new sea star species from the Sea of Japan. PeerJ 2017, 5, e2863. [Google Scholar] [CrossRef]
- Madsen, F.J. The Henricia sanguinolenta complex (Echinodermata, Asteroidea) of the Norwegian Sea and adjacent waters. A re-evaluation, with notes on related species. Steenstrupia 1987, 13, 201–268. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; de Waard, J.R. Biological identifications through DNA barcodes. Proc. Royal Soc. B-Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The barcode of life data system (http://www.barcodinglife.org). Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Ward, R.D.; Holmes, B.H.; O’Hara, T.D. DNA barcoding discriminates echinoderm species. Mol. Ecol. Resour. 2008, 8, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Hoareau, T.B.; Boissin, E. Design of phylum-specific hybrid primers for DNA barcoding: Addressing the need for efficient COI amplification in the Echinodermata. Mol. Ecol. Resour. 2010, 10, 960–967. [Google Scholar] [CrossRef]
- Ubagan, M.D.; Shin, S. Newly recorded sea star Henricia oculata (Asteroidea: Spinulosida: Echinasteridae) in the East Sea, Korea. Korean J. Environ. Biol. 2020, 38, 563–566. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Mah, C.; World Asteroidea Database [Internet]. World Register of Marine Species. 2022. Available online: http://www.marinespecies.org (accessed on 22 August 2022).
- Bratova, O.; Paskerova, G.G. Henricia spp. (Echinodermata: Asteroidea: Echinasteridae) of the White Sea: Morphology, morphometry, and synonymy. Can. J. Zool. 2017, 96, 341–355. [Google Scholar] [CrossRef]
- Layton, K.K.S.; Corstorphine, E.A.; Hubert, P.D.N. Exploring Canadian Echinoderm Diversity through DNA Barcodes. PLoS ONE 2016, 11, e0166118. [Google Scholar] [CrossRef]
- Wakita, D.; Fujita, T.; Kajihara, H. Molecular systematics and morphological analyses of the subgenus Setihenricia (Echinodermata: Asteroidea: Henricia) from Japan. Species Divers. 2019, 24, 119–135. [Google Scholar] [CrossRef]
- Lopes, E.M.; Pérez-Portela, R.; Paiva, P.C.; Ventura, C.R.R. The molecular phylogeny of the sea star Echinaster (Asteroidea: Echinasteridae) provides insights for genus taxonomy. Invertebr. Biol. 2016, 135, 235–244. [Google Scholar] [CrossRef]
- Kim, D.; Kim, M.; Shin, S. A newly recorded sea star of the genus Luidia (Asteroidea: Paxillosida: Luidiidae) from the Korea Strait, Korea. Animal Syst. Evol. Divers. 2017, 33, 131–135. [Google Scholar] [CrossRef]
- Xiao, N.; Liu, R.; Yuan, S.; Sha, Z. A preliminary phylogenetic analysis of Luidia (Paxillosida: Luidiidae) from Chinese waters with cytochrome oxidase subunit I (COI) sequences. J. Ocean. Univ. China 2013, 12, 459–468. [Google Scholar] [CrossRef]
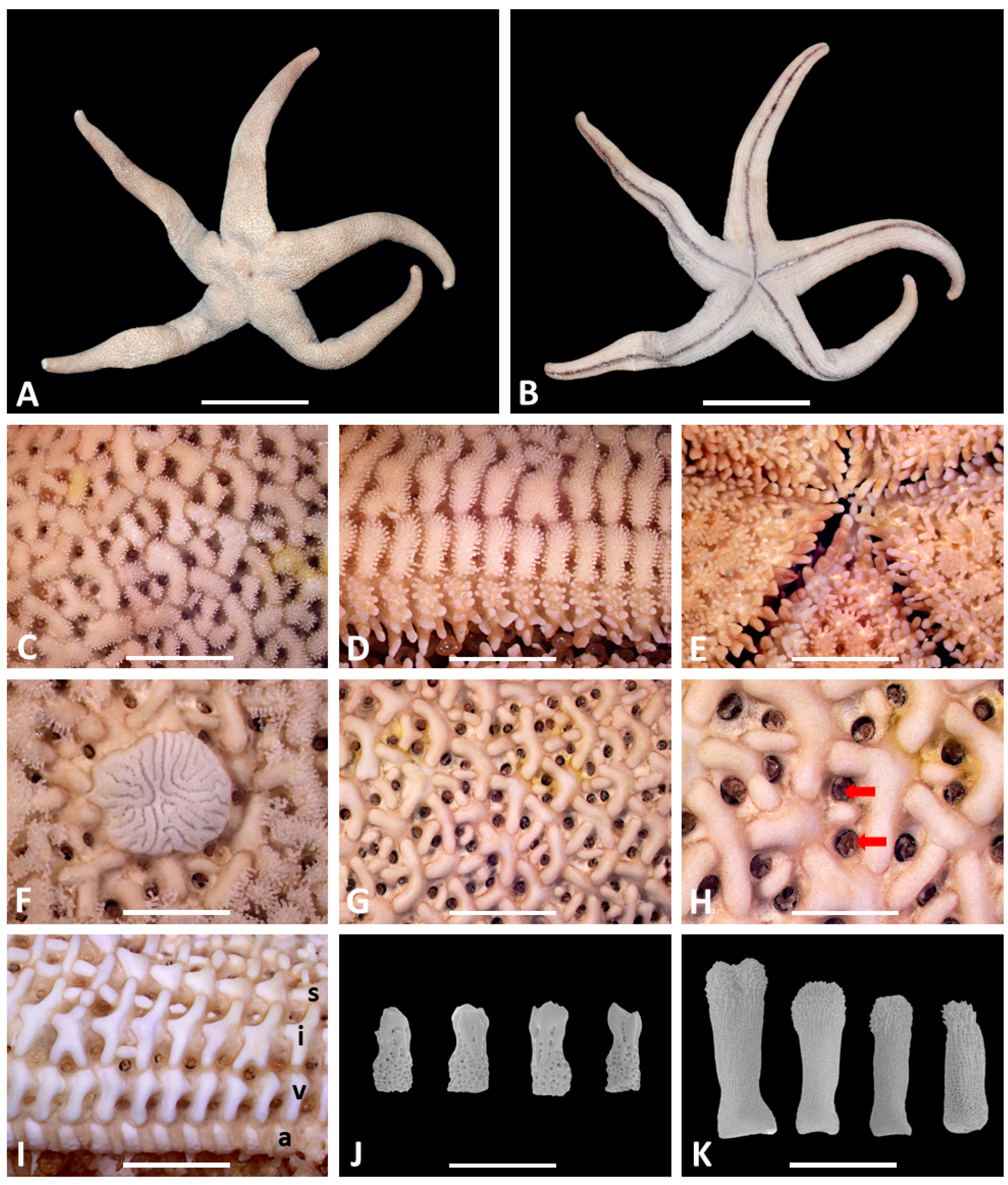

| Characters | H. djakonovi (Our Specimen) | H. leviuscula (Our Specimen) | H. reniossa (Hayashi, 1940) | H. sanguinolenta (Our Specimen) |
|---|---|---|---|---|
| Range of R/r (Max R) | 5.5 | 6.0 | 7.8–8.0 | 4.1–4.5 |
| Arm | wide arm base tapering to tip | slender, tapering to tip | slender, tapering to tip | thick arm base, tapering to tip |
| Number of abactinal papula | 1–3 | 1 or 2 | 1 or 2 | 1–5 |
| Shape of abactinal papular area | narrow | narrow | narrow | wide |
| Number of abactinal spine | 126–144 | 40–60 | 40–60 or more | 7–16 |
| Shape of abactinal spine | pillar | granuliform | slender | club shape |
| Shape of abactinal plate | crescent, lobed | roundish or elliptical | reniform | crescent, rod-like |
| Shape of inferomarginal plate | elongated cross, rounded cross | rounded cross, rod-like | elongated cross, rounded cross | elongated cross, rounded cross |
| Number of adambulacral spine | 13–16 | 7–10 | 15–25 | 11–17 |
| Pattern of adambulacral furrow + near ventrolateral plate | 1–3 flat tip + 4–16 shorter | 1 long, slender + 2–10 slender bluntly pointed tip | 1–3 long, slender + 4–25 shorter | 1–3 flat tip + 4–17 shorter, bluntly pointed tip |
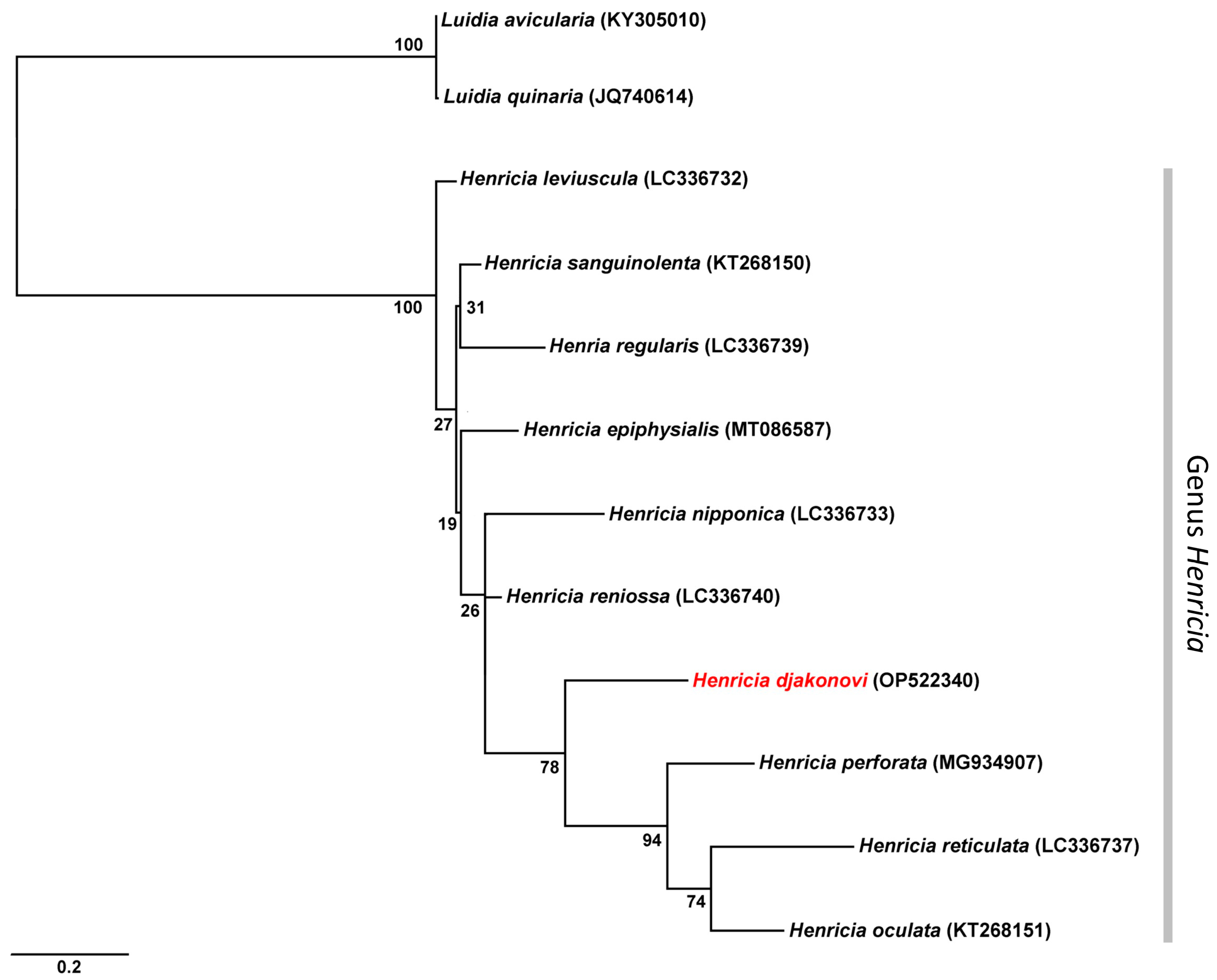
| Species | GenBank Accession No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | H. djakonovi | OP522340 | Present study | ||||||||||||
| 2 | H. epiphysialis | MT086587 | 13.8 | [8] | |||||||||||
| 3 | H. leviuscula | LC336732 | 13.3 | 6.4 | [27] | ||||||||||
| 4 | H. nipponica | LC336733 | 15.0 | 9.5 | 10.2 | [27] | |||||||||
| 5 | H. oculata | KT268151 | 13.3 | 14.5 | 14.5 | 15.7 | [28] | ||||||||
| 6 | H. perforata | MG934907 | 15.0 | 13.8 | 13.6 | 15.0 | 10.2 | unpublished | |||||||
| 7 | H. regularis | LC336739 | 14.0 | 8.6 | 8.1 | 10.2 | 16.7 | 14.8 | [27] | ||||||
| 8 | H. reniossa | LC336740 | 11.0 | 7.1 | 6.0 | 8.6 | 14.5 | 13.1 | 7.9 | [27] | |||||
| 9 | H. reticulata | LC336737 | 16.4 | 14.3 | 14.3 | 16.4 | 11.0 | 12.6 | 15.5 | 15.0 | [27] | ||||
| 10 | H. sanguinolenta | KT268150 | 12.6 | 6.0 | 5.0 | 9.8 | 15.5 | 14.3 | 7.1 | 5.2 | 15.0 | [28] | |||
| 11 | L. avicularia | KY305010 | 24.3 | 21.9 | 21.2 | 20.5 | 24.5 | 24.3 | 21.4 | 21.4 | 22.6 | 21.4 | [29] | ||
| 12 | L. quinaria | JQ740614 | 24.5 | 22.1 | 21.4 | 20.7 | 24.8 | 24.5 | 21.7 | 21.7 | 22.9 | 21.7 | 0.2 | [30] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ubagan, M.D.; Alboasud, M.S.A.; Lee, T. Morphological Description and Molecular Analysis of Newly Recorded Asteroid, Henricia djakonovi Chichvarkhin, 2017 (Asteroidea: Spinulosida: Echinasteridae), from Dokdo Island, Korea. Taxonomy 2023, 3, 46-54. https://doi.org/10.3390/taxonomy3010004
Ubagan MD, Alboasud MSA, Lee T. Morphological Description and Molecular Analysis of Newly Recorded Asteroid, Henricia djakonovi Chichvarkhin, 2017 (Asteroidea: Spinulosida: Echinasteridae), from Dokdo Island, Korea. Taxonomy. 2023; 3(1):46-54. https://doi.org/10.3390/taxonomy3010004
Chicago/Turabian StyleUbagan, Michael Dadole, Mariya Shihab Ahmed Alboasud, and Taekjun Lee. 2023. "Morphological Description and Molecular Analysis of Newly Recorded Asteroid, Henricia djakonovi Chichvarkhin, 2017 (Asteroidea: Spinulosida: Echinasteridae), from Dokdo Island, Korea" Taxonomy 3, no. 1: 46-54. https://doi.org/10.3390/taxonomy3010004
APA StyleUbagan, M. D., Alboasud, M. S. A., & Lee, T. (2023). Morphological Description and Molecular Analysis of Newly Recorded Asteroid, Henricia djakonovi Chichvarkhin, 2017 (Asteroidea: Spinulosida: Echinasteridae), from Dokdo Island, Korea. Taxonomy, 3(1), 46-54. https://doi.org/10.3390/taxonomy3010004






