Phylogenomic Analysis Supports the Transfer of 20 Pathovars from Xanthomonas campestris into Xanthomonas euvesicatoria
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Genome Sequencing and Assembly
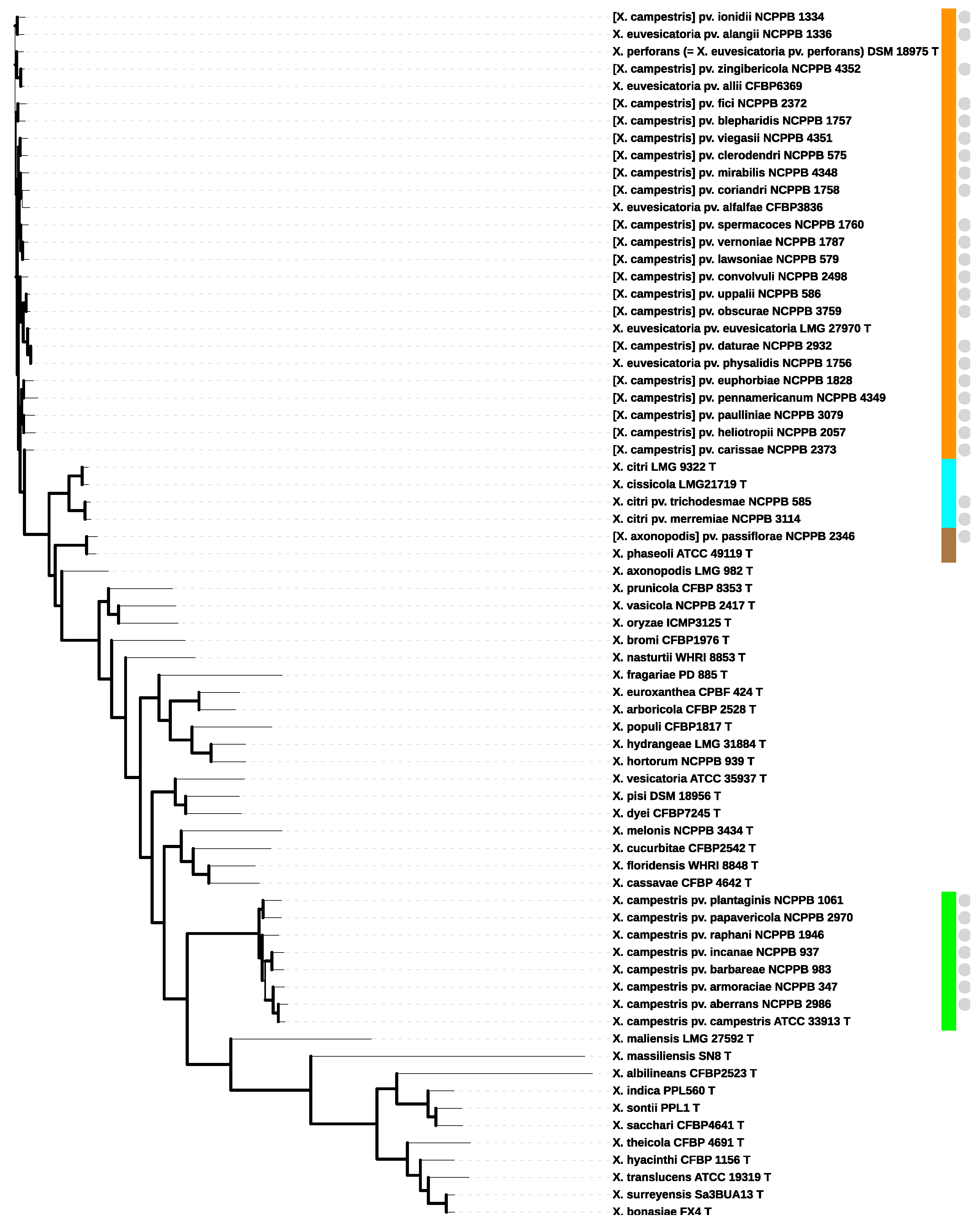
3.2. Phylogenomic Reconstruction
3.3. Species Delineation Based on Average Nucleotide Identity
| Pathovar and NCPPB Number | X. euvesicatoria LMG 27970 T | X. campestris ATCC 33913 | X. citri LMG 9322 | X. phaseoli 49119 |
|---|---|---|---|---|
| X. campestris pv. aberrans 2986 | 85.80 | 98.66 | 85.65 | 85.88 |
| X. campestris pv. armoraciae 347 | 85.74 | 98.16 | 85.79 | 85.88 |
| X. campestris pv. barbareae 983 | 85.79 | 97.31 | 85.84 | 85.95 |
| X. campestris pv. incanae 937 | 85.78 | 97.29 | 85.79 | 85.89 |
| X. campestris pv. papavericola 2970 | 85.66 | 96.49 | 85.60 | 85.78 |
| X. campestris pv. plantaginis 1061 | 85.88 | 96.45 | 85.78 | 85.79 |
| X. campestris pv. raphani 1946 | 85.83 | 97.23 | 85.72 | 85.80 |
| X. phaseoli pv. passiflorae 2346 | 93.86 | 85.96 | 93.65 | 98.16 |
| X. citri pv. merremiae 3114 | 93.85 | 85.81 | 96.20 | 93.91 |
| X. citri pv. trichodesmae 585 | 93.77 | 85.83 | 96.16 | 93.82 |
| X. euvesicatoria pv. alangii 1336 | 98.01 | 85.86 | 94.33 | 94.18 |
| [X. campestris] pv. blepharidis 1757 | 97.90 | 85.86 | 94.45 | 94.11 |
| [X. campestris] pv. carissae 2373 | 97.54 | 85.80 | 94.73 | 94.12 |
| [X. campestris] pv. clerodendri 575 | 97.98 | 85.85 | 94.31 | 94.08 |
| [X. campestris] pv. convolvuli 2498 | 98.13 | 85.93 | 94.50 | 94.14 |
| [X. campestris] pv. coriandri 1758 | 97.98 | 85.89 | 94.47 | 94.10 |
| [X. campestris] pv. daturae 2932 | 98.73 | 85.85 | 94.27 | 94.01 |
| [X. campestris] pv. euphorbiae 1828 | 97.57 | 85.91 | 94.41 | 94.10 |
| [X. campestris] pv. fici 2372 | 98.00 | 85.91 | 94.40 | 94.19 |
| [X. campestris] pv. heliotropii 2057 | 97.53 | 85.97 | 94.31 | 94.06 |
| [X. campestris] pv. ionidii 1334 | 97.92 | 85.88 | 94.38 | 94.08 |
| [X. campestris] pv. lawsoniae 579 | 98.04 | 85.90 | 94.45 | 94.12 |
| [X. campestris] pv. mirabilis 4348 | 98.00 | 85.84 | 94.36 | 94.17 |
| [X. campestris] pv. obscurae 3759 | 98.42 | 85.93 | 94.37 | 94.11 |
| [X. campestris] pv. paulliniae 3079 | 97.69 | 85.85 | 94.36 | 94.37 |
| [X. campestris] pv. pennamericanum 4349 | 97.45 | 85.80 | 94.27 | 94.04 |
| X. euvesicatoria pv. physalidis 1756 | 98.74 | 85.92 | 94.31 | 94.10 |
| [X. campestris] pv. spermacoces 1760 | 97.83 | 85.86 | 94.50 | 94.10 |
| [X. campestris] pv. uppalii 586 | 98.38 | 85.95 | 94.37 | 94.09 |
| [X. campestris] pv. vernoniae 1787 | 98.10 | 85.95 | 94.41 | 94.18 |
| [X. campestris] pv. viegasii 4351 | 98.05 | 85.94 | 94.31 | 94.18 |
| [X. campestris] pv. zingibericola 4352 | 98.10 | 85.88 | 94.47 | 94.23 |
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| GenBank Accession | Taxon | Reference |
|---|---|---|
| GCA_019201075.1 | [X. axonopodis] pv. passiflorae NCPPB 2346 | This study |
| GCA_019201485.1 | [X. campestris] pv. blepharidis NCPPB 1757 | This study |
| GCA_019201365.1 | [X. campestris] pv. carissae NCPPB 2373 | This study |
| GCA_019201225.1 | [X. campestris] pv. clerodendri NCPPB 575 | This study |
| GCA_019201245.1 | [X. campestris] pv. convolvuli NCPPB 2498 | This study |
| GCA_019201305.1 | [X. campestris] pv. coriandri NCPPB 1758 | This study |
| GCA_019201325.1 | [X. campestris] pv. daturae NCPPB 2932 | This study |
| GCA_019201425.1 | [X. campestris] pv. euphorbiae NCPPB 1828 | This study |
| GCA_019201375.1 | [X. campestris] pv. fici NCPPB 2372 | This study |
| GCA_019201405.1 | [X. campestris] pv. heliotropii NCPPB 2057 | This study |
| GCA_019201465.1 | [X. campestris] pv. ionidii NCPPB 1334 | This study |
| GCA_019201185.1 | [X. campestris] pv. lawsoniae NCPPB 579 | This study |
| GCA_019201545.1 | [X. campestris] pv. merremiae NCPPB 3114 | This study |
| GCA_019201145.1 | [X. campestris] pv. mirabilis NCPPB 4348 | This study |
| GCA_019201105.1 | [X. campestris] pv. obscurae NCPPB 3759 | This study |
| GCA_019201285.1 | [X. campestris] pv. paulliniae NCPPB 3079 | This study |
| GCA_019201165.1 | [X. campestris] pv. pennamericanum NCPPB 4349 | This study |
| GCA_019201445.1 | [X. campestris] pv. spermacoces NCPPB 1760 | This study |
| GCA_019201525.1 | [X. campestris] pv. trichodesmae NCPPB 585 | This study |
| GCA_019201115.1 | [X. campestris] pv. uppalii NCPPB 586 | This study |
| GCA_019201345.1 | [X. campestris] pv. vernoniae NCPPB 1787 | This study |
| GCA_019201265.1 | [X. campestris] pv. viegasii NCPPB 4351 | This study |
| GCA_019201205.1 | [X. campestris] pv. zingibericola NCPPB 4352 | This study |
| GCA_002939705.1 | X. albilineans CFBP 2523 T | - |
| GCA_001013475.1 | X. arboricola CFBP 2528 T | [81] |
| GCA_001401595.1 | X. axonopodis LMG 982 T | - |
| GCA_017163705.1 | X. bonasiae FX4 T | [82] |
| GCA_002939755.1 | X. bromi CFBP 1976 T | [83] |
| GCA_020813115.1 | X. campestris pv. aberrans NCPPB 2986 | This study |
| GCA_020731405.1 | X. campestris pv. armoraciae NCPPB 347 | This study |
| GCA_020813315.1 | X. campestris pv. barbareae NCPPB 983 | This study |
| GCA_000007145.1 | X. campestris pv. campestris ATCC 33913 T | [62] |
| GCA_020813295.1 | X. campestris pv. incanae NCPPB 937 | This study |
| GCA_020813015.1 | X. campestris pv. papavericola NCPPB 2970 | This study |
| GCA_020813005.1 | X. campestris pv. plantaginis NCPPB 1061 | This study |
| GCA_020813075.1 | X. campestris pv. raphani NCPPB 1946 | This study |
| GCA_000454545.1 | X. cassavae CFBP 4642 T | [84] |
| GCA_002019225.1 | X. cissicola LMG 21719 T | [11] |
| GCA_002018575.1 | X. citri LMG 9322 T | [85] |
| GCA_002939885.1 | X. cucurbitae CFBP 2542 T | - |
| GCA_002939865.1 | X. dyei CFBP 7245 T | - |
| GCA_900476395.1 | X. euroxanthea CPBF 424 T | [86] |
| GCA_019193005.1 | X. euvesicatoria pv. alangii NCPPB 1336 | This study |
| GCA_017724035.1 | X. euvesicatoria pv. alfalfae CFBP3836 | [87] |
| GCA_000730305.1 | X. euvesicatoria pv. allii CFBP6369 | [88] |
| GCA_001401555.1 | X. euvesicatoria pv. euvesicatoria LMG 27970 T | - |
| GCA_019192985.1 | X. euvesicatoria pv. physalidis NCPPB1756 | This study |
| GCA_001642575.1 | X. floridensis WHRI 8848 T | [89] |
| GCA_900380235.1 | X. fragariae PD 885 T | [90] |
| GCA_003064105.1 | X. hortorum NCPPB 939 T | [91] |
| GCA_009769165.1 | X. hyacinthi CFBP 1156 T | [92] |
| GCA_905142475.1 | X. hydrangeae LMG 31884 T | [93] |
| GCA_022669045.1 | X. indica PPL560 T | [94] |
| GCA_009192945.1 | X. maliensis LMG 27592 T | [95] |
| GCA_900018785.1 | X. massiliensis SN8 T | [96] |
| GCA_020783655.1 | X. melonis NCPPB 3434 T | - |
| GCA_001660815.1 | X. nasturtii WHRI 8853 T | [89] |
| GCA_004136375.1 | X. oryzae ICMP 3125 T | [97] |
| GCA_013112235.1 | X. perforans DSM 18975 T | - |
| GCA_022749655.1 | X. phaseoli ATCC 49119 T | - |
| GCA_001010415.1 | X. pisi DSM 18956 T | [98] |
| GCA_002940065.1 | X. populi CFBP 1817 T | - |
| GCA_002846205.1 | X. prunicola CFBP 8353 T | [99] |
| GCA_002940085.1 | X. sacchari CFBP 4641 T | - |
| GCA_008119715.1 | X. sontii PPL1 T | [100] |
| GCA_014836395.1 | X. surreyensis Sa3BUA13 T | [101] |
| GCA_014236795.1 | X. theicola CFBP 4691 T | [102] |
| GCA_020880735.1 | X. translucens ATCC 19319 T | [103] |
| GCA_000772705.2 | X. vasicola NCPPB 2417 T | [12] |
| GCA_001908725.1 | X. vesicatoria ATCC 35937 T | [104] |
Appendix B
| GenBank Accession | Pathovar | Completeness (%) | Contamination (%) |
|---|---|---|---|
| GCA_020813115.1 | aberrans | 99.89 | 0.03 |
| GCA_019193005.1 | alangii | 99.94 | 0.61 |
| GCA_020731405.1 | armoraciae | 99.89 | 0.03 |
| GCA_020813315.1 | barbareae | 99.85 | 0.03 |
| GCA_019201485.1 | blepharidis | 99.89 | 0.03 |
| GCA_020813135.1 | campestris | 99.77 | 0 |
| GCA_019201365.1 | carissae | 99.94 | 0.41 |
| GCA_019201225.1 | clerodendri | 99.94 | 0.36 |
| GCA_019201245.1 | convolvuli | 99.94 | 0.03 |
| GCA_019201305.1 | coriandri | 99.89 | 0.3 |
| GCA_019201325.1 | daturae | 99.87 | 0.53 |
| GCA_019201425.1 | euphorbiae | 99.89 | 0.03 |
| GCA_019201375.1 | fici | 99.94 | 0.68 |
| GCA_019201405.1 | heliotropii | 99.82 | 0.32 |
| GCA_020813295.1 | incanae | 99.64 | 0.53 |
| GCA_019201465.1 | ionidii | 99.89 | 0.15 |
| GCA_019201185.1 | lawsoniae | 99.64 | 0.03 |
| GCA_019201545.1 | merremiae | 99.81 | 0.34 |
| GCA_019201145.1 | mirabilis | 99.94 | 0.22 |
| GCA_019201105.1 | obscurae | 99.94 | 0.07 |
| GCA_020813015.1 | papavericola | 99.89 | 1.71 |
| GCA_019201075.1 | passiflorae | 99.89 | 0.42 |
| GCA_019201285.1 | paulliniae | 99.89 | 0.03 |
| GCA_019201165.1 | pennamericanum | 99.89 | 0 |
| GCA_019192985.1 | physalidis | 99.94 | 0.03 |
| GCA_020813005.1 | plantaginis | 99.89 | 0.53 |
| GCA_020813075.1 | raphani | 99.89 | 0.11 |
| GCA_019201445.1 | spermacoces | 99.89 | 0.03 |
| GCA_019201525.1 | trichodesmae | 99.76 | 0.93 |
| GCA_019201115.1 | uppalii | 99.62 | 0.25 |
| GCA_019201345.1 | veroniae | 99.89 | 0.37 |
| GCA_019201265.1 | viegasii | 99.94 | 0.03 |
| GCA_019201205.1 | zingibericola | 99.89 | 0.18 |
References
- Timilsina, S.; Potnis, N.; Newberry, E.A.; Liyanapathiranage, P.; Iruegas-Bocardo, F.; White, F.F.; Goss, E.M.; Jones, J.B. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 2020, 18, 415–427. [Google Scholar] [CrossRef]
- Jacques, M.-A.; Arlat, M.; Boulanger, A.; Boureau, T.; Carrère, S.; Cesbron, S.; Chen, N.W.G.; Cociancich, S.; Darrasse, A.; Denancé, N.; et al. Using ecology, physiology, and genomics to understand host specificity in Xanthomonas. Annu. Rev. Phytopathol. 2016, 54, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Dye, D.W.; Bradbury, J.F.; Goto, M.; Hayward, A.C.; Lelliott, R.A.; Schroth, M.N. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev. Plant Pathol. 1980, 59, 153–168. [Google Scholar]
- Parkinson, N.; Cowie, C.; Heeney, J.; Stead, D. Phylogenetic structure of Xanthomonas determined by comparison of gyrB sequences. Int. J. Syst. Evol. Microbiol. 2009, 59, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Elliott, C. Manual of Bacterial Plant Pathogens, 2nd ed.; Chronica Botanica Company: Waltham, MA, USA, 1951. [Google Scholar]
- Dye, D.D.W.; Lelliott, R.A.R. Genus II. Xanthomonas Dowson 1939. In Bergey’s Manual of Determinative Bacteriology; Buchanan, R.E., Gibbons, N.E., Eds.; Williams & Wilkins Co.: Baltimore, MD, USA, 1974; pp. 243–249. [Google Scholar]
- Young, J.M.; Dye, D.W.; Bradbury, J.F.; Panagopoulos, C.G.; Robbs, C.F. A proposed nomenclature and classification for plant pathogenic bacteria. New Zeal. J. Agric. Res. 1978, 21, 153–177. [Google Scholar] [CrossRef]
- Vauterin, L.; Hoste, B.; Kersters, K.; Swings, J. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 1995, 45, 472–489. [Google Scholar] [CrossRef]
- Yang, P.; Vauterin, L.; Vancanneyt, M.; Swings, J.; Kersters, K. Application of Fatty Acid Methyl Esters for the Taxonomic Analysis of the Genus Xanthomonas. Syst. Appl. Microbiol. 1993, 16, 47–71. [Google Scholar] [CrossRef]
- Constantin, E.C.; Cleenwerck, I.; Maes, M.; Baeyen, S.; Van Malderghem, C.; De Vos, P.; Cottyn, B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016, 65, 792–806. [Google Scholar] [CrossRef]
- Bansal, K.; Kumar, S.; Patil, P.B. Phylo-taxonogenomics supports revision of taxonomic status of twenty Xanthomonas pathovars to Xanthomonas citri. Phytopathology 2021. [Google Scholar] [CrossRef]
- Studholme, D.J.; Wicker, E.; Abrare, S.M.; Aspin, A.; Bogdanove, A.; Broders, K.; Dubrow, Z.; Grant, M.; Jones, J.B.; Karamura, G.; et al. Transfer of Xanthomonas campestris pv. arecae and X. campestris pv. musacearum to X. vasicola (Vauterin) as X. vasicola pv. arecae comb. nov. and X. vasicola pv. musacearum comb. nov. and description of X. vasicola pv. vasculorum pv. nov. Phytopathology 2020, 110, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [PubMed]
- Knösel, D. Eine an Kohl blattfleckenerzeugende Varietas von Xanthomonas campestris (Pammel) Dowson. Z. Pflanzenkrankh. Pflanzenschutz 1961, 68, 1–6. [Google Scholar]
- Sahin, F.; Miller, S.A. A new pathotype of Xanthomonas campestris pv. armoraciae that causes bacterial leaf spot of radish. Plant Dis. 1997, 81, 1334. [Google Scholar] [CrossRef] [PubMed]
- Mcculloch, L. A bacterial leaf spot of horse-radish caused by Bacterium campestre var. armoraciae, n. var. J. Agric. Res. 1929, 38, 269–287. [Google Scholar]
- Starr, M.P.; Burkholder, W.H. Lipolytic activity of phytopathogenic bacteria determined by means of spirit blue agar and its taxonomic significance. Phytopathology 1942, 32, 598–604. [Google Scholar]
- Burkholder, W.H. The black rot of Barbarea vulgaris. Phytopathology 1941, 31, 347–348. [Google Scholar]
- Kendrick, J.; Baker, K.F. Bacterial blight of garden stocks and its control by hot-water seed treatment. Bull. Calif. Agric. Exp. Stn. 1942, 665, 1–23. [Google Scholar]
- Mary, K.; McWhorter, F.P.; Smythe, C.V.; Williams, H.H. Bacterial blight of poppy caused by Bacterium papavericola, sp. nov. J. Agric. Res. 1930, 40, 1–9. [Google Scholar]
- Thornberry, H.H.; Anderson, H.W. Some bacterial diseases of plants in Illinois. Phytopathology 1937, 27, 946–949. [Google Scholar]
- White, H.E. Bacterial spot of radish and turnip. Phytopathology 1930, 20, 653–662. [Google Scholar]
- Padhya, A.C.; Patel, M.K. A new bacterial leaf-spot on Alangium lamarckii Thw. Curr. Sci. 1962, 31, 196–197. [Google Scholar]
- Srinivasan, M.C.; Patel, M.K.; Thirumalachar, M.J. Two bacterial leaf-spot diseases on Physalis minima and studies on their relationship to Xanthomonas vesicatoria (Doidge) Dowson. Proc. Natl. Acad. Sci. India 1962, 56, 93–96. [Google Scholar] [CrossRef]
- Pant, N.M.; Kulkarni, Y.S. Bacterial leaf-spot of Merremia gangetica (L.) Cufod. Biovigyanam 1976, 2, 207–208. [Google Scholar]
- Patel, M.K.; Kulkarni, Y.S.; Dhande, G.W. Some new bacterial diseases in plants. Curr. Sci. 1952, 21, 345–346. [Google Scholar]
- Pereira, A.L.G. Uma nova doença bacteriana do maracujá (Passiflora edulis, Sims) causada por Xanthomonas passiflorae n. sp. Arq. Inst. Biológico São Paulo 1967, 36, 163–174. [Google Scholar]
- Srinivasan, M.C.; Patel, M.K.; Thirumalachar, M.J. Three undescribed Species of Xanthomonas. Curr. Sci. 1956, 11, 366–367. [Google Scholar]
- Moniz, L.; Sabley, J.E.; More, W.D. A new bacterial canker of Carissa congesta in Maharashtra. Indian Phytopathol. 1973, 3, 105. [Google Scholar]
- Patel, M.K.K.; Kulkarni, Y.S.S.; Dhande, G.W. Two new bacterial diseases of plants. Curr. Sci. 1952, 21, 74–75. [Google Scholar]
- Nagarkoti, M.S.; Banerjee, A.K.; Swarup, J. Xanthomonas convolvuli spec. nov. causing leaf spot of Convolvulus arvensis in India. Indian J. Mycol. Plant Pathol. 1973, 3, 105. [Google Scholar]
- Srinivasan, M.C.; Patel, M.K.; Thirumalachar, M.J. A bacterial blight disease of coriander. Proc. Indian Acad. Sci.-Sect. B 1961, 53, 298–301. [Google Scholar] [CrossRef]
- Jain, K.L.; Dange, S.R.S.; Siradhana, B.S. Bacterial leaf spot of Datura metel caused by Xanthomonas campestris f. sp. daturi f. spec. nov. Curr. Sci. 1975, 44, 447. [Google Scholar]
- Sabet, K.A.; Ishag, F.; Khalil, O. Studies on the bacterial diseases of Sudan crops: VII. New records. Ann. Appl. Biol. 1969, 63, 357–369. [Google Scholar] [CrossRef]
- Cavara, F. Bacteriosi del fico. Atti dell’Accademia Gioenia Sci. Nat. Catania Ser. 4 1905, 14, 1–17. [Google Scholar]
- Padhya, A.C.; Patel, M.K. A new bacterial leaf-spot on Ionidium heterophyllum Vent. Indian Phytopathol. 1963, 16, 98–99. [Google Scholar]
- Patel, M.K.; Bhatt, V.V.; Kulkarni, Y.S. Three new bacterial diseases of plants from Bombay. Curr. Sci. 1951, 20, 326–327. [Google Scholar]
- Durgapal, J.C.; Trivedi, B.M. Bacterial blight of “Four-O’Clock”—A new disease in India. Curr. Sci. 1976, 45, 111–112. [Google Scholar]
- Chand, R.; Singh, P.N. Xanthomonas campestris pv. obscurae pv. nov. causal agent of leaf blight of Ipomoea obscura in India / Xanthomonas campestris pv. obscurae pv. nov. als Verursacher einer Blattfleckenkrankheit an Ipomoea obscura in Indien. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 1994, 101, 590–593. [Google Scholar]
- Robbs, C.F.; Medeiros, A.G.; Kimura, O. Mancha bacteriana das folhas do guaranazeiro causada por um novo patovar de Xanthomonas campestris. Arq. Univ. Fed. Rural Rio Janeiro 1982, 5, 195–201. [Google Scholar]
- Qhobela, M.; Claflin, L.E. Characterization of Xanthomonas campestris pv. pennamericanum pv. nov., causal agent of bacterial leaf streak of pearl millet. Int. J. Syst. Bacteriol. 1988, 38, 362–366. [Google Scholar] [CrossRef]
- Patel, M.K. Xanthomonas uppalii sp. nov. pathogenic on Ipomoea muricata. Indian Phytopathol. 1948, 1, 67–69. [Google Scholar]
- Patel, M.K.; Desai, S.G.; Patel, A.J. A new bacterial leaf-spot on Vernonia cinerea Less. Sci. Cult. 1968, 34, 220–221. [Google Scholar]
- Robbs, C.F.; Rodrigues Neto, J.; Malavolta Junior, V.A.; Kimura, O. Bacterial spot and blight of yellow-shrimp (Pachystachys lutea) caused by a new pathovar of Xanthomonas campestris. Summa Phytopathol. 1989, 15, 174–179. [Google Scholar]
- Ren, X.-Z.; Fang, Z.-D. Identification of the causal organism of the bacterial wilt of ginger. Acta Phytopathol. Sin. 1981, 11, 51–56. [Google Scholar]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Ahmed, S.A.; Davenport, K.W.; Flynn, M.C.; Lo, C.C.; Chain, P.S.G. Standardized phylogenetic and molecular evolutionary analysis applied to species across the microbial tree of life. Sci. Rep. 2020, 10, 1723. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 1–4. [Google Scholar] [CrossRef]
- Gonçalves, E.R.; Rosato, Y.B. Genotypic characterization of xanthomonad strains isolated from passion fruit plants (Passiflora spp.) and their relatedness to different Xanthomonas species. Int. J. Syst. Evol. Microbiol. 2000, 50, 811–821. [Google Scholar] [CrossRef]
- Cerqueira Melo, R.D.C.; Dorea Braganca, C.A.; Nogueira Pestana, K.; da Silva, H.S.A.; Ferreira, C.F.; Alves Santos de Oliveira, S. Improvement of the specific detection of Xanthomonas phaseoli pv. manihotis based on the pthB gene. Acta Sci. Agron. 2019, 41, e42708. [Google Scholar] [CrossRef]
- Kodama, Y.; Shumway, M.; Leinonen, R. The Sequence Read Archive: Explosive growth of sequencing data. Nucleic Acids Res. 2012, 40, D54–D56. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.-S.; Park, S.-C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- da Silva, A.C.R.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.C.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef]
- Bansal, K.; Midha, S.; Kumar, S.; Patil, P.B. Ecological and evolutionary insights into Xanthomonas citri pathovar diversity. Appl. Environ. Microbiol. 2017, 83, e02993-16. [Google Scholar] [CrossRef] [PubMed]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Parkinson, N.; Aritua, V.; Heeney, J.; Cowie, C.; Bew, J.; Stead, D. Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2881–2887. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, J.M.; Pesce, C.; Lefeuvre, P.; Koebnik, R. Comparative genomics of a cannabis pathogen reveals insight into the evolution of pathogenicity in Xanthomonas. Front. Plant Sci. 2015, 6, 431. [Google Scholar] [CrossRef] [PubMed]
- Dubrow, Z.E.; Bogdanove, A.J. Genomic insights advance the fight against black rot of crucifers. J. Gen. Plant Pathol. 2021, 87, 127–136. [Google Scholar] [CrossRef]
- Vicente, J.G.; Everett, B.; Roberts, S.J. Identification of isolates that cause a leaf spot disease of brassicas as Xanthomonas campestris pv. raphani and pathogenic and genetic comparison with related pathovars. Phytopathology 2006, 96, 735–745. [Google Scholar] [CrossRef]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Schaad, N.W. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef]
- List of new names and new combinations previously effectively, but not validly, published. Int. J. Syst. Evol. Microbiol. 2006, 56, 925–927. [CrossRef]
- Ah-You, N.; Gagnevin, L.; Grimont, P.A.D.D.; Brisse, S.; Nesme, X.; Chiroleu, F.; Bui Thi Ngoc, L.; Jouen, E.; Lefeuvre, P.; Vernière, C.; et al. Polyphasic characterization of xanthomonads pathogenic to members of the Anacardiaceae and their relatedness to species of Xanthomonas. Int. J. Syst. Evol. Microbiol. 2009, 59, 306–318. [Google Scholar] [CrossRef]
- Bradbury, J.F. Guide to Plant Pathogenic Bacteria; CAB International: Wallingford, UK, 1986; ISBN 0851985572. [Google Scholar]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; McVeigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- Bull, C.T.; De Boer, S.H.; Denny, T.P.; Firrao, G.; Saux, M.F.-L.; Saddler, G.S.; Scortichini, M.; Stead, D.E.; Takikawa, Y. Comprehensive List of names of plant pathogenic bacteria, 1980-2007. J. Plant Pathol. 2010, 92, 551–592. [Google Scholar]
- Young, J.M.; Bull, C.T.; De Boer, S.H.; Firrao, G.; Saddler, G.E.; Stead, D.E.; Takikawa, Y. Names of plant pathogenic bacteria, 1864–2004. Rev. Plant Pathol. 2004, 75, 721–763. [Google Scholar]
- Duff, J. Angular leaf spot of ornamental Ficus spp. due to Xanthomonas campestris pv. fici. Australas. Plant Pathol. 1991, 20, 1–2. [Google Scholar] [CrossRef]
- Jindal, J.K.; Patel, P.N. Angular leaf spot of pipal (Ficus religiosa) due to Xanthomonas fici sp. nov. Indian Phytopathol. 1972, 15, 164–166. [Google Scholar]
- Young, J.M.; Bradbury, J.F.; Davis, R.E.; Dickey, R.S.; Ercolani, G.L.; Hayward, A.C.; Vidaver, A.K. Nomenclatural revisions of plant pathogenic bacteria and list of names 1980–1988. Rev. Plant Pathol. 1991, 70, 211–221. [Google Scholar]
- Smith, E.F. Description of Bacillus phaseoli n. sp. Bot. Gaz. 1897, 24, 192. [Google Scholar]
- Gabriel, D.W.; Kingsley, M.T.; Hunter, J.E.; Gottwald, T. Reinstatement of Xanthomonas citri (ex Hasse) and X. phaseoli (ex Smith) to species and reclassification of all X. campestris pv. citri strains. Int. J. Syst. Bacteriol. 1989, 39, 14–22. [Google Scholar] [CrossRef]
- Garita-Cambronero, J.; Palacio-Bielsa, A.; López, M.M.; Cubero, J. Comparative genomic and phenotypic characterization of pathogenic and non-pathogenic strains of Xanthomonas arboricola reveals insights into the infection process of bacterial spot disease of sone fruits. PLoS ONE 2016, 11, e0161977. [Google Scholar] [CrossRef]
- Mafakheri, H.; Taghavi, S.M.; Zarei, S.; Portier, P.; Dimkić, I.; Koebnik, R.; Kuzmanović, N.; Osdaghi, E. Xanthomonas bonasiae sp. nov. and Xanthomonas youngii sp. nov., isolated from crown gall tissues. Int. J. Syst. Evol. Microbiol. 2022, 72, 005418. [Google Scholar] [CrossRef]
- Hersemann, L.; Wibberg, D.; Blom, J.; Widmer, F.; Kölliker, R. Draft Genome sequence of the Xanthomonas bromi type strain LMG 947. Genome Announc. 2016, 4, e00961-16. [Google Scholar] [CrossRef] [PubMed]
- Bolot, S.; Munoz Bodnar, A.; Cunnac, S.; Ortiz, E.; Szurek, B.; Noël, L.D.; Arlat, M.; Jacques, M.-A.; Gagnevin, L.; Portier, P.; et al. Draft Genome sequence of the Xanthomonas cassavae type strain CFBP 4642. Genome Announc. 2013, 1, e00679-13. [Google Scholar] [CrossRef]
- Gordon, J.L.; Lefeuvre, P.; Escalon, A.; Barbe, V.; Cruveiller, S.; Gagnevin, L.; Pruvost, O. Comparative genomics of 43 strains of Xanthomonas citri pv. citri reveals the evolutionary events giving rise to pathotypes with different host ranges. BMC Genom. 2015, 16, 1098. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, C.; Martins, L.; Teixeira, M.; Blom, J.; Pothier, J.F.; Fonseca, N.A.; Tavares, F. Comparative genomics of Xanthomonas euroxanthea and Xanthomonas arboricola pv. juglandis strains isolated from a single walnut host tree. Microorganisms 2021, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Jacques, M.A.; Bolot, S.; Charbit, E.; Darrasse, A.; Briand, M.; Arlat, M.; Gagnevin, L.; Koebnik, R.; Noël, L.D.; Portier, P.; et al. High-quality draft genome sequence of Xanthomonas alfalfae subsp. alfalfae strain CFBP 3836. Genome Announc. 2013, 1, e01035-13. [Google Scholar] [CrossRef]
- Gagnevin, L.; Bolot, S.; Gordon, J.L.; Pruvost, O.; Vernière, C.; Robène, I.; Arlat, M.; Noël, L.D.; Carrère, S.; Jacques, M.-A.; et al. Draft genome sequence of Xanthomonas axonopodis pv. allii strain CFBP 6369. Genome Announc. 2014, 2, e00727-14. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.G.; Rothwell, S.; Holub, E.B.; Studholme, D.J. Pathogenic, phenotypic and molecular characterisation of Xanthomonas nasturtii sp. nov. and Xanthomonas floridensis sp. nov., new species of Xanthomonas associated with watercress production in Florida. Int. J. Syst. Evol. Microbiol. 2017, 67, 3645–3654. [Google Scholar] [CrossRef]
- Gétaz, M.; Krijger, M.; Rezzonico, F.; Smits, T.H.M.; van der Wolf, J.M.; Pothier, J.F. Genome-based population structure analysis of the strawberry plant pathogen Xanthomonas fragariae reveals two distinct groups that evolved independently before its species description. Microb. Genom. 2018, 4, e000189. [Google Scholar] [CrossRef]
- Michalopoulou, V.A.; Vicente, J.G.; Studholme, D.J.; Sarris, P.F. Draft genome sequences of pathotype strains for three pathovars belonging to three Xanthomonas species. Microbiol. Resour. Announc. 2018, 7, 4–5. [Google Scholar] [CrossRef]
- Cohen, S.P.; Luna, E.K.; Lang, J.M.; Ziegle, J.; Chang, C.; Leach, J.E.; Le-Saux, M.F.; Portier, P.; Koebnik, R.; Jacobs, J.M. High-Quality Genome resource of Xanthomonas hyacinthi generated via long-read sequencing. Plant Dis. 2020, 104, 1011–1012. [Google Scholar] [CrossRef]
- Dia, N.C.; Van Vaerenbergh, J.; Van Malderghem, C.; Blom, J.; Smits, T.H.M.; Cottyn, B.; Pothier, J.F. Xanthomonas hydrangeae sp. nov., a novel plant pathogen isolated from Hydrangea arborescens. Int. J. Syst. Evol. Microbiol. 2021, 71, 005163. [Google Scholar] [CrossRef]
- Rana, R.; Madhavan, V.N.; Saroha, T.; Bansal, K.; Kaur, A.; Sonti, R.V.; Patel, H.K.; Patil, P.B. Xanthomonas indica sp. nov., a novel member of non-pathogenic Xanthomonas community from healthy rice seeds. Curr. Microbiol. 2022, 79, 304. [Google Scholar] [CrossRef] [PubMed]
- Triplett, L.R.; Verdier, V.; Campillo, T.; Van Malderghem, C.; Cleenwerck, I.; Maes, M.; Deblais, L.; Corral, R.; Koita, O.; Cottyn, B.; et al. Characterization of a novel clade of Xanthomonas isolated from rice leaves in Mali and proposal of Xanthomonas maliensis sp. nov. Antonie Van Leeuwenhoek 2015, 107, 869–881. [Google Scholar] [CrossRef]
- Ndongo, S.; Beye, M.; Dubourg, G.; Nguyen, T.T.; Couderc, C.; Fabrizio, D.P.; Fournier, P.-E.; Raoult, D.; Angelakis, E. Genome analysis and description of Xanthomonas massiliensis sp. nov., a new species isolated from human faeces. New Microbes New Infect. 2018, 26, 63–72. [Google Scholar] [CrossRef]
- Mücke, S.; Reschke, M.; Erkes, A.; Schwietzer, C.-A.; Becker, S.; Streubel, J.; Morgan, R.D.; Wilson, G.G.; Grau, J.; Boch, J. Transcriptional Reprogramming of rice cells by Xanthomonas oryzae TALEs. Front. Plant Sci. 2019, 10, 162. [Google Scholar] [CrossRef] [PubMed]
- Naushad, S.; Adeolu, M.; Wong, S.; Sohail, M.; Schellhorn, H.E.; Gupta, R.S. A phylogenomic and molecular marker based taxonomic framework for the order Xanthomonadales: Proposal to transfer the families Algiphilaceae and Solimonadaceae to the order Nevskiales ord. nov. and to create a new family within the order Xanthomonadales. Antonie Van Leeuwenhoek 2015, 107, 467–485. [Google Scholar] [CrossRef]
- López, M.M.; Lopez-Soriano, P.; Garita-Cambronero, J.; Beltrán, C.; Taghouti, G.; Portier, P.; Cubero, J.; Fischer-Le Saux, M.; Marco-Noales, E. Xanthomonas prunicola sp. nov., a novel pathogen that affects nectarine (Prunus persica var. nectarina) trees. Int. J. Syst. Evol. Microbiol. 2018, 68, 1857–1866. [Google Scholar] [CrossRef]
- Bansal, K.; Kaur, A.; Midha, S.; Kumar, S.; Korpole, S.; Patil, P.B. Xanthomonas sontii sp. nov., a non-pathogenic bacterium isolated from healthy basmati rice (Oryza sativa) seeds from India. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2021, 114, 1935–1947. [Google Scholar] [CrossRef]
- Gilroy, R.; Ravi, A.; Getino, M.; Pursley, I.; Horton, D.L.; Alikhan, N.-F.; Baker, D.; Gharbi, K.; Hall, N.; Watson, M.; et al. Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ 2021, 9, e10941. [Google Scholar] [CrossRef]
- Koebnik, R.; Burokiene, D.; Bragard, C.; Chang, C.; Saux, M.F.-L.; Kölliker, R.; Lang, J.M.; Leach, J.E.; Luna, E.K.; Portier, P.; et al. The Complete Genome Sequence of Xanthomonas theicola, the causal agent of canker on tea plants, reveals novel secretion systems in Clade-1 xanthomonads. Phytopathology 2021, 111, 611–616. [Google Scholar] [CrossRef]
- Jaenicke, S.; Bunk, B.; Wibberg, D.; Spröer, C.; Hersemann, L.; Blom, J.; Winkler, A.; Schatschneider, S.; Albaum, S.P.; Kölliker, R.; et al. Complete genome sequence of the barley pathogen Xanthomonas translucens pv. translucens DSM 18974T (ATCC 19319T). Genome Announc. 2016, 4, e01334-16. [Google Scholar] [CrossRef] [PubMed]
- Richard, D.; Boyer, C.; Lefeuvre, P.; Canteros, B.I.; Beni-Madhu, S.; Portier, P.; Pruvost, O. Complete genome sequences of six copper-resistant Xanthomonas strains causing bacterial spot of solaneous plants, belonging to X. gardneri, X. euvesicatoria, and X. vesicatoria, using long-read technology. Genome Announc. 2017, 5, e01693-16. [Google Scholar] [CrossRef] [PubMed]
| Pathovar | Strain NCPPB Number | Host | FAME Cluster | References |
|---|---|---|---|---|
| Controls: X. campestris | ||||
| aberrans | 2986 | Brassica oleracea var. capitata | 2 | [14] |
| armoraciae | 347 | Iberis sp. | 1 | [15,16,17] |
| barbareae | 983 | Barbarea vulgaris | 7 | [18] |
| incanae | 937 | Matthiola sp. | 2 | [19] |
| papavericola | 2970 | Papaver rhoeas | 2 | [20] |
| plantaginis | 1061 | Plantago lanceolata | 3 | [21] |
| raphani | 1946 | Raphanus sativus | 2 | [22] |
| Controls: X. euvesicatoria | ||||
| alangii | 1336 | Alangium lamarckii | 1 | [10,23] |
| physalidis | 1756 | Physalis minima | 1 | [10,24] |
| Controls: X. citri | ||||
| merremiae | 3114 | Merremia gangetica | 1 | [11,25] |
| trichodesmae | 585 | Trichodesma zeylanicum | 1 | [11,26] |
| Control: X. phaseoli | ||||
| passiflorae | 2346 | Passiflora edulis | 1 | [27] |
| Transfer to Xanthomonas euvesicatoria | ||||
| blepharidis | 1757 | Blepharis boarhaavifolia | 1 | [28] |
| carissae | 2373 | Carissa carandas | 1 | [29] |
| clerodendri | 575 | Clerodendron sp. | 1 | [30] |
| convolvuli | 2498 | Convolvulus arvensis | (1) | [31] |
| coriandri | 1758 | Coriandrum sativum | 1 | [32] |
| daturae | 2932 | Datura metel | N.d. | [33] |
| euphorbiae | 1828 | Euphorbia acalyphoides | 1 | [34] |
| fici | 2372 | Ficus religiosa | (1) | [35] |
| heliotropii | 2057 | Heliotropium sudanicum | [1] | [34] |
| ionidii | 1334 | Ionidium heterophyllum | 1 | [36] |
| lawsoniae | 579 | Lawsonia inermis | [1] | [37] |
| mirabilis | 4348 | Mirabilis jalapa | N.d. | [38] |
| obscurae | 3759 | Ipomea obscura | N.d. | [39] |
| paulliniae | 3079 | Paullinia cupana | (1) | [40] |
| pennamericanum | 4349 | Pennisetum americanum | N.d. | [41] |
| spermacoces | 1760 | Spermacoce hispida | 14 | [28] |
| uppalii | 586 | Ipomoea muricata | (1) | [42] |
| vernoniae | 1787 | Vernonia cinerea | (1) | [43] |
| viegasii | 4351 | Pachystachys lutea | N.d. | [44] |
| zingibericola | 4352 | Zingiber officinale | (1) | [45] |
| Assembly Accession | Strain (NCPPB Number) | Length (b.p.) | Number of Contigs | N50 Length (b.p.) |
|---|---|---|---|---|
| GCA_020813115.1 | aberrans 2986 | 5,136,506 | 75 | 145,536 |
| GCA_020731405.1 | armoraciae 347 | 5,063,577 | 72 | 149,552 |
| GCA_020813315.1 | barbareae 983 | 4,982,365 | 62 | 208,499 |
| GCA_020813295.1 | incanae 937 | 4,916,318 | 50 | 191,313 |
| GCA_020813015.1 | papavericola 2970 | 5,509,044 | 46 | 220,536 |
| GCA_020813005.1 | plantaginis 1061 | 5,214,263 | 76 | 153,285 |
| GCA_020813075.1 | raphani 1946 | 4,898,270 | 53 | 196,893 |
| GCA_019193005.1 | alangii 1336 | 4,984,130 | 20 | 399,322 |
| GCA_019192985.1 | physalidis 1756 | 5,125,394 | 37 | 354,850 |
| GCA_019201545.1 | merremiae 3114 | 5,092,415 | 53 | 310,143 |
| GCA_019201525.1 | trichodesmae 585 | 5,572,310 | 82 | 194,166 |
| GCA_019201075.1 | passiflorae 2346 | 5,022,320 | 68 | 164,613 |
| GCA_019201485.1 | blepharidis 1757 | 4,969,587 | 58 | 167,765 |
| GCA_019201365.1 | carissae 2373 | 5,089,141 | 136 | 93,135 |
| GCA_019201225.1 | clerodendri 575 | 5,097,109 | 39 | 312,137 |
| GCA_019201245.1 | convolvuli 2498 | 4,982,289 | 54 | 201,220 |
| GCA_019201305.1 | coriandri 1758 | 5,049,250 | 34 | 522,126 |
| GCA_019201325.1 | daturae 2932 | 5,173,277 | 46 | 333,897 |
| GCA_019201425.1 | euphorbiae 1828 | 4,926,792 | 26 | 995,918 |
| GCA_019201375.1 | fici 2372 | 4,880,201 | 87 | 109,976 |
| GCA_019201405.1 | heliotropii 2057 | 5,161,499 | 120 | 107,075 |
| GCA_019201465.1 | ionidii 1334 | 4,974,037 | 50 | 264,595 |
| GCA_019201185.1 | lawsoniae 579 | 5,050,397 | 112 | 114,657 |
| GCA_019201145.1 | mirabilis 4348 | 5,091,739 | 25 | 745,972 |
| GCA_019201105.1 | obscurae 3759 | 5,007,609 | 61 | 242,019 |
| GCA_019201285.1 | paulliniae 3079 | 4,806,543 | 18 | 540,544 |
| GCA_019201165.1 | pennamericanum 4349 | 5,014,174 | 87 | 133,002 |
| GCA_019201445.1 | spermacoces 1760 | 5,070,626 | 45 | 344,807 |
| GCA_019201115.1 | uppalii 586 | 4,954,104 | 37 | 305,235 |
| GCA_019201345.1 | vernoniae 1787 | 4,961,493 | 98 | 150,860 |
| GCA_019201265.1 | viegasii 4351 PT | 5,007,865 | 38 | 232,093 |
| GCA_019201205.1 | zingibericola 4352 | 4,926,474 | 31 | 267,819 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harrison, J.; Hussain, R.M.F.; Aspin, A.; Grant, M.R.; Vicente, J.G.; Studholme, D.J. Phylogenomic Analysis Supports the Transfer of 20 Pathovars from Xanthomonas campestris into Xanthomonas euvesicatoria. Taxonomy 2023, 3, 29-45. https://doi.org/10.3390/taxonomy3010003
Harrison J, Hussain RMF, Aspin A, Grant MR, Vicente JG, Studholme DJ. Phylogenomic Analysis Supports the Transfer of 20 Pathovars from Xanthomonas campestris into Xanthomonas euvesicatoria. Taxonomy. 2023; 3(1):29-45. https://doi.org/10.3390/taxonomy3010003
Chicago/Turabian StyleHarrison, Jamie, Rana M. F. Hussain, Andrew Aspin, Murray R. Grant, Joana G. Vicente, and David J. Studholme. 2023. "Phylogenomic Analysis Supports the Transfer of 20 Pathovars from Xanthomonas campestris into Xanthomonas euvesicatoria" Taxonomy 3, no. 1: 29-45. https://doi.org/10.3390/taxonomy3010003
APA StyleHarrison, J., Hussain, R. M. F., Aspin, A., Grant, M. R., Vicente, J. G., & Studholme, D. J. (2023). Phylogenomic Analysis Supports the Transfer of 20 Pathovars from Xanthomonas campestris into Xanthomonas euvesicatoria. Taxonomy, 3(1), 29-45. https://doi.org/10.3390/taxonomy3010003







