Acinetobacter nematophilus sp. nov., Alcaligenes nematophilus sp. nov., Enterobacter nematophilus sp. nov., and Kaistia nematophila sp. nov., Isolated from Soil-Borne Nematodes and Proposal for the Elevation of Alcaligenes faecalis subsp. faecalis, Alcaligenes faecalis subsp. parafaecalis, and Alcaligenes faecalis subsp. phenolicus to the Species Level
Abstract
1. Introduction
2. Material and Methods
2.1. Bacteria Isolation
2.2. Bacteria Molecular Characterization
2.3. 16S rRNA Gene Sequencing
2.4. Whole Genome Sequencing
2.5. Whole Genome and Whole Proteome-Based Phylogenies
2.6. Physiological, Biochemical and Morphological Characterization
3. Results and Discussion
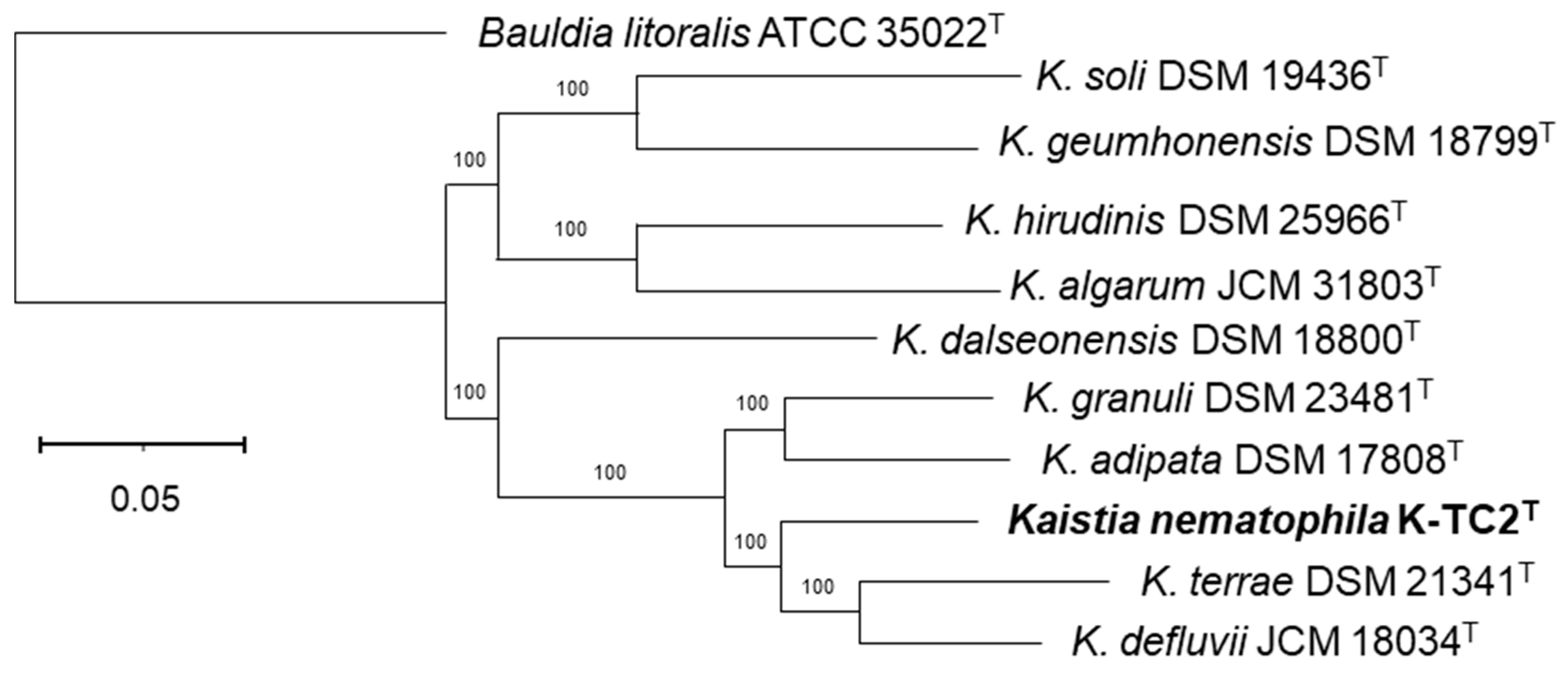
3.1. Phylogenetic and Phylogenomic Reconstructions and Sequence Comparisons
3.2. Genomic Features
3.3. Physiological and Biochemical Characteristics
3.4. Ecology
3.5. Conclusions
3.6. Protologues
- Emended description of Alcaligenes faecalis
- Description of Acinetobacter nematophilus sp. nov.
- Description of Alcaligenes nematophilus sp. nov.
- Description of Alcaligenes parafaecalis sp. nov.
- Description of Alcaligenes phenolicus sp. nov.
- Description of Enterobacter nematophilus sp. nov.
- Description of Kaistia nematophila sp. nov.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ogier, J.-C.; Frayssinet, M.; Gaudriault, S. Entomopathogenic nematode-associated microbiota: From monoxenic paradigm to pathobiome. Microbiome 2020, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Shokoohi, E.; Mashela, P.W.; Machado, R.A.R. Bacterial communities associated with Zeldia punctata, a bacterivorous soil-borne nematode. Int. Microbiol. 2022, 25, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.; Kuhnert, P.; Erb, M.; Machado, R.A.R. Identification of Photorhabdus symbionts by MALDI-TOF MS. Microbiology 2020, 166, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.A.R.; Thönen, L.; Arce, C.; Theepan, V.; Prada, F.; Wüthrich, D.; Robert, C.A.M.; Vogiatzaki, E.; Shi, Y.-M.; Schaeren, O.P. Engineering bacterial symbionts of nematodes improves their biocontrol potential to counter the western corn rootworm. Nat. Biotechnol. 2020, 38, 600–608. [Google Scholar] [CrossRef]
- Waterfield, N.R.; Ciche, T.; Clarke, D. Photorhabdus and a host of hosts. Annu. Rev. Microbiol. 2009, 63, 557–574. [Google Scholar] [CrossRef]
- Zhou, X.; Kaya, H.K.; Heungens, K.; Goodrich-Blair, H. Response of ants to a deterrent factor (s) produced by the symbiotic bacteria of entomopathogenic nematodes. Appl. Environ. Microbiol. 2002, 68, 6202–6209. [Google Scholar] [CrossRef] [PubMed]
- Murfin, K.E.; Dillman, A.R.; Foster, J.M.; Bulgheresi, S.; Slatko, B.E.; Sternberg, P.W.; Goodrich-Blair, H. Nematode-bacterium symbioses—Cooperation and conflict revealed in the “Omics” age. Biol. Bull. 2012, 223, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Rafaluk-Mohr, C.; Ashby, B.; Dahan, D.A.; King, K.C. Mutual fitness benefits arise during coevolution in a nematode-defensive microbe model. Evol. Lett. 2018, 2, 246–256. [Google Scholar] [CrossRef]
- Jones, R.S.; Fenton, A.; Speed, M.P.; Mappes, J. Investment in multiple defences protects a nematode-bacterium symbiosis from predation. Anim. Behav. 2017, 129, 1–8. [Google Scholar] [CrossRef]
- Hodgkin, J.; Kuwabara, P.E.; Corneliussen, B. A novel bacterial pathogen, Microbacterium nematophilum, induces morphological change in the nematode C. elegans. Curr. Biol. 2000, 10, 1615–1618. [Google Scholar] [CrossRef]
- Emelianoff, V.; Chapuis, E.; Le Brun, N.; Chiral, M.; Moulia, C.; Ferdy, J.-B. A survival-reproduction trade-off in entomopathogenic nematodes mediated by their bacterial symbionts. Evol. Int. J. Org. Evol. 2008, 62, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Machado, R.A.R.; van Doan, C.; Arce, C.C.M.; Hu, L.; Robert, C.A.M. Entomopathogenic nematodes increase predation success by inducing cadaver volatiles that attract healthy herbivores. Elife 2019, 8, e46668. [Google Scholar] [CrossRef]
- Loulou, A.; M’saad Guerfali, M.; Muller, A.; Bhat, A.H.; Abolafia, J.; Machado, R.A.R.; Kallel, S. Potential of Oscheius tipulae nematodes as biological control agents against Ceratitis capitata. PLoS ONE 2022, 17, e0269106. [Google Scholar] [CrossRef]
- Loulou, A.; Mastore, M.; Caramella, S.; Bhat, A.; Brivio, M.; Machado, R.; Kallel, S. Entomopathogenic potential of bacteria associated with soil-borne nematodes and insect immune responses to their infection. PLoS ONE 2023, 18, e0280675. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.H.; Loulou, A.; Abolafia, J.; Machado, R.A.R.; Kallel, S. Comparative morphological and molecular analyses of Acrobeloides bodenheimeri and A. tricornis Cobb, 1924 (Rhabditida, Cephalobidae) from Tunisia. Nematology 2023, 25, 207–226. [Google Scholar] [CrossRef]
- Brisou, J.; Prevot, A.R. Études de systématique bactérienne. 10. Révision des espèces réunies dans le genre Achromobacter. In Annales de l’Institut Pasteur; MASSON EDITEUR: Paris, France, 1954; Volume 86, pp. 722–728. [Google Scholar]
- Elnar, A.G.; Kim, M.-G.; Lee, J.-E.; Han, R.-H.; Yoon, S.-H.; Lee, G.-Y.; Yang, S.-J.; Kim, G.B. Acinetobacter pullorum sp. nov., Isolated from chicken meat. J. Microbiol. Biotechnol. 2020, 30, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microbes Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef]
- Li, W.; Zhang, D.; Huang, X.; Qin, W. Acinetobacter harbinensis sp. nov., isolated from river water. Int. J. Syst. Evol. Microbiol. 2014, 64, 1507–1513. [Google Scholar] [CrossRef]
- Nishimura, Y.; Ino, T.; Iizuka, H. Acinetobacter radioresistens sp. nov. isolated from cotton and soil. Int. J. Syst. Evol. Microbiol. 1988, 38, 209–211. [Google Scholar] [CrossRef]
- Nemec, A.; De Baere, T.; Tjernberg, I.; Vaneechoutte, M.; van der Reijden, T.J.; Dijkshoorn, L. Acinetobacter ursingii sp. nov. and Acinetobacter schindleri sp. nov., isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 2001, 51, 1891–1899. [Google Scholar] [CrossRef]
- Nemec, A.; Musilek, M.; Maixnerova, M.; De Baere, T.; van der Reijden, T.; Vaneechoutte, M.; Dijkshoorn, L. Acinetobacter beijerinckii sp nov and Acinetobacter gyllenbergii sp nov., haemolytic organisms isolated from humans. Int. J. Syst. Evol. Microbiol. 2009, 59, 118–124. [Google Scholar] [CrossRef]
- Castellani, A.; Chalmers, A.J. Manual of Tropical Medicine, 3rd ed.; Williams Wood and Co.: New York, NY, USA, 1919. [Google Scholar]
- van Trappen, S.; Tan, T.-L.; Samyn, E.; Vandamme, P. Alcaligenes aquatilis sp. nov., a novel bacterium from sediments of the Weser Estuary, Germany, and a salt marsh on Shem Creek in Charleston Harbor, USA. Int. J. Syst. Evol. Microbiol. 2005, 55, 2571–2575. [Google Scholar] [CrossRef]
- Schroll, G.; Busse, H.-J.; Parrer, G.; Rölleke, S.; Lubitz, W.; Denner, E.B.M. Alcaligenes faecalis subsp. parafaecalis subsp. nov., a bacterium accumulating poly-β-hydroxybutyrate from acetone-butanol bioprocess residues. Syst. Appl. Microbiol. 2001, 24, 37–43. [Google Scholar]
- Lu, C.-Y.; Li, Y.-Q.; Tian, Y.; Han, M.-X.; Rao, M.P.N.; Li, Y.-R.; Zhu, Z.-N.; Wei, D.-Q.; An, D.-D.; Li, W.-J. Alcaligenes endophyticus sp. nov., isolated from roots of Ammodendron bifolium. Int. J. Syst. Evol. Microbiol. 2017, 67, 939–943. [Google Scholar] [CrossRef]
- Abbas, S.; Ahmed, I.; Kudo, T.; Iida, T.; Ali, G.M.; Fujiwara, T.; Ohkuma, M. Heavy metal-tolerant and psychrotolerant bacterium Acinetobacter pakistanensis sp. nov. isolated from a textile dyeing wastewater treatment pond. Pak. J. Agric. Sci. 2014, 51, 593–606. [Google Scholar]
- Rehfuss, M.; Urban, J. Alcaligenes faecalis subsp. phenolicus subsp. nov. a phenol-degrading, denitrifying bacterium isolated from a graywater bioprocessor. Syst. Appl. Microbiol. 2005, 28, 421–429. [Google Scholar]
- Hormaeche, E.; Edwards, P.R. A proposed genus Enterobacter. Int. Bull. Bacteriol. Nomencl. Taxon. 1960, 10, 71–74. [Google Scholar] [CrossRef]
- Cho, G.-S.; Stein, M.; Fiedler, G.; Igbinosa, E.O.; Koll, L.P.; Brinks, E.; Rathje, J.; Neve, H.; Franz, C.M. Polyphasic study of antibiotic-resistant enterobacteria isolated from fresh produce in Germany and description of Enterobacter vonholyi sp. nov. isolated from marjoram and Enterobacter dykesii sp. nov. isolated from mung bean sprout. Syst. Appl. Microbiol. 2021, 44, 126174. [Google Scholar] [CrossRef]
- García-González, T.; Sáenz-Hidalgo, H.K.; Silva-Rojas, H.V.; Morales-Nieto, C.; Vancheva, T.; Koebnik, R.; Ávila-Quezada, G.D. Enterobacter cloacae, an emerging plant-pathogenic bacterium affecting chili pepper seedlings. Plant Pathol. J. 2018, 34, 1–10. [Google Scholar] [CrossRef]
- Wang, C.; Wu, W.; Wei, L.; Feng, Y.; Kang, M.; Xie, Y.; Zong, Z. Enterobacter wuhouensis sp. nov. and Enterobacter quasihormaechei sp. nov. recovered from human sputum. Int. J. Syst. Evol. Microbiol. 2020, 70, 874–881. [Google Scholar] [CrossRef]
- Wu, W.; Feng, Y.; Zong, Z. Enterobacter sichuanensis sp. nov., recovered from human urine. Int. J. Syst. Evol. Microbiol. 2018, 68, 3922–3927. [Google Scholar] [CrossRef]
- Wu, W.; Wei, L.; Feng, Y.; Kang, M.; Zong, Z. Enterobacter huaxiensis sp. nov. and Enterobacter chuandaensis sp. nov., recovered from human blood. Int. J. Syst. Evol. Microbiol. 2019, 69, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Hu, M.; Chen, S.; Hu, A.; Li, S.; Han, H.; Lu, G.; Zeng, L.; Zhou, J. Enterobacter asburiae and Pantoea ananatis causing rice bacterial blight in China. Plant Dis. 2021, 105, 2078–2088. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Feng, Y.; Zong, Z. Precise species identification for Enterobacter: A genome sequence-based study with reporting of two novel species, Enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. Msystems 2020, 5, e00527-20. [Google Scholar] [CrossRef]
- Im, W.-T.; Yokota, A.; Kim, M.-K.; Lee, S.-T. Kaistia adipata gen. nov., sp. nov., a novel α-proteobacterium. J. Gen. Appl. Microbiol. 2004, 50, 249–254. [Google Scholar] [CrossRef]
- Jin, L.; Kim, K.K.; Lee, H.-G.; Ahn, C.-Y.; Oh, H.-M. Kaistia defluvii sp. nov., isolated from river sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 2878–2882. [Google Scholar] [CrossRef]
- Jin, L.; Kim, K.K.; Baek, S.-H.; Lee, S.-T. Kaistia geumhonensis sp. nov. and Kaistia dalseonensis sp. nov., two members of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 2011, 61, 2577–2581. [Google Scholar] [CrossRef]
- Lee, H.-W.; Yu, H.-S.; Liu, Q.; Jung, H.-M.; An, D.-S.; Im, W.-T.; Jin, F.-X.; Lee, S.-T. Kaistia granuli sp. nov., isolated from anaerobic granules in an upflow anaerobic sludge blanket reactor. Int. J. Syst. Evol. Microbiol. 2007, 57, 2280–2283. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Weon, H.-Y.; Kim, Y.-S.; Anandham, R.; Yoo, S.-H.; Park, I.-C.; Kwon, S.-W. Kaistia terrae sp. nov., isolated from a wetland in Korea. Int. J. Syst. Evol. Microbiol. 2010, 60, 949–952. [Google Scholar] [CrossRef]
- Weon, H.-Y.; Lee, C.-M.; Hong, S.-B.; Kim, B.-Y.; Yoo, S.-H.; Kwon, S.-W.; Go, S.-J. Kaistia soli sp. nov., isolated from a wetland in Korea. Int. J. Syst. Evol. Microbiol. 2008, 58, 1522–1524. [Google Scholar] [CrossRef]
- Glaeser, S.P.; Galatis, H.; Martin, K.; Kämpfer, P. Kaistia hirudinis sp. nov., isolated from the skin of Hirudo verbana. Int. J. Syst. Evol. Microbiol. 2013, 63, 3209–3213. [Google Scholar] [CrossRef] [PubMed]
- Orozco, R.A.; Lee, M.-M.; Stock, S.P. Soil sampling and isolation of entomopathogenic nematodes (Steinernematidae, Heterorhabditidae). JoVE (J. Vis. Exp.) 2014, 89, e52083. [Google Scholar]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Dymock, D.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA Sequencing, 1st ed.; John Wiley and Sons: New York, NY, USA, 1991. [Google Scholar]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; pp. 95–98. [Google Scholar]
- Seemann, T. Barrnap 0.7: Rapid Ribosomal RNA Prediction. 2013. Available online: https://github.com/tseemann/barrnap (accessed on 5 June 2022).
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: Oxford, UK, 2000; ISBN 0195350510. [Google Scholar]
- Chevenet, F.; Brun, C.; Bañuls, A.-L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Machado, R.A.R.; Bhat, A.H.; Abolafia, J.; Muller, A.; Bruno, P.; Fallet, P.; Arce, C.C.M.; Turlings, T.C.J.; Bernal, J.S.; Kajuga, J. Multi-locus phylogenetic analyses uncover species boundaries and reveal the occurrence of two new entomopathogenic nematode species, Heterorhabditis ruandica n. sp. and Heterorhabditis zacatecana n. sp. J. Nematol. 2021, 53, 1–42. [Google Scholar] [CrossRef]
- Machado, R.A.R.; Somvanshi, V.S.; Muller, A.; Kushwah, J.; Bhat, C.G. Photorhabdus hindustanensis sp. nov., Photorhabdus akhurstii subsp. akhurstii subsp. nov., and Photorhabdus akhurstii subsp. bharatensis subsp. nov., isolated from Heterorhabditis entomopathogenic nematodes. Int. J. Syst. Evol. Microbiol. 2021, 71, 4998. [Google Scholar]
- Petit, R.A.; Read, T.D. Bactopia: A flexible pipeline for complete analysis of bacterial genomes. Msystems 2020, 5, e00190-20. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Van Dongen, S.M. Graph Clustering by Flow Simulation [Internet]. Ph. D Thesis, University of Utrecht, Utrecht, The Netherlands, 2000. [Google Scholar]
- van Dongen, S.; Abreu-Goodger, C. Using MCL to extract clusters from networks. In Bacterial Molecular Networks; Springer: Berlin/Heidelberg, Germany, 2012; pp. 281–295. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef]
- Pan, H.; Li, J.; Liu, H.-H.; Lu, X.-Y.; Zhang, Y.-F.; Tian, Y. Acinetobacter tibetensis sp. nov., isolated from a soil under a greenhouse in Tibet. Curr. Microbiol. 2023, 80, 51. [Google Scholar] [CrossRef]
- Carvalheira, A.; Gonzales-Siles, L.; Salvà-Serra, F.; Lindgren, Å.; Svensson-Stadler, L.; Thorell, K.; Piñeiro-Iglesias, B.; Karlsson, R.; Silva, J.; Teixeira, P. Acinetobacter portensis sp. nov. and Acinetobacter guerrae sp. nov., isolated from raw meat. Int. J. Syst. Evol. Microbiol. 2020, 70, 4544–4554. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.; Murray, R.; Stackebrandt, E. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, C.O. Kaistia algarum sp. nov., isolated from a freshwater green alga Paulinella chromatophora. Int. J. Syst. Evol. Microbiol. 2018, 68, 3028–3033. [Google Scholar] [CrossRef]
- Nemec, A.; Musílek, M.; Šedo, O.; de Baere, T.; Maixnerova, M.; van der Reijden, T.J.K.; Zdráhal, Z.; Vaneechoutte, M.; Dijkshoorn, L. Acinetobacter bereziniae sp. nov. and Acinetobacter guillouiae sp. nov., to accommodate Acinetobacter genomic species 10 and 11, respectively. Int. J. Syst. Evol. Microbiol. 2010, 60, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Kersters, K. Genus Alcaligenes Castellani and Chalmers 1919. Bergey’s Man. Syst. Bacteriol. 1984, 1, 361–373. [Google Scholar]
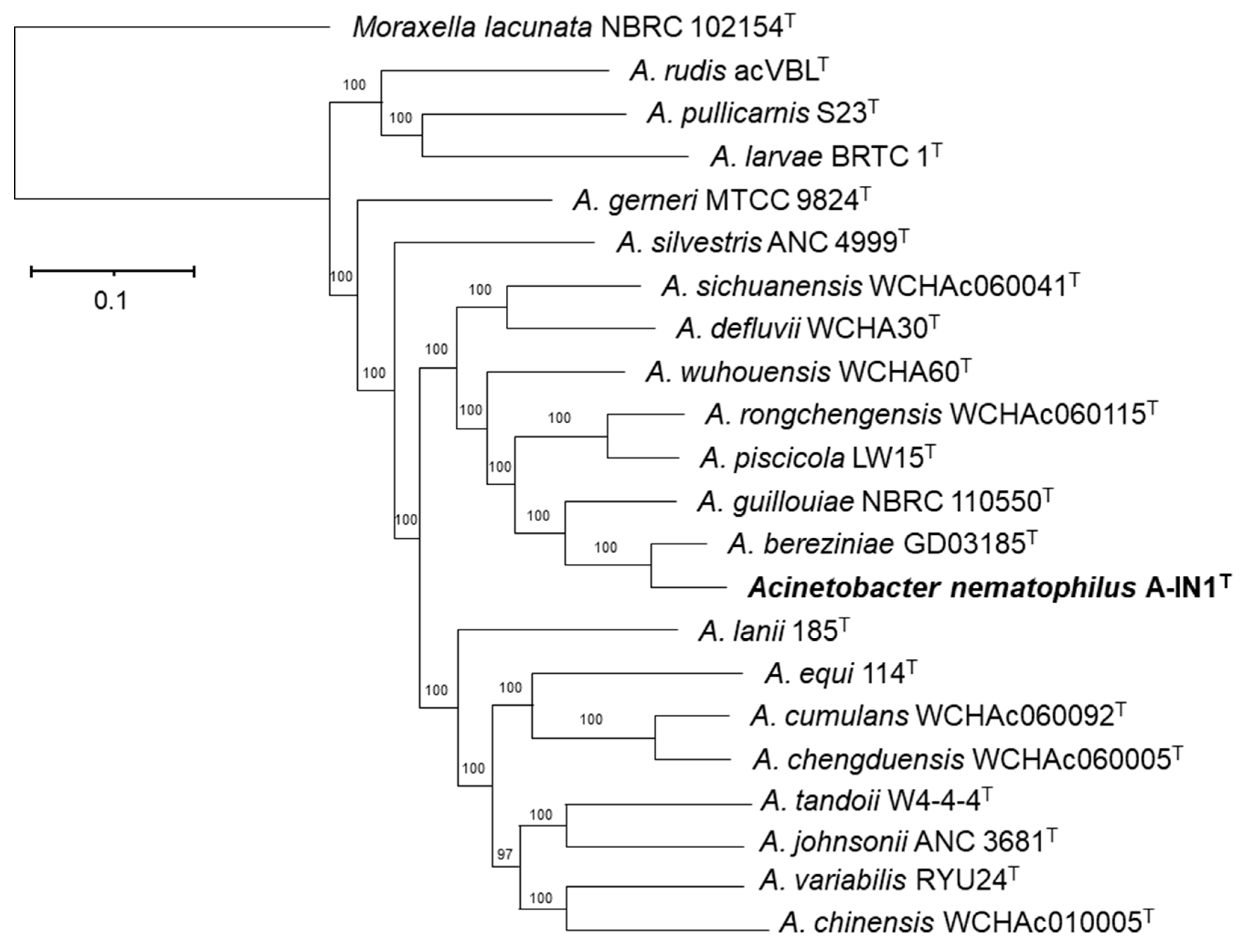


| A. rudis DSM 24031T | A. pullicarnis S23T | A. larvae BRTC-1T | A. gerneri MTCC 9824T | A. silvestris ANC 4999T | A. sichuanensis WCHAc060041T | A. defluvii WCHA30T | A. wuhouensis WCHA60T | A. rongchengensis WCHAc060115T | A. piscicola LW15T | A. guillouiae DSM 590T | A. bereziniae LMG 1003T | A. nematophilus sp. nov. A-IN1T | A. lanii 185T | A. equi 114T | A. cumulans WCHAc060092T | A. chengduensis WCHAc060005T | A. tandoii DSM 14970T | A. johnsonii DSM 69633T | A. variabilis NIPH 2171T | A. chinensis WCHAc010005T | |
| A. rudis DSM 24031T | ID | 21.2 | 22.0 | 21.9 | 20.9 | 21.3 | 21.4 | 21.7 | 21.0 | 21.3 | 21.5 | 21.7 | 20.8 | 21.4 | 21.5 | 21.3 | 21.1 | 20.8 | 20.9 | 21.1 | 21.3 |
| A. pullicarnis S23T | 21.2 | ID | 22.0 | 22.7 | 21.0 | 22.1 | 22.8 | 22.7 | 24.9 | 21.7 | 22.2 | 22.5 | 21.0 | 22.0 | 21.8 | 22.6 | 21.7 | 23.0 | 22.0 | 24.1 | 22.7 |
| A. larvae BRTC-1T | 22.0 | 22.0 | ID | 22.2 | 20.8 | 20.9 | 22.1 | 21.8 | 21.8 | 21.6 | 22.0 | 21.9 | 21.1 | 22.0 | 21.9 | 21.4 | 20.8 | 21.3 | 21.1 | 21.7 | 21.5 |
| A. gerneri MTCC 9824T | 21.9 | 22.7 | 22.2 | ID | 21.6 | 22.2 | 23.1 | 23.1 | 22.9 | 22.2 | 22.3 | 22.7 | 21.5 | 22.1 | 22.3 | 23.0 | 22.8 | 23.1 | 21.9 | 23.1 | 22.6 |
| A. silvestris ANC 4999T | 20.9 | 21.0 | 20.8 | 21.6 | ID | 21.5 | 21.6 | 22.0 | 22.0 | 22.1 | 22.1 | 22.0 | 21.5 | 21.7 | 21.7 | 21.2 | 21.0 | 21.0 | 20.7 | 20.3 | 20.8 |
| A. sichuanensis WCHAc060041T | 21.3 | 22.1 | 20.9 | 22.2 | 21.5 | ID | 27.0 | 24.2 | 27.1 | 22.9 | 23.1 | 26.6 | 22.5 | 22.0 | 22.1 | 23.2 | 22.5 | 22.0 | 21.4 | 21.6 | 23.2 |
| A. defluvii WCHA30T | 21.4 | 22.8 | 22.1 | 23.1 | 21.6 | 27.0 | ID | 25.4 | 28.0 | 23.2 | 23.4 | 28.4 | 22.7 | 22.4 | 22.7 | 23.7 | 22.4 | 22.3 | 22.0 | 22.4 | 24.7 |
| A. wuhouensis WCHA60T | 21.7 | 22.7 | 21.8 | 23.1 | 22.0 | 24.2 | 25.4 | ID | 24.3 | 24.2 | 24.5 | 24.4 | 23.3 | 22.1 | 22.7 | 24.6 | 22.9 | 22.0 | 22.3 | 22.6 | 27.5 |
| A. rongchengensis WCHAc060115T | 21.0 | 24.9 | 21.8 | 22.9 | 22.0 | 27.1 | 28.0 | 24.3 | ID | 36.4 | 24.5 | 27.1 | 23.2 | 21.8 | 21.9 | 22.5 | 21.8 | 23.2 | 21.7 | 22.4 | 22.9 |
| A. piscicola LW15T | 21.3 | 21.7 | 21.6 | 22.2 | 22.1 | 22.9 | 23.2 | 24.2 | 36.4 | ID | 25.2 | 24.2 | 23.6 | 21.3 | 22.3 | 22.0 | 21.6 | 21.8 | 21.3 | 21.3 | 22.2 |
| A. guillouiae DSM 590T | 21.5 | 22.2 | 22.0 | 22.3 | 22.1 | 23.1 | 23.4 | 24.5 | 24.5 | 25.2 | ID | 26.2 | 25.1 | 22.0 | 22.4 | 22.2 | 21.9 | 22.4 | 21.9 | 22.9 | 22.1 |
| A. bereziniae LMG 1003T | 21.7 | 22.5 | 21.9 | 22.7 | 22.0 | 26.6 | 28.4 | 24.4 | 27.1 | 24.2 | 26.2 | ID | 39.6 | 22.0 | 22.4 | 22.4 | 21.5 | 21.6 | 21.8 | 22.9 | 23.0 |
| A. nematophilus sp. nov. A-IN1T | 20.8 | 21.0 | 21.1 | 21.5 | 21.5 | 22.5 | 22.7 | 23.3 | 23.2 | 23.6 | 25.1 | 39.6 | ID | 21.2 | 21.6 | 21.3 | 20.9 | 20.7 | 20.8 | 20.8 | 21.6 |
| A. lanii 185T | 21.4 | 22.0 | 22.0 | 22.1 | 21.7 | 22.0 | 22.4 | 22.1 | 21.8 | 21.3 | 22.0 | 22.0 | 21.2 | ID | 22.5 | 22.9 | 22.2 | 21.8 | 21.7 | 22.4 | 21.7 |
| A. equi 114T | 21.5 | 21.8 | 21.9 | 22.3 | 21.7 | 22.1 | 22.7 | 22.7 | 21.9 | 22.3 | 22.4 | 22.4 | 21.6 | 22.5 | ID | 22.6 | 22.5 | 22.0 | 21.6 | 21.5 | 21.8 |
| A. cumulans WCHAc060092T | 21.3 | 22.6 | 21.4 | 23.0 | 21.2 | 23.2 | 23.7 | 24.6 | 22.5 | 22.0 | 22.2 | 22.4 | 21.3 | 22.9 | 22.6 | ID | 33.8 | 21.6 | 22.3 | 22.4 | 23.5 |
| A. chengduensis WCHAc060005T | 21.1 | 21.7 | 20.8 | 22.8 | 21.0 | 22.5 | 22.4 | 22.9 | 21.8 | 21.6 | 21.9 | 21.5 | 20.9 | 22.2 | 22.5 | 33.8 | ID | 21.2 | 21.3 | 21.9 | 22.2 |
| A. tandoii DSM 14970T | 20.8 | 23.0 | 21.3 | 23.1 | 21.0 | 22.0 | 22.3 | 22.0 | 23.2 | 21.8 | 22.4 | 21.6 | 20.7 | 21.8 | 22.0 | 21.6 | 21.2 | ID | 22.4 | 22.5 | 20.6 |
| A. johnsonii DSM 69633T | 20.9 | 22.0 | 21.1 | 21.9 | 20.7 | 21.4 | 22.0 | 22.3 | 21.7 | 21.3 | 21.9 | 21.8 | 20.8 | 21.7 | 21.6 | 22.3 | 21.3 | 22.4 | ID | 22.0 | 21.1 |
| A. variabilis NIPH 2171T | 21.1 | 24.1 | 21.7 | 23.1 | 20.3 | 21.6 | 22.4 | 22.6 | 22.4 | 21.3 | 22.9 | 22.9 | 20.8 | 22.4 | 21.5 | 22.4 | 21.9 | 22.5 | 22.0 | ID | 22.2 |
| A. chinensis WCHAc010005T | 21.3 | 22.7 | 21.5 | 22.6 | 20.8 | 23.2 | 24.7 | 27.5 | 22.9 | 22.2 | 22.1 | 23.0 | 21.6 | 21.7 | 21.8 | 23.5 | 22.2 | 20.6 | 21.1 | 22.2 | ID |
| A. endophyticus DSM 100498T | A. pakistanensis KCTC 42083T | A. parafaecalis sp. nov. DSM 13975T | A. aquatilis BU33N | A. faecalis DSM 30030T | A. phenolicus sp. nov. DSM 16503T | A. nematophilus sp. nov. A-TC9T | |
| A. endophyticus DSM 100498T | ID | 18.2 | 18.1 | 18.2 | 18.0 | 18.0 | 18.2 |
| A. pakistanensis KCTC 42083T | 18.2 | ID | 32.7 | 29.5 | 30.8 | 30.5 | 30.4 |
| A. parafaecalis sp. nov. DSM 13975T | 18.1 | 32.7 | ID | 34.1 | 36.8 | 35.7 | 35.5 |
| A. aquatilis BU33N | 18.2 | 29.5 | 34.1 | ID | 41.6 | 39.0 | 38.4 |
| A. faecalis DSM 30030T | 18.0 | 30.8 | 36.8 | 41.6 | ID | 49.1 | 47.0 |
| A. phenolicus sp. nov. DSM 16503T | 18.0 | 30.5 | 35.7 | 39.0 | 49.1 | ID | 66.3 |
| A. nematophilus sp. nov. A-TC9T | 18.2 | 30.4 | 35.5 | 38.4 | 47.0 | 66.3 | ID |
| E. cancerogenus ATCC33241T | E. quasihormaechei WCHEQ120003T | E. hormaechei ATCC 49162T | E. xiangfangensis LMG27195T | E. hoffmannii DSM 14563T | E. oligotrophicus CCA6T | E. soli LMG 25861T | E. wuhouensis WCHEW120002T | E. huaxiensis WCHEHu090008T | E. quasimori 090044T | E. mori LMG 25706T | E. ludwigii EN-119T | E. dissolvens ATCC 23373T | E. cloacae DSM 30054T | E. kobei DSM 13645T | E. chuandaensis 090028T | E. bugandensis EB-247T | E. nematophilus sp. nov. E-TC7T | E. sichuanensis WCHECL1597T | E. chengduensis WCHECl-C4T | E. asburiae ATCC 35953T | E. dykesii E1T | E. vonholyi E13T | E. roggenkampii DSM 16690T | E. quasiroggenkampii WCHECL1060T | |
| E. cancerogenus ATCC33241T | ID | 31.7 | 31.6 | 31.5 | 31.5 | 32.1 | 29.5 | 32.0 | 32.1 | 32.3 | 31.9 | 30.4 | 30.7 | 30.8 | 31.2 | 31.7 | 31.8 | 31.8 | 31.2 | 32.0 | 32.4 | 31.8 | 31.5 | 32.0 | 31.5 |
| E. quasihormaechei WCHEQ120003T | 31.7 | ID | 51.6 | 52.8 | 53.5 | 33.5 | 29.8 | 33.9 | 33.7 | 34.9 | 34.0 | 31.4 | 32.1 | 32.3 | 33.4 | 35.3 | 36.7 | 35.3 | 32.4 | 33.9 | 34.2 | 33.7 | 33.3 | 34.3 | 33.6 |
| E. hormaechei ATCC 49162T | 31.6 | 51.6 | ID | 60.0 | 58.0 | 33.8 | 30.2 | 33.9 | 33.8 | 35.0 | 34.2 | 31.6 | 32.4 | 32.9 | 33.7 | 35.4 | 35.7 | 35.3 | 32.6 | 34.5 | 34.6 | 33.8 | 33.3 | 34.3 | 33.7 |
| E. xiangfangensis LMG27195T | 31.5 | 52.8 | 60.0 | ID | 66.6 | 33.8 | 30.2 | 33.8 | 33.8 | 35.0 | 34.2 | 31.5 | 32.5 | 32.6 | 33.4 | 35.2 | 35.7 | 35.2 | 32.8 | 34.1 | 34.8 | 33.9 | 33.7 | 34.7 | 34.0 |
| E. hoffmannii DSM 14563T | 31.5 | 53.5 | 58.0 | 66.6 | ID | 33.8 | 30.0 | 33.7 | 33.8 | 34.8 | 34.1 | 31.4 | 32.1 | 32.5 | 33.4 | 35.0 | 35.5 | 34.9 | 32.5 | 34.0 | 34.4 | 33.5 | 33.1 | 34.2 | 33.5 |
| E. oligotrophicus CCA6T | 32.1 | 33.5 | 33.8 | 33.8 | 33.8 | ID | 31.6 | 34.0 | 34.5 | 34.9 | 34.7 | 32.7 | 33.3 | 33.2 | 33.3 | 34.1 | 34.1 | 33.9 | 32.9 | 33.7 | 34.8 | 33.9 | 33.7 | 34.4 | 33.9 |
| E. soli LMG 25861T | 29.5 | 29.8 | 30.2 | 30.2 | 30.0 | 31.6 | ID | 30.7 | 30.8 | 31.0 | 30.8 | 30.3 | 30.4 | 30.3 | 30.2 | 30.6 | 30.5 | 30.7 | 30.3 | 30.7 | 31.3 | 31.0 | 30.5 | 30.8 | 30.8 |
| E. wuhouensis WCHEW120002T | 32.0 | 33.9 | 33.9 | 33.8 | 33.7 | 34.0 | 30.7 | ID | 39.6 | 38.0 | 37.9 | 32.8 | 33.3 | 33.2 | 33.9 | 36.2 | 35.8 | 36.3 | 34.1 | 35.9 | 37.7 | 36.6 | 35.6 | 36.0 | 35.6 |
| E. huaxiensis WCHEHu090008T | 32.1 | 33.7 | 33.8 | 33.8 | 33.8 | 34.5 | 30.8 | 39.6 | ID | 37.6 | 37.4 | 32.1 | 32.8 | 32.7 | 33.5 | 35.2 | 35.0 | 35.1 | 33.6 | 35.2 | 36.4 | 35.3 | 34.5 | 35.1 | 34.6 |
| E. quasimori 090044T | 32.3 | 34.9 | 35.0 | 35.0 | 34.8 | 34.9 | 31.0 | 38.0 | 37.6 | ID | 66.8 | 33.0 | 34.6 | 34.4 | 36.1 | 38.5 | 38.1 | 38.5 | 36.4 | 38.9 | 40.9 | 40.0 | 39.3 | 39.8 | 40.2 |
| E. mori LMG 25706T | 31.9 | 34.0 | 34.2 | 34.2 | 34.1 | 34.7 | 30.8 | 37.9 | 37.4 | 66.8 | ID | 32.6 | 34.0 | 34.0 | 35.4 | 37.7 | 37.0 | 37.5 | 35.2 | 37.3 | 39.9 | 38.9 | 37.6 | 37.5 | 37.3 |
| E. ludwigii EN-119T | 30.4 | 31.4 | 31.6 | 31.5 | 31.4 | 32.7 | 30.3 | 32.8 | 32.1 | 33.0 | 32.6 | ID | 34.4 | 34.2 | 34.6 | 34.7 | 34.8 | 34.7 | 34.6 | 35.0 | 36.1 | 35.5 | 35.0 | 35.5 | 34.9 |
| E. dissolvens ATCC 23373T | 30.7 | 32.1 | 32.4 | 32.5 | 32.1 | 33.3 | 30.4 | 33.3 | 32.8 | 34.6 | 34.0 | 34.4 | ID | 62.1 | 34.9 | 35.6 | 35.5 | 35.8 | 37.1 | 35.5 | 36.7 | 36.1 | 35.6 | 36.1 | 36.0 |
| E. cloacae DSM 30054T | 30.8 | 32.3 | 32.9 | 32.6 | 32.5 | 33.2 | 30.3 | 33.2 | 32.7 | 34.4 | 34.0 | 34.2 | 62.1 | ID | 35.7 | 35.6 | 35.6 | 35.5 | 36.9 | 36.2 | 37.2 | 35.8 | 35.5 | 36.3 | 35.8 |
| E. kobei DSM 13645T | 31.2 | 33.4 | 33.7 | 33.4 | 33.4 | 33.3 | 30.2 | 33.9 | 33.5 | 36.1 | 35.4 | 34.6 | 34.9 | 35.7 | ID | 42.4 | 43.1 | 43.5 | 39.2 | 43.3 | 42.7 | 41.5 | 40.9 | 41.3 | 40.6 |
| E. chuandaensis 090028T | 31.7 | 35.3 | 35.4 | 35.2 | 35.0 | 34.1 | 30.6 | 36.2 | 35.2 | 38.5 | 37.7 | 34.7 | 35.6 | 35.6 | 42.4 | ID | 53.9 | 56.1 | 39.7 | 42.7 | 45.6 | 44.5 | 43.1 | 43.4 | 42.7 |
| E. bugandensis EB-247T | 31.8 | 36.7 | 35.7 | 35.7 | 35.5 | 34.1 | 30.5 | 35.8 | 35.0 | 38.1 | 37.0 | 34.8 | 35.5 | 35.6 | 43.1 | 53.9 | ID | 63.7 | 39.9 | 42.8 | 45.1 | 44.1 | 43.3 | 44.2 | 43.2 |
| E. nematophilus sp. nov. E-TC7T | 31.8 | 35.3 | 35.3 | 35.2 | 34.9 | 33.9 | 30.7 | 36.3 | 35.1 | 38.5 | 37.5 | 34.7 | 35.8 | 35.5 | 43.5 | 56.1 | 63.7 | ID | 40.2 | 42.9 | 45.7 | 44.9 | 43.6 | 43.8 | 42.7 |
| E. sichuanensis WCHECL1597T | 31.2 | 32.4 | 32.6 | 32.8 | 32.5 | 32.9 | 30.3 | 34.1 | 33.6 | 36.4 | 35.2 | 34.6 | 37.1 | 36.9 | 39.2 | 39.7 | 39.9 | 40.2 | ID | 42.6 | 45.2 | 44.2 | 44.2 | 45.5 | 44.6 |
| E. chengduensis WCHECl-C4T | 32.0 | 33.9 | 34.5 | 34.1 | 34.0 | 33.7 | 30.7 | 35.9 | 35.2 | 38.9 | 37.3 | 35.0 | 35.5 | 36.2 | 43.3 | 42.7 | 42.8 | 42.9 | 42.6 | ID | 52.7 | 50.9 | 49.7 | 48.9 | 49.5 |
| E. asburiae ATCC 35953T | 32.4 | 34.2 | 34.6 | 34.8 | 34.4 | 34.8 | 31.3 | 37.7 | 36.4 | 40.9 | 39.9 | 36.1 | 36.7 | 37.2 | 42.7 | 45.6 | 45.1 | 45.7 | 45.2 | 52.7 | ID | 69.3 | 57.2 | 51.7 | 51.7 |
| E. dykesii E1T | 31.8 | 33.7 | 33.8 | 33.9 | 33.5 | 33.9 | 31.0 | 36.6 | 35.3 | 40.0 | 38.9 | 35.5 | 36.1 | 35.8 | 41.5 | 44.5 | 44.1 | 44.9 | 44.2 | 50.9 | 69.3 | ID | 57.4 | 51.1 | 51.5 |
| E. vonholyi E13T | 31.5 | 33.3 | 33.3 | 33.7 | 33.1 | 33.7 | 30.5 | 35.6 | 34.5 | 39.3 | 37.6 | 35.0 | 35.6 | 35.5 | 40.9 | 43.1 | 43.3 | 43.6 | 44.2 | 49.7 | 57.2 | 57.4 | ID | 56.0 | 56.4 |
| E. roggenkampii DSM 16690T | 32.0 | 34.3 | 34.3 | 34.7 | 34.2 | 34.4 | 30.8 | 36.0 | 35.1 | 39.8 | 37.5 | 35.5 | 36.1 | 36.3 | 41.3 | 43.4 | 44.2 | 43.8 | 45.5 | 48.9 | 51.7 | 51.1 | 56.0 | ID | 65.1 |
| E. quasiroggenkampii WCHECL1060T | 31.5 | 33.6 | 33.7 | 34.0 | 33.5 | 33.9 | 30.8 | 35.6 | 34.6 | 40.2 | 37.3 | 34.9 | 36.0 | 35.8 | 40.6 | 42.7 | 43.2 | 42.7 | 44.6 | 49.5 | 51.7 | 51.5 | 56.4 | 65.1 | ID |
| K. soli DSM 19436T | K. geumhonensis DSM 18799T | K. hirudinis DSM 25966T | K. algarum JCM 31803T | K. dalseonensis DSM 18800T | K. granuli DSM 23481T | K. adipata DSM 17808T | K. nematophila sp. nov. K-TC2T | K. terrae DSM 21341T | K. defluvii JCM 18034T | |
| K. soli DSM 19436T | ID | 22.2 | 21.0 | 21.0 | 21.3 | 20.8 | 20.9 | 21.2 | 20.7 | 21.0 |
| K. geumhonensis DSM 18799T | 22.2 | ID | 21.5 | 20.8 | 21.3 | 21.0 | 21.3 | 21.5 | 20.7 | 21.3 |
| K. hirudinis DSM 25966T | 21.0 | 21.5 | ID | 24.1 | 21.8 | 21.2 | 21.4 | 21.5 | 20.8 | 21.4 |
| K. algarum JCM 31803T | 21.0 | 20.8 | 24.1 | ID | 21.4 | 21.0 | 20.9 | 21.2 | 20.9 | 21.1 |
| K. dalseonensis DSM 18800T | 21.3 | 21.3 | 21.8 | 21.4 | ID | 21.8 | 21.9 | 22.2 | 21.8 | 22.1 |
| K. granuli DSM 23481T | 20.8 | 21.0 | 21.2 | 21.0 | 21.8 | ID | 30.0 | 27.8 | 27.1 | 27.4 |
| K. adipata DSM 17808T | 20.9 | 21.3 | 21.4 | 20.9 | 21.9 | 30.0 | ID | 28.0 | 25.7 | 27.1 |
| K. nematophila sp. nov. K-TC2T | 21.2 | 21.5 | 21.5 | 21.2 | 22.2 | 27.8 | 28.0 | ID | 29.2 | 30.9 |
| K. terrae DSM 21341T | 20.7 | 20.7 | 20.8 | 20.9 | 21.8 | 27.1 | 25.7 | 29.2 | ID | 30.7 |
| K. defluvii JCM 18034T | 21.0 | 21.3 | 21.4 | 21.1 | 22.1 | 27.4 | 27.1 | 30.9 | 30.7 | ID |
| Bacterial Strain | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acinetobacter | Alcaligenes | Enterobacter | Kaistia | |||||||||||||||
| Biochemical test | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 |
| β-Galactosidase | − | − | − | − | − | − | − | + | + | + | + | + | + | + | − | + | + | + |
| Arginine dihydrolase | + | + | − | + | + | + | + | + | + | + | + | + | − | + | − | + | + | − |
| Lysine decarboxylase | − | − | − | − | + | + | + | + | + | − | + | + | − | + | − | + | − | − |
| Ornithine decarboxylase | − | − | − | + | − | + | + | + | + | + | + | + | − | + | − | + | + | − |
| Citrate utilization | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + |
| H2S production | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Urease | − | − | − | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + |
| Tryptophan deaminase | − | − | − | − | + | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Indole production | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − |
| Acetoin production | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | − | − |
| Gelatinase | − | − | − | − | − | − | − | + | + | − | + | + | − | − | − | − | − | − |
| Glucose oxidation | + | − | + | − | − | − | − | − | + | + | + | + | − | − | − | − | − | − |
| Mannitol oxidation | − | − | − | − | − | − | − | + | + | + | + | + | − | − | − | − | − | − |
| Inositol oxidation | − | − | − | − | − | − | − | + | + | − | − | + | − | − | − | − | − | − |
| Sorbitol oxidation | − | − | − | − | − | − | − | − | + | − | + | + | − | − | − | − | − | − |
| Rhamnose oxidation | + | − | + | − | − | − | − | + | + | + | + | + | − | − | − | − | + | − |
| Sucrose oxidation | − | − | − | − | − | − | − | − | + | + | + | − | − | − | − | − | − | − |
| Melibiose oxidation | + | − | + | − | − | − | − | − | + | − | + | − | − | − | − | − | − | − |
| Amygdalin oxidation | − | − | − | − | − | − | − | + | + | + | + | + | − | − | − | − | − | − |
| Arabinose oxidation | + | − | + | − | − | − | − | − | + | + | + | + | − | − | − | − | + | − |
| (Cytochrome) oxidase | − | − | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Catalase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| NO2 production | − | − | − | − | − | − | − | − | − | − | + | − | − | + | − | − | − | − |
| NO2 reduction to N2 gas | − | − | − | − | − | − | − | + | + | + | − | + | − | − | − | + | + | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machado, R.A.R.; Loulou, A.; Bhat, A.H.; Mastore, M.; Terrettaz, C.; Brivio, M.F.; Kallel, S. Acinetobacter nematophilus sp. nov., Alcaligenes nematophilus sp. nov., Enterobacter nematophilus sp. nov., and Kaistia nematophila sp. nov., Isolated from Soil-Borne Nematodes and Proposal for the Elevation of Alcaligenes faecalis subsp. faecalis, Alcaligenes faecalis subsp. parafaecalis, and Alcaligenes faecalis subsp. phenolicus to the Species Level. Taxonomy 2023, 3, 148-168. https://doi.org/10.3390/taxonomy3010012
Machado RAR, Loulou A, Bhat AH, Mastore M, Terrettaz C, Brivio MF, Kallel S. Acinetobacter nematophilus sp. nov., Alcaligenes nematophilus sp. nov., Enterobacter nematophilus sp. nov., and Kaistia nematophila sp. nov., Isolated from Soil-Borne Nematodes and Proposal for the Elevation of Alcaligenes faecalis subsp. faecalis, Alcaligenes faecalis subsp. parafaecalis, and Alcaligenes faecalis subsp. phenolicus to the Species Level. Taxonomy. 2023; 3(1):148-168. https://doi.org/10.3390/taxonomy3010012
Chicago/Turabian StyleMachado, Ricardo A. R., Ameni Loulou, Aashaq Hussain Bhat, Maristella Mastore, Céline Terrettaz, Maurizio Francesco Brivio, and Sadreddine Kallel. 2023. "Acinetobacter nematophilus sp. nov., Alcaligenes nematophilus sp. nov., Enterobacter nematophilus sp. nov., and Kaistia nematophila sp. nov., Isolated from Soil-Borne Nematodes and Proposal for the Elevation of Alcaligenes faecalis subsp. faecalis, Alcaligenes faecalis subsp. parafaecalis, and Alcaligenes faecalis subsp. phenolicus to the Species Level" Taxonomy 3, no. 1: 148-168. https://doi.org/10.3390/taxonomy3010012
APA StyleMachado, R. A. R., Loulou, A., Bhat, A. H., Mastore, M., Terrettaz, C., Brivio, M. F., & Kallel, S. (2023). Acinetobacter nematophilus sp. nov., Alcaligenes nematophilus sp. nov., Enterobacter nematophilus sp. nov., and Kaistia nematophila sp. nov., Isolated from Soil-Borne Nematodes and Proposal for the Elevation of Alcaligenes faecalis subsp. faecalis, Alcaligenes faecalis subsp. parafaecalis, and Alcaligenes faecalis subsp. phenolicus to the Species Level. Taxonomy, 3(1), 148-168. https://doi.org/10.3390/taxonomy3010012







