GPU-Enabled Volume Renderer for Use with MATLAB
Abstract
1. Introduction
1.1. Rendering Equation
1.1.1. Emission
1.1.2. Absorption
1.1.3. Discretization
2. Theoretical Basis
2.1. Rendering Pipeline
2.1.1. Sampling
2.1.2. Illumination
2.1.3. Compositing
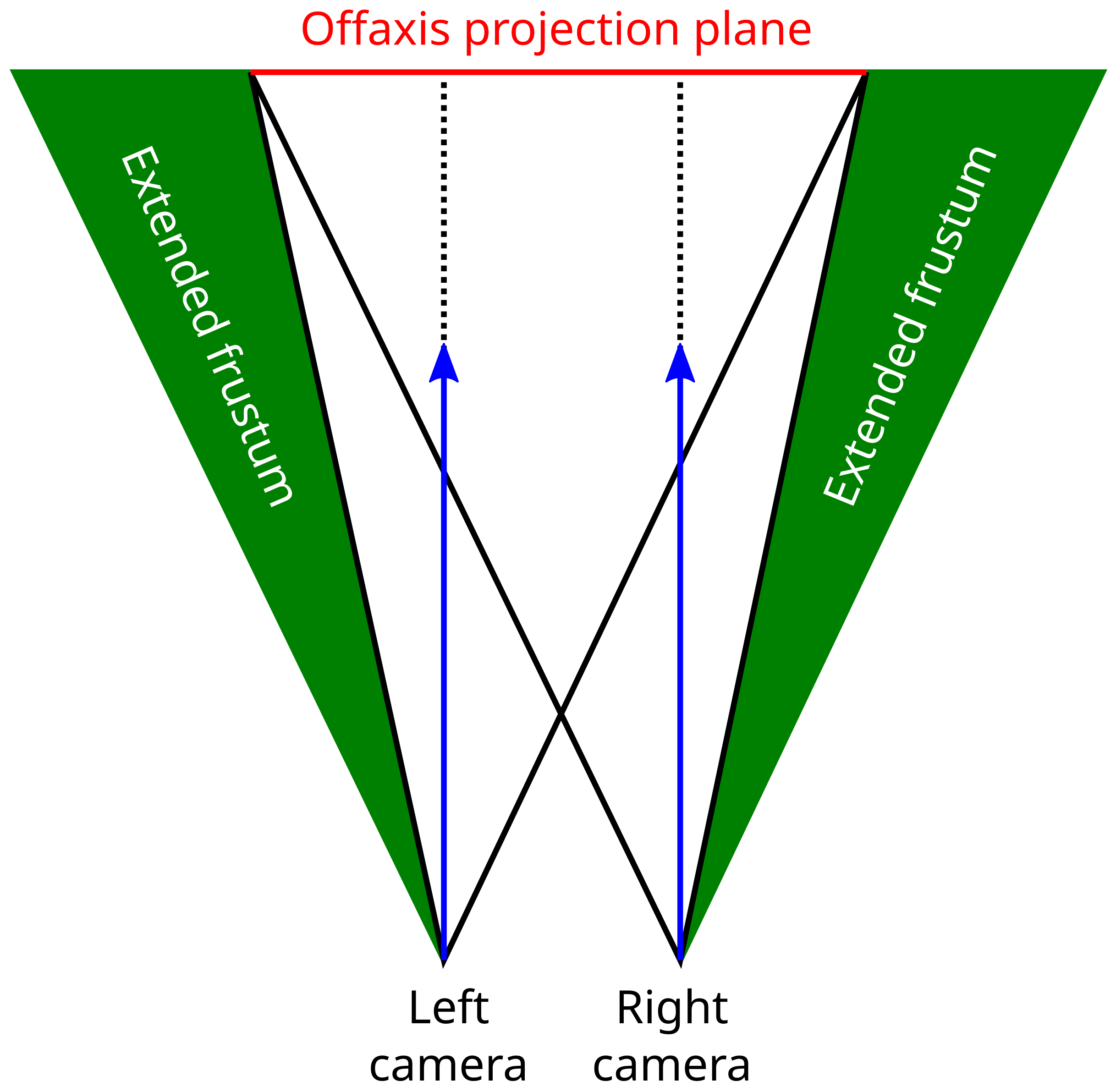
2.2. Stereo Rendering
3. Materials and Methods
3.1. Performance Realization
3.1.1. GPU Architecture and CUDA Implementation
3.1.2. Ray Casting on GPU
3.1.3. Memory Management
3.1.4. Illumination Model and LUT Optimization
Henyey–Greenstein
| Algorithm 1 Pseudo-code for the ‘shade’ function of our renderer. This function calculates the illumination at a given voxel position by evaluating the contributions of multiple light sources, considering the surface normal, view direction, and reflection properties. The algorithm iteratively processes each light source using texture lookups for reflection and illumination to determine the light contribution to the voxel’s appearance. |
| Require: , , , |
| Require: , , , |
| Ensure: Calculated light contribution at the given voxel position |
| 1: |
| 2: for each in do |
| 3: |
| 4: |
| 5: |
| 6: |
| 7: |
| 8: |
| 9: |
| 10: |
| 11: |
| 12: |
| 13: |
| 14: |
| 15: end for |
| 16: return |
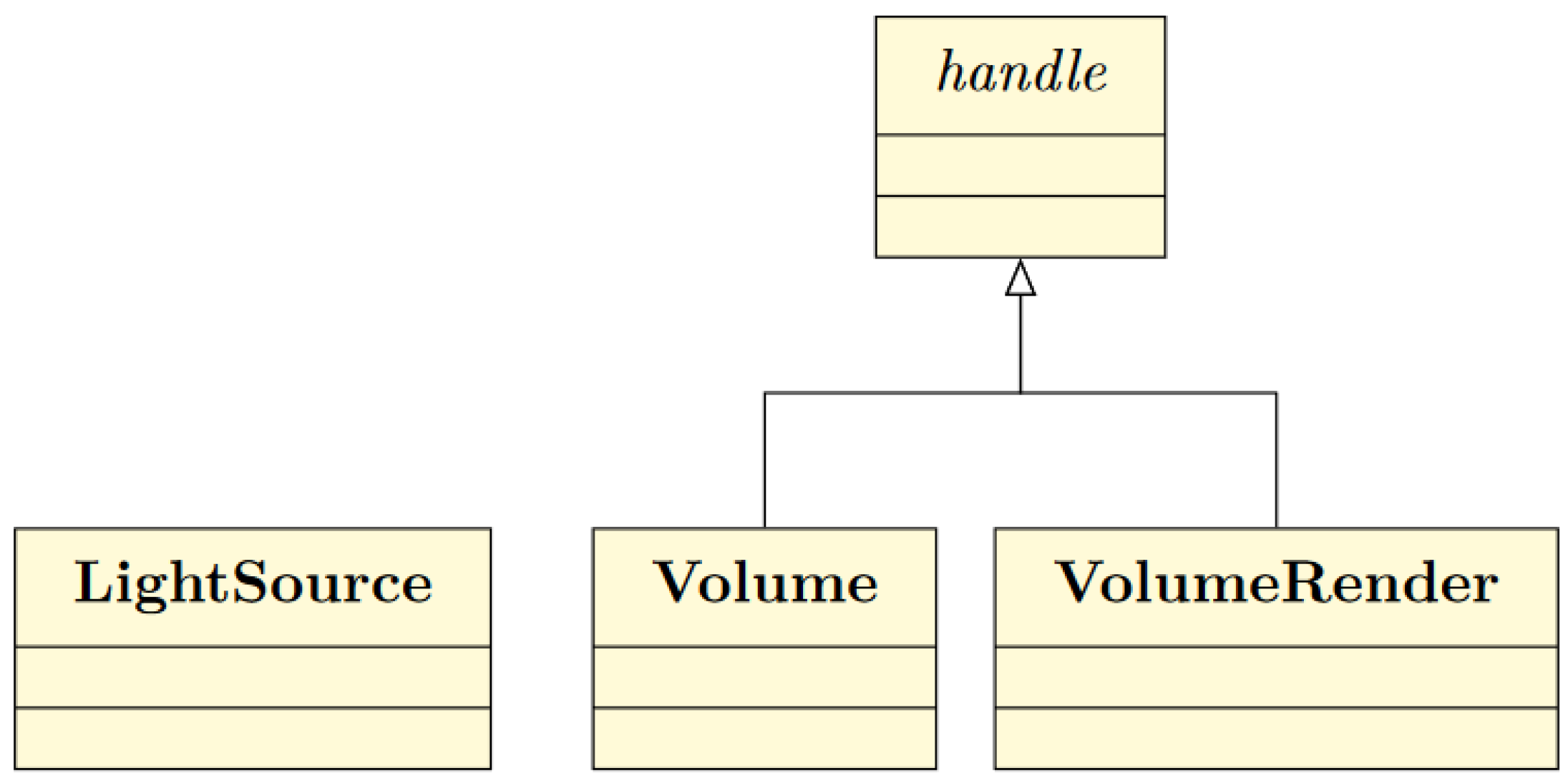
3.2. MATLAB® Interface
Handle Superclass
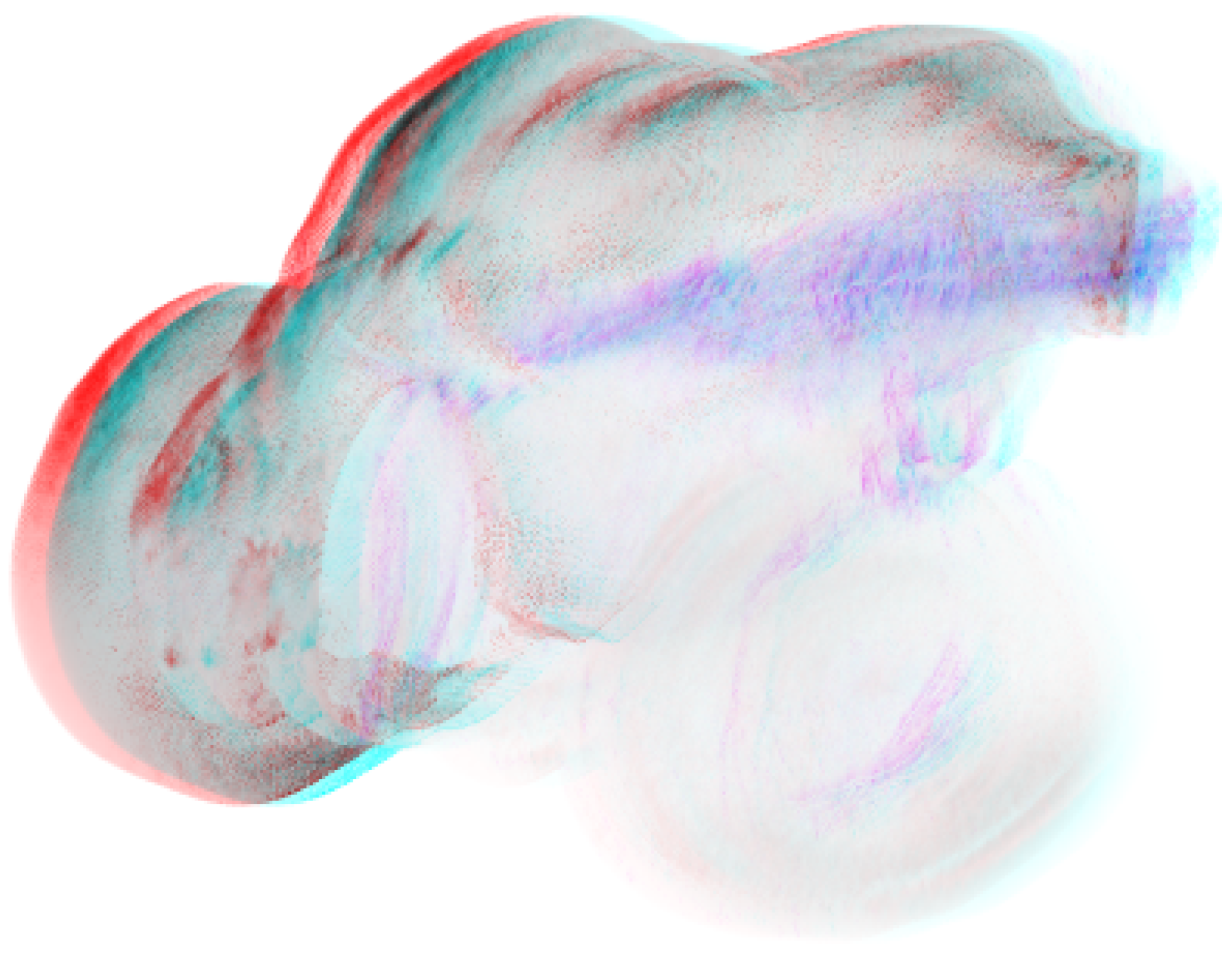
3.3. Case Study
3.3.1. Dataset
3.3.2. Scene
- Rotation of the average brain by 150° (30 image frames);
- Fading out of the interior and one-half while rotating 900° (180 image frames);
- Rotation of half of the average brain shell by 150° (30 image frames);
- Rendering of the 3A10-marked neural structure with full rotation of 1200° (240 image frames).
4. Results
4.1. Implementation
4.2. Volume Design
4.3. Case Study
5. Discussion
5.1. Principal Findings
5.2. Limitations
5.3. Future Work
6. Conclusions
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaufman, A.E. 43—Volume Visualization in Medicine. In Handbook of Medical Imaging; Bankman, I.N., Ed.; Biomedical Engineering; Academic Press: San Diego, CA, USA, 2000; pp. 713–730. [Google Scholar] [CrossRef]
- de Oliveira Santos, B.F.; da Costa, M.D.S.; Centeno, R.S.; Cavalheiro, S.; de Paiva Neto, M.A.; Lawton, M.T.; Chaddad-Neto, F. Clinical Application of an Open-Source 3D Volume Rendering Software to Neurosurgical Approaches. World Neurosurg. 2018, 110, e864–e872. [Google Scholar] [CrossRef] [PubMed]
- Smit, N.; Bruckner, S. Towards Advanced Interactive Visualization for Virtual Atlases. In Biomedical Visualisation: Volume 3; Rea, P.M., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 85–96. [Google Scholar] [CrossRef]
- Hernandez-Cortés, K.S.; Mesa-Pujals, A.A.; García-Gómez, O.; Montoya-Arquímedes, P. Brain morphometry in adult: Volumetric visualization as a tool in image processing. Rev. Mex. Neurocienc. 2021, 22. [Google Scholar] [CrossRef]
- Huang, S.H.; Irawati, N.; Chien, Y.F.; Lin, J.Y.; Tsai, Y.H.; Wang, P.Y.; Chu, L.A.; Li, M.L.; Chiang, A.S.; Tsia, K.K.; et al. Optical volumetric brain imaging: Speed, depth, and resolution enhancement. J. Phys. Appl. Phys. 2021, 54, 323002. [Google Scholar] [CrossRef]
- Zhou, L.; Fan, M.; Hansen, C.; Johnson, C.R.; Weiskopf, D. A Review of Three-Dimensional Medical Image Visualization. Health Data Sci. 2022, 2022, 9840519. [Google Scholar] [CrossRef]
- Dickie, D.A.; Shenkin, S.D.; Anblagan, D.; Lee, J.; Blesa Cabez, M.; Rodriguez, D.; Boardman, J.P.; Waldman, A.; Job, D.E.; Wardlaw, J.M. Whole Brain Magnetic Resonance Image Atlases: A Systematic Review of Existing Atlases and Caveats for Use in Population Imaging. Front. Neuroinform. 2017, 11, 1. [Google Scholar] [CrossRef]
- Diao, B.; Bagayogo, N.A.; Carreras, N.P.; Halle, M.; Ruiz-Alzola, J.; Ungi, T.; Fichtinger, G.; Kikinis, R. The use of 3D digital anatomy model improves the communication with patients presenting with prostate disease: The first experience in Senegal. PLoS ONE 2022, 17, e0277397. [Google Scholar] [CrossRef]
- Kikinis, R.; Pieper, S.D.; Vosburgh, K.G. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy; Jolesz, F.A., Ed.; Springer: New York, NY, USA, 2014; pp. 277–289. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Royer, L.A.; Weigert, M.; Günther, U.; Maghelli, N.; Jug, F.; Sbalzarini, I.F.; Myers, E.W. ClearVolume: Open-source live 3D visualization for light-sheet microscopy. Nat. Methods 2015, 12, 480–481. [Google Scholar] [CrossRef]
- Jarrett, T.; Comrie, A.; Sivitilli, A.; Pretorius, P.C.; Vitello, F.; Marchetti, L. iDaVIE: Immersive Data Visualisation Interactive Explorer. 2024. Available online: https://zenodo.org/records/13752029 (accessed on 18 June 2024).
- Selvamanikkam, M.; Noga, M.; Khoo, N.S.; Punithakumar, K. High-Resolution Stereoscopic Visualization of Pediatric Echocardiography Data on Microsoft HoloLens 2. IEEE Access 2024, 12, 9776–9783. [Google Scholar] [CrossRef]
- Fujii, Y.; Azumi, T.; Nishio, N.; Kato, S.; Edahiro, M. Data Transfer Matters for GPU Computing. In Proceedings of the 2013 International Conference on Parallel and Distributed Systems, Seoul, Republic of Korea, 15–18 December 2013; pp. 275–282. [Google Scholar] [CrossRef]
- Gao, H.; Liu, Y.; Cao, F.; Wu, H.; Xu, F.; Zhong, S. VIDAR: Data Quality Improvement for Monocular 3D Reconstruction through In-situ Visual Interaction. In Proceedings of the 2024 IEEE International Conference on Robotics and Automation (ICRA), Yokohama, Japan, 13–17 May 2024; pp. 7895–7901. [Google Scholar] [CrossRef]
- Hu, J.; Fan, Q.; Hu, S.; Lyu, S.; Wu, X.; Wang, X. UMedNeRF: Uncertainty-Aware Single View Volumetric Rendering For Medical Neural Radiance Fields. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–4, ISSN 1945-8452. [Google Scholar] [CrossRef]
- Dhawan, A.P. Medical Image Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Reyes-Aldasoro, C.C. Biomedical Image Analysis Recipes in MATLAB: For Life Scientists and Engineers; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Mathotaarachchi, S.; Wang, S.; Shin, M.; Pascoal, T.A.; Benedet, A.L.; Kang, M.S.; Beaudry, T.; Fonov, V.S.; Gauthier, S.; Labbe, A.; et al. VoxelStats: A MATLAB Package for Multi-Modal Voxel-Wise Brain Image Analysis. Front. Neuroinform. 2016, 10, 20. [Google Scholar] [CrossRef]
- Wait, E.; Winter, M.; Cohen, A.R. Hydra image processor: 5-D GPU image analysis library with MATLAB and python wrappers. Bioinformatics 2019, 35, 5393–5395. [Google Scholar] [CrossRef] [PubMed]
- Interactively Explore, Label, and Publish Animations of 2-D or 3-D Medical Image Data-MATLAB-MathWorks Deutschland. Available online: https://de.mathworks.com/help/medical-imaging/ref/medicalimagelabeler-app.html (accessed on 14 September 2024).
- Explore 3-D Volumetric Data with Volume Viewer App-MATLAB & Simulink-MathWorks Deutschland. Available online: https://de.mathworks.com/help/images/explore-3-d-volumetric-data-with-volume-viewer-app.html (accessed on 30 August 2022).
- Kroon, D.J. Volume Render. Available online: https://de.mathworks.com/matlabcentral/fileexchange/19155-volume-render (accessed on 30 August 2022).
- Robertson, S. Ray Tracing Volume Renderer. Available online: https://de.mathworks.com/matlabcentral/fileexchange/37381-ray-tracing-volume-renderer (accessed on 30 August 2022).
- Röttger, S.; Kraus, M.; Ertl, T. Hardware-Accelerated Volume and Isosurface Rendering Based on Cell-Projection; ACM, Inc.: New York, NY, USA, 2000. [Google Scholar]
- Wiki, O. Vertex Rendering—OpenGL Wiki. 2022. Available online: https://www.khronos.org/opengl/wiki/vertex_Rendering (accessed on 23 May 2024).
- Lorensen, W.E.; Cline, H.E. Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput. Graph. 1987, 21, 163–169. [Google Scholar] [CrossRef]
- Levoy, M. Display of Surfaces from Volume Data. IEEE Comput. Graph. Appl. 1988, 8, 29–37. [Google Scholar] [CrossRef]
- Max, N.L. Optical Models for Direct Volume Rendering. IEEE Trans. Vis. Comput. Graph. 1995, 1, 99–108. [Google Scholar] [CrossRef]
- Wikipedia. Volume Ray Casting—Wikipedia, The Free Encyclopedia. 2023. Available online: http://en.wikipedia.org/w/index.php?title=Volume%20ray%20casting&oldid=1146671341 (accessed on 18 April 2023).
- Shannon, C.E. Communication in the presence of noise. Proc. Inst. Radio Eng. IRE 1949, 37, 10–21. [Google Scholar] [CrossRef]
- Henyey, L.; Greenstein, J. Diffuse radiation in the galaxy. Astrophys. J. 1941, 93, 70–83. [Google Scholar] [CrossRef]
- Porter, T.; Duff, T. Compositing digital images. In Proceedings of the 11th Annual Conference on Computer Graphics and Interactive Techniques, SIGGRAPH ’84, New York, NY, USA, 23–27 July 1984; pp. 253–259. [Google Scholar] [CrossRef]
- Bavoil, L.; Myers, K. Order independent transparency with dual depth peeling. NVIDIA OpenGL SDK 2008, 1, 2–4. [Google Scholar]
- Ikits, M.; Kniss, J.; Lefohn, A.; Hansen, C. Rendering. In GPU GEMS Chapter 39, Volume Rendering Techniques, 5th ed.; Addison Wesley: Boston, MA, USA, 2007; Chapter 39.4.3. [Google Scholar]
- Bourke, P. Calculating Stereo Pairs. 1999. Available online: http://paulbourke.net/stereographics/stereorender (accessed on 8 September 2022).
- Williams, A.; Barrus, S.; Morley, R.K.; Shirley, P. An Efficient and Robust Ray-Box Intersection Algorithm. J. Graph. Gpu Game Tools 2005, 10, 49–54. [Google Scholar] [CrossRef]
- NVIDIA Corporation. NVIDIA CUDA C Programming Guide. Version 8.0. 2017. Available online: https://docs.nvidia.com/cuda/archive/8.0/pdf/CUDA_C_Programming_Guide.pdf (accessed on 8 September 2022).
- The MathWorks, I. Comparison of Handle and Value Classes-MATLAB & Simulink -MathWorks Deutschland. Available online: https://de.mathworks.com/help/matlab/matlab_oop/comparing-handle-and-value-classes.html (accessed on 18 September 2024).
- Ronneberger, O.; Liu, K.; Rath, M.; Rueß, D.; Mueller, T.; Skibbe, H.; Drayer, B.; Schmidt, T.; Filippi, A.; Nitschke, R.; et al. ViBE-Z: A framework for 3D virtual colocalization analysis in zebrafish larval brains. Nat. Methods 2012, 9, 735–742. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2019, arXiv:1811.02629. [Google Scholar]
- Devkota, S.; Pattanaik, S. Deep Learning based Super-Resolution for Medical Volume Visualization with Direct Volume Rendering. arXiv 2022, arXiv:2210.08080. [Google Scholar]
- Weiss, J.; Navab, N. Deep Direct Volume Rendering: Learning Visual Feature Mappings From Exemplary Images. arXiv 2021, arXiv:2106.05429. [Google Scholar]
- Hu, J.; Yu, C.; Liu, H.; Yan, L.; Wu, Y.; Jin, X. Deep Real-time Volumetric Rendering Using Multi-feature Fusion. In Proceedings of the ACM SIGGRAPH 2023 Conference, SIGGRAPH ’23, New York, NY, USA, 6–10 August 2023; pp. 1–10. [Google Scholar] [CrossRef]
- Ikits, M.; Kniss, J.; Lefohn, A.; Hansen, C. Volumetric Lighting. In GPU GEMS Chapter 39, Volume Rendering Techniques, 5th ed.; Addison Wesley: Boston, MA, USA, 2007; Chapter 39.5.1. [Google Scholar]
- Ament, M.; Dachsbacher, C. Anisotropic Ambient Volume Shading. IEEE Trans. Vis. Comput. Graph. 2016, 22, 1015–1024. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scheible, R. GPU-Enabled Volume Renderer for Use with MATLAB. Digital 2024, 4, 990-1007. https://doi.org/10.3390/digital4040049
Scheible R. GPU-Enabled Volume Renderer for Use with MATLAB. Digital. 2024; 4(4):990-1007. https://doi.org/10.3390/digital4040049
Chicago/Turabian StyleScheible, Raphael. 2024. "GPU-Enabled Volume Renderer for Use with MATLAB" Digital 4, no. 4: 990-1007. https://doi.org/10.3390/digital4040049
APA StyleScheible, R. (2024). GPU-Enabled Volume Renderer for Use with MATLAB. Digital, 4(4), 990-1007. https://doi.org/10.3390/digital4040049





