Abstract
Background/Objectives: Ginsenosides, one of the most pharmaceutically valuable chemical compounds in Panax ginseng, are synthesized with several enzymes, including UGTs. UGTs determine absorbability and physiological function upon consumption. Thus, understanding the functional residues of ginsenoside biosynthesis-associated UGTs is crucial for enhancing the production of valuable ginsenoside varieties. Methods: We collected the UGT homologs of high sequence similarity from two rate-limiting steps of the biosynthetic pathway. The 3D structures of these proteins were predicted using the AlphaFold3 model. The ligand-binding interactions of these UGTs were examined using SwissDock and CB-Dock2. Enzyme kinetics were analyzed with MPEK. Using these tools, we performed in silico mutagenic analyses to identify the functional residues of UGTs in detail. Results: We elucidated the molecular mechanisms of experimentally verified functional residues in UGTs, many of which were associated with optimal ligand interaction angles that expose target carbons. We also identified putatively important amino acid residues that mediate ligand interactions and modulate reaction kinetics by more than 25%. In this study, residues at positions 62, 224, 397, and 398 were shown to significantly influence enzyme kinetics. Conclusions: Our study provides the first structural analysis of the functional residues of ginsenoside biosynthetic UGTs based on their 3D structures. We identified several key amino acid residues essential for proper ginsenoside biosynthesis: (1) residues determining ligand interactions, (2) residues modulating ligand binding angles, and (3) residues affecting reaction kinetics. Our findings demonstrate an effective approach to identifying functional residues in plant enzymes and present valuable UGT candidates for future experimental validation.
1. Introduction
Ginsenosides are the triterpenoid compounds accumulated in Panax spp., and possess many of the pharmaceutical effects of these plants [1]. In detail, ginsenosides species include protopanaxadiol (PPD), protopanaxatriol (PPT), ocotillol (OCT), and oleanane (OA)-type saponins [2]. Of these, Panax ginseng primarily accumulates PPD- and PPT-type ginsenosides. These ginsenoside skeleton structures are formed from squalene through squalene epoxidase (SE) and are subsequently cyclized by dammarendiol-II synthase (DDS) [2]. Next, dammarendiols are converted by CYPs (cytochrome P450) into protopanaxadiol and protopanaxatriol, which are the direct precursors of PPD- and PPT-type ginsenosides [2]. From this step, ginsenoside compounds undergo glycosylation by various UGTs (UDP-dependent glycosyltransferases) [2]. UGTs further catalyze ginsenosides by attaching sugar moieties onto the C3, C6, and C20 positions. PPD can be catalyzed into CK, Rh2, F2, Rg3, Rd, Rb1, Rb2, Rb3, and Rc through a cascade of UGTs [2]. PPT can be catalyzed into Rh1, F1, Rf, and Rg1 through a cascade of UGTs [2].
UGT-based modification mediates the conversion of ginsenosides into various molecules with numerous pharmaceutical benefits. Ginsenoside Rb2 has been reported to possess anti-cancer, anti-diabetic, cardio-protective, anti-inflammatory, anti-oxidative, anti-osteoporosis, antiviral, and anti-photoaging effects [3]. Ginsenoside Rg3 has also been shown to exhibit anti-inflammatory and anti-oxidative effects, thereby contributing to an anti-allergic effect [4]. In addition to these, ginsenoside Re has been reported to possess anti-diabetes mellitus, anti-Parkinson’s disease, anti-inflammatory, and anti-cancer effects; ginsenoside Rg1 has demonstrated anti-hyperglycemic and anti-depressive effects; ginsenoside Rc has been reported to have anti-alcoholic liver disease, anti-myocardial necrosis, and anti-endothelial insulin resistance effects; and compound K has shown anti-tumor, anti-inflammatory, and hepatoprotective effects [5]. One previous study indicated that sugar moiety compositions of PPT-type ginsenosides modulate binding affinity to the glucocorticoid receptor (GR) [6]. Another line of evidence showed that sugar moieties attached only to the C3 position possess a potent inhibitory effect on Na+/K+-ATPase activity, while this effect is diminished when a monosaccharide is linked to the C6 or C20 position [7]. Thus, modulating the sugar moiety of ginsenoside molecules significantly alters their pharmaceutical activity.
Therefore, many efforts have been dedicated to produce these valuable ginsenosides. Treatment using methyl jasmonate on the root tissue of P. ginseng has been shown to enhance the expression of UGT genes, thus increasing ginsenoside Rb1, Rg1, Rh2, and Rg3 (Korean Patent Number 10-2014-0041261). Using a yeast system (Saccharomyces cerevisiae) with transgenic expression of UGTs and CYP enzymes, compound K, ginsenoside F1, and ginsenoside Rh1 were successfully synthesized [8,9]. In the case of ginsenoside Rh2, factory-level production was shown to be successfully achieved [10]. Enzymatic assessment was also performed for ginsenoside production. Cellulase KN was shown to produce the ginsenoside F1 [11]. A mutant form of glycosyltransferase BS-YjiC has been reported to catalyze PPD into ginsenoside F12 and Rh2 (US12134789B2). A new UGT named UGT109A1 from Bacillus subtilis has been reported to glycosylate the C3 and C12 position, leading to the production of unnatural ginsenoside variants [12].
As reviewed above, the production of ginsenosides has attracted great interest. Thus, assessing the enzymatic activity or specificity of UGTs and understanding them could greatly advance productivity. To do that, we aimed to identify important amino acid residues and reveal their potential roles. Homologs of high sequence similarity (over 99%) were identified as potentially active UGTs in the ginsenoside biosynthetic process. Next, sequence alignment was performed to reveal potentially important amino acids. Through 3D structure modeling with AlphaFold3, we applied a series of AI-based models to assess protein–ligand interaction or catalytic activity, using SwissDock, CB-Dock2, and MPEK with in silico mutated proteins. Through these comprehensive analyses, we identified the first potent amino acid residue in UGTs that may alter their reaction kinetics and several other residues that potentially change ligand interactivity.
2. Materials and Methods
2.1. Identification of Target Sequences
Among many reactions associated with ginsenoside biosynthesis, we focused on two rate-limiting steps: PPD-to-ginsenoside CK and PPT-to-ginsenoside F1. UGTs involved in these reactions were identified using BLASTp with an e-value cutoff of and a similarity of 99% [13,14]. Sequences were aligned with MUSCLE pipeline, revealing several variable regions [15]. To assess sequence conservation, aligned sequences were analyzed with Jalview version 2.11.4.0, and the quality score metric was used [16]. Based on these criteria, we identified eight amino acid residues with potential functional relevance in ginsenoside biosynthesis. To evaluate the applicability of our in-silico approach, we also tested seven amino acid residues previously shown to have functional relevance in ginsenoside biosynthesis [9]. Mutagenesis was performed by substituting residues with naturally occurring counterparts or alanine.
2.2. 3D Structure-Based Protein–Ligand Interactivity and Enzyme Catalytic Activity Assay
3D structures of all proteins were predicted with AlphaFold3, and the most probable structural model for each was used [17]. We utilized AlphaFold3 to predict the 3D structures of the target UGTs, and obtained confidence scores above 0.8 across entire sequences (Figure S1). AlphaFold3 generated highly reliable structural models, as confirmed by comparison between the predicted structure of a representative UGT and the experimentally determined 3D structure of a homologous UGT (Figure S2). To examine UGT–ligand interactivity, we used SwissDock and CB-Dock2 [18,19,20]. To predict catalytic activity, MPEK was applied to each UGT–ligand pair [21].
2.3. Mutagenic Analysis to Reveal Functional Amino Acid Residue
First, we performed multiple sequence alignment with MUSCLE to identify regions with sequence variation [15]. The 3D structures predicted by AlphaFold3 were then manually analyzed with ChimeraX to identify target sites for mutagenesis [17,22]. Eight amino acid residues were selected for this study. Residue numbering was standardized based on the multiple sequence alignment, and mutant UGT sequences were generated manually. All sequences were then modeled with AlphaFold3, and the most probable structures were used for further analysis [17]. Using these predicted structures or amino acid sequences as input, we performed SwissDock, CB-Dock2, and MPEK with default settings [18,19,20,21]. Specifically, we analyzed the AC_score from SwissDock to estimate protein–ligand affinity, inspected CB-Dock2-predicted structures with ChimeraX, and used MPEK to predict kcat values [18,19,20,21,22]. MPEK can predict both kcat and Km values with any protein–ligand pair, and thus was applied in our analysis as well [21]. In applying these models to plant enzymes, we noted the following limitations or considerations for each AI tool: AlphaFold3 showed no discernible limitations; SwissDock was unable to predict the exact binding site; CB-Dock2 predictions of binding affinity were unreliable due to excessive variation; and MPEK reported kcat values and Km values that did not always correspond to kcat results.
2.4. Data Visualization
Heatmaps were generated with TB-tools II [23]. Centered Z-scores for ligand affinity and enzyme activity were calculated, setting the wild-type value to zero. To visualize ligand orientation, we examined the detailed structures to determine whether the ligand exposed the C3, C6, or C20 positions toward the sugar donor. Each prediction was repeated until at least three independent runs yielded consistent results.
3. Results
3.1. Identification of Potential Amino Acids of Functional Relevance in Ginsenoside Biosynthesis
Among the many biochemical reactions involved in the ginsenoside biosynthetic process, we analyzed UGTs associated with two rate-limiting steps: PPD-to-CK and PPT-to-F1 (Figure 1A; Table 1). Previous studies identified four UGTs that catalyze the conversion of PPD or PPT into CK or F1 (Table 1). To identify amino acids potentially correlated with functionality, we collected homologs with high sequence similarity (over 99%) (Table 2). Sequence similarity analysis identified a total of 27 homologous proteins, including the functional UGTs involved in the target reactions (Table 2). Using these homologs, we sought to identify amino acids with potential functional relevance in ginsenoside biosynthesis.
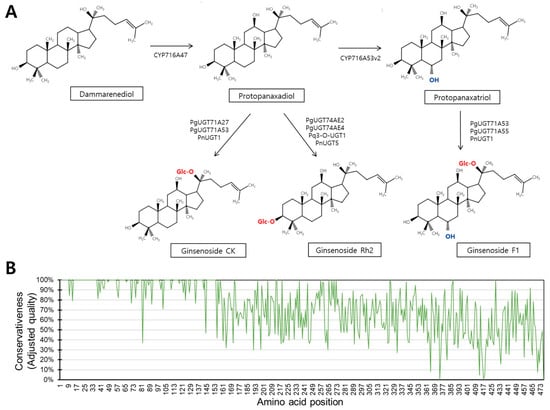
Figure 1.
Target reaction and amino acid conservativeness. (A) Schematic diagram presenting UGT-mediated reactions catalyzing PPD and PPT into ginsenoside CK, ginsenoside Rh2, and ginsenoside F1. (B) Amino acid conservativeness plot made by comparing all close homologs.

Table 1.
List of target enzymes in this study.

Table 2.
List of target UGTs that share high sequence similarity with UGTs in Table 1.
Comparison of the sequences of the 27 homologous UGTs revealed variable regions in the amino acid sequences (Figure 1B). Regions with low sequence similarity were located in C-terminus regions (Figure 1B). Previous studies reported functionally relevant amino acids in the N-terminal region, with only one residue identified outside this region (Table 3) [9]. In this study, we aimed to determine how these functional amino acids contribute to catalytic activity and to identify novel residues that affect different aspects of catalytic function, including ligand interaction, the ligand interaction angle, and reaction kinetics.

Table 3.
List of functional amino acids that were experimentally shown to alter catalytic activities in the ginsenoside biosynthetic process.
3.2. Functional Amino Acid Residues from the Previous Study Mainly Alter Ligand Interactivity
We established a strategy to gain detailed functional insights using a mutagenesis approach (Figure 2). In short, we predicted the 3D structures of all UGTs, manually analyzed their structures and sequences, and identified amino acid residues with potential functional relevance (Figure 2). We then performed in silico mutagenesis of these UGTs and analyzed their kinetic constants, ligand interactions, and ligand specificities using different deep-learning models (Figure 2). To validate our strategy, we first examined experimentally proven amino acid sites known to have functional relevance [9].
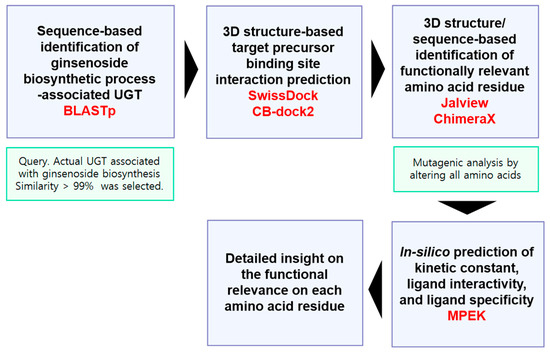
Figure 2.
In-silico procedure to predict the specific functionality of each amino acid residue.
A previous study identified seven amino acids that can alter UGT functionality [9]. These residues were reported to affect ligand specificity or interaction, but the available data did not include information on catalytic activity [9]. The majority of these residues were located near the ligand-interacting helix (A10, I13, H82, A142, H144, and L185 (Figure 3A). SwissDock was used to evaluate the relative interaction between each UGT and either PPD or PPT (Figure 3B). Many mutant UGTs showed moderately altered ligand affinity, whereas mutations at H82 or A142 caused significant changes (> 25%) in binding affinity to PPD, and mutations at I13, H82, and A142 significantly affected binding affinity to PPT (Figure 3B).
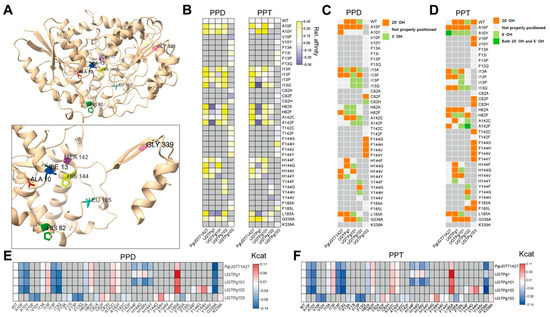
Figure 3.
In-silico identification of detailed functional contribution of each amino acid residue. (A) Representative visualization of functional amino acid residues in 3D structure. UGTPg1 was selected to visualize physical locations of amino acids. (B) Ligand interactivity analysis of each UGT. Relative ligand affinity was visualized by normalizing affinity with Z-score by setting affinity of wild-type UGT as 0. Vertical axis indicates amino acid mutant variants of each UGT annotated in horizontal axis. (C,D) Ligand interacting angle was visualized with heatmap. By manually analyzing UGT–ligand structure, we determined directionality of ligands. (C) Ligand interacting angle of PPD with each UGT. (D) Ligand interacting angle of PPT with each UGT. (E,F) Predicted catalytic activity of all analyzed UGTs. Kcat value was normalized into log2(wild-type centered Z-score) and visualized with heatmap. (E) Catalytic activity of UGTs with PPD were visualized. (F) Catalytic activity of UGTs with PPT were visualized.
We analyzed the ligand interaction angle using CB-Dock2 and found that the majority of mutants altered the interacting angle with PPD or PPT (Figure 3C,D). Catalytic activity was then assessed using MPEK, which revealed that these mutations modulated catalytic activity (Figure 3E,F). However, none of these mutations produced a predicted change in catalytic activity greater than 25% (Figure 3E,F). These results are consistent with experimental findings and confirm the validity of our pipeline, allowing us to proceed with the identification of novel amino acid residues important for the detailed functionality of UGTs.
3.3. Eight Amino Acid Residues with Functional Relevance
We manually analyzed the structures and sequences of all 27 UGTs and curated eight putative amino acid residues based on three criteria: (1) residues showing sequence variation among the 27 UGTs; (2) residues located within ligand-interacting regions; and (3) residues that have not been previously reported (Figure 4A). Many of these residues were located near UDP-sugar binding socket regions (positions 176, 224, 397, and 398), while others were dispersed throughout the structure (Figure 4A). To determine the detailed functional contributions of these amino acids in the ginsenoside biosynthetic process, we generated mutant versions of these residues and analyzed them using our established pipeline (Figure 2 and Figure 4B). First, we assessed the relative affinity of each UGT for PPD or PPT and observed the differential effects of amino acid residues on PPD or PPT affinity (Figure 4B). Notably, almost all amino acid positions altered affinity toward PPD and PPT by at least 25% in at least one enzyme (Figure 4B).
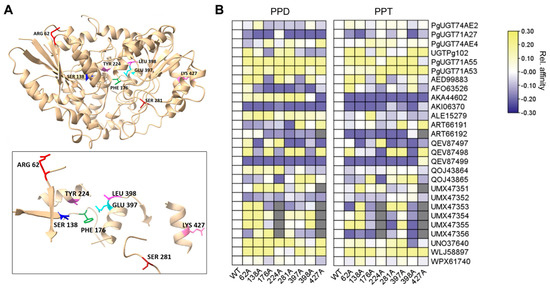
Figure 4.
In-silico identification of relative affinity with PPD or PPT of noble amino acid residues. (A) Representative visualization of functional amino acid residues in 3D structure. PgUGT71A53 was selected to visualize physical location of amino acids. (B) Ligand interactivity analysis of each UGT. Relative ligand affinity was visualized by normalizing affinity with Z-score by setting affinity of wild-type UGT as 0. Horizontal axis indicates amino acid mutant variants of each UGTs annotated in vertical axis.
All eight amino acids were capable of altering the ligand interaction orientation to some extent (Figure 5A,B). Specifically, ALE15279 and WPX61740 were unaffected by any of the mutations analyzed in this study (Figure 5A,B). In addition, five UGTs (ART66192, UMX47353, UMX47354, UMX47355, and UMX47356) interacted with PPD and PPT in orientations where sugar attachment was impossible, regardless of mutation—for example, in many cases the ligand was buried inside amino acid regions, as shown by superposition analyses (Figure 5A,B).
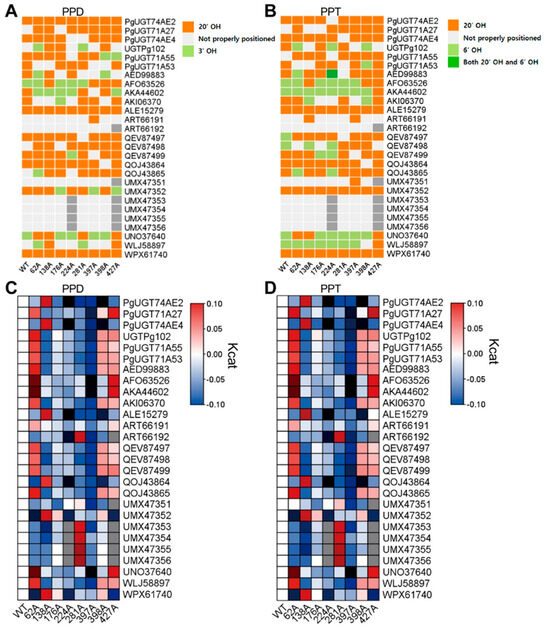
Figure 5.
In-silico identification of detailed functional contribution of noble amino acid residues. (A,B) Ligand interacting angle was visualized with heatmap. By manually analyzing UGT–ligand structure, we determined directionality of ligands. (A) Ligand interacting angle of PPD with each UGT. (B) Ligand interacting angle of PPT with each UGT. (C,D) Predicted catalytic activity of all analyzed UGTs. Kcat value was normalized into log2(wild-type centered Z-score) and visualized with heatmap. (C) Catalytic activity of UGTs with PPD were visualized. (D) Catalytic activity of UGTs with PPT were visualized.
Next, we analyzed the catalytic activities of all mutant UGTs with PPD or PPT (Figure 5C,D). Catalytic activity varied among mutant variants for the eight amino acid residues, and four positions (62, 224, 397, and 398) were found to modulate activity by more than 25% (Figure 5C,D). Among these, position 62 acted as an inhibitory site for catalytic activity, whereas the others enhanced the catalytic activity of UGTs (Figure 5C,D).
In summary, we developed an easy-to-use pipeline for assessing the functional contributions of amino acid residues in plant enzymes (Figure 2). Notably, the confidence scores in regions near three functional sites and ligand-binding pockets exceeded 0.9 (Figure S1). Thus, we identified three promising amino acid positions that can determine ligand affinity, the ligand interaction angle, and reaction kinetics (Figure 6).
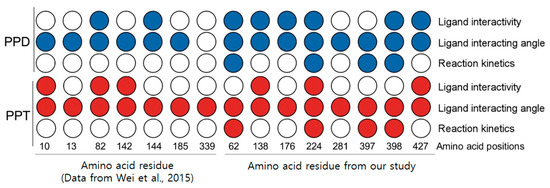
Figure 6.
Summary of this study. From left, seven amino acids revealed from previous study [9]; eight amino acids on right side were analyzed in this study.
4. Discussion
Our study took advantage of an in-silico pipeline to analyze the specific functional roles of individual amino acid residues in plant enzymes (Figure 2). Through our approach, we identified functionally relevant amino acid residues in UGTs associated with ginsenoside biosynthesis. Specifically, we confirmed that amino acids determining ligand specificity were also predicted, in our pipeline, to influence ligand interactivity and interaction angles [9] (Figure 6). We then revealed eight putative functionally relevant amino acids and applied our pipeline to them (Figure 2, Figure 4 and Figure 5). As a result, we discovered three amino acid residues that are conserved among 27 UGTs and alter all aspects of UGT function, including kinetics (Figure 6). These three residues have potential value for future industrial applications.
Ginsenoside biosynthetic and modification processes involve not only UGTs, but also other enzyme classes, such as cellulases. For example, cellulase KN was previously shown to catalyze the conversion of ginsenoside Re into F1 [11]. Specifically, the reaction proceeded via ginsenoside Rg1 as an intermediate [11]. This insight suggests an approach similar to ours could be applied to cellulase family members to identify functionally relevant amino acids in cellulase KN (Figure 2). However, the amino acid sequence of cellulase KN is currently unavailable. Therefore, it is necessary to search public databases for cellulase proteins predicted to interact with ginsenosides and exhibit catalytic activity.
A mutant form of glycosyltransferase BS-YjiC was shown to catalyze the conversion of PPD into ginsenoside F12 and Rh2 (US12134789B2). Mutations at positions 125, 178, and 313 were tested, and the most stable variants were double mutants K125I/N178I and K125I/P313W (US12134789B2). The major difference between our analysis and this patent is that catalytic activity in the patented mutants increased due to enhanced stability, whereas our analysis predicted increased kcat values (Figure 6). This study adapted an insilico methodology from our ongoing research aimed at identifying novel plant enzymes associated with various enzymatic reactions (Jung et al., in revision). Although the core pipeline is the same, the present study included targeted amino acid selection and mutagenic analysis. The increased enzyme activity predicted in our analysis could also be partially explained by enhanced stability during the reaction process, although other possibilities cannot be excluded. One plausible explanation for increased enzyme activity is that these altered amino acids may improve the flexibility of the catalytic domain, thereby accelerating the reaction. However, the exact reason for these increased kcat values remains to be elucidated, as our pipeline cannot predict it.
UGT109A1 from Bacillus subtilis was shown to catalyze glycosylation at the C3, C12, and C20 positions rather than the C3, C6, and C20 positions [12]. UGT109A1 could recognize dammarendiol-II, PPD, and PPT as substrates and convert them [12]. It produced novel ginsenosides such as 3β,12β-Di-O-Glc-PPD from PPD and 3β,12β-Di-O-Glc-PPT from PPT [12]. In cell line experiments, 3β,12β-Di-O-Glc-PPD demonstrated higher anti-lung cancer activity than Rg3 [12]. Considering these findings, our predictions of “not properly located” ligand orientations may need to be revisited (Figure 3C,D and Figure 5A,B). Nevertheless, the majority of these orientations severely disrupted the UDP-sugar binding pockets and failed to expose hydroxyl groups toward the UDP-sugar binding site. However, a few orientations could potentially produce alternative products (Figure 3C,D and Figure 5A,B). Although our primary focus was the two reactions of interest, these additional predictions could be investigated in future studies.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biochem5040036/s1, Figure S1: Confidence map of 3D structures predicted from two representative UGTs. Red box indicates the ligand binding amino acid and nearby region. Yellow arrow indicates the identified functional residue sites. A. Confidence map drawn with PgUGT71A27. B. Confidence map drawn with PgUGT74AE2; Figure S2: Structure comparison between alphafold3-predicted 3D structure from PgUGT71A27 and experimentally determined 3D structure of MtUGT71G1 (PDB:2ACW). RMSD was annotated to quantitatively show the similarity. Only monomer structure was aligned and shown.
Author Contributions
Conceptualization, J.K.; methodology, K.J.; software, J.K.; validation, K.J., N.K. and C.P.; formal analysis, K.J.; investigation, J.K.; data curation, K.J.; writing—original draft preparation, J.K.; writing—review and editing, J.K.; visualization, K.J. and J.K.; supervision, J.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Korea National University of Education.
Data Availability Statement
Data are available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| UGT | UDP-Glycosyltransferase |
| PPD | Protopanaxadiol |
| PPT | Protopanaxatriol |
References
- Leung, K.W.; Wong, A.S. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef]
- Hou, M.; Wang, R.; Zhao, S.; Wang, Z. Ginsenosides in Panax genus and their biosynthesis. Acta Pharm. Sin. B 2021, 11, 1813–1834. [Google Scholar] [CrossRef]
- Miao, L.; Yang, Y.; Li, Z.; Fang, Z.; Zhang, Y.; Han, C.-C. Ginsenoside Rb2: A review of pharmacokinetics and pharmacological effects. J. Ginseng Res. 2022, 46, 206–213. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Huang, T.-H.; Yeh, K.-W.; Chen, Y.-L.; Shen, S.-C.; Liou, C.-J. Ginsenoside Rg3 ameliorates allergic airway inflammation and oxidative stress in mice. J. Ginseng Res. 2021, 45, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Mou, C.; Hu, Y.; He, Z.; Cho, J.Y.; Kim, J.H. In vivo metabolism, pharmacokinetics, and pharmacological activities of ginsenosides from ginseng. J. Ginseng Res. 2025, 49, 479–487. [Google Scholar] [CrossRef]
- Li, W.; Zheng, L.; Ma, X.; Xia, J.; Sheng, J.; Ge, P.; Yuan, Y.; Fan, Y.; Zhou, Y. The sugar moiety in protopanaxadiol ginsenoside affects its ability to target glucocorticoid receptor to regulate lipid metabolism. Bioorganic Chem. 2024, 153, 107885. [Google Scholar] [CrossRef]
- Chen, R.J.Y.; Chung, T.-Y.; Li, F.-Y.; Lin, N.-H.; Tzen, J.T.C. Effect of sugar positions in ginsenosides and their inhibitory potency on Na+/K+-ATPase activity. Acta Pharmacol. Sin. 2009, 30, 61–69. [Google Scholar] [CrossRef]
- Yan, X.; Fan, Y.; Wei, W.; Wang, P.; Liu, Q.; Wei, Y.; Zhang, L.; Zhao, G.; Yue, J.; Zhou, Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014, 24, 770–773. [Google Scholar] [CrossRef]
- Wei, W.; Wang, P.; Wei, Y.; Liu, Q.; Yang, C.; Zhao, G.; Yue, J.; Yan, X.; Zhou, Z. Characterization of Panax ginseng UDP-Glycosyltransferases Catalyzing Protopanaxatriol and Biosyntheses of Bioactive Ginsenosides F1 and Rh1 in Metabolically Engineered Yeasts. Mol. Plant 2015, 8, 1412–1424. [Google Scholar] [CrossRef]
- Wang, P.; Wei, W.; Ye, W.; Li, X.; Zhao, W.; Yang, C.; Li, C.; Yan, X.; Zhou, Z. Synthesizing ginsenoside Rh2 in Saccharomyces cerevisiae cell factory at high-efficiency. Cell Discov. 2019, 5, 5. [Google Scholar] [CrossRef]
- Wang, Y.; Choi, K.-D.; Yu, H.; Jin, F.; Im, W.-T. Production of ginsenoside F1 using commercial enzyme Cellulase KN. J. Ginseng Res. 2016, 40, 121–126. [Google Scholar] [CrossRef]
- Liang, H.; Hu, Z.; Zhang, T.; Gong, T.; Chen, J.; Zhu, P.; Li, Y.; Yang, J. Production of a bioactive unnatural ginsenoside by metabolically engineered yeasts based on a new UDP-glycosyltransferase from Bacillus subtilis. Metab. Eng. 2017, 44, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270–W277. [Google Scholar] [CrossRef]
- Bugnon, M.; Röhrig, U.F.; Goullieux, M.; Perez, M.A.; Daina, A.; Michielin, O.; Zoete, V. SwissDock 2024: Major enhancements for small-molecule docking with Attracting Cavities and AutoDock Vina. Nucleic Acids Res. 2024, 52, W324–W332. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.-X.; Cao, Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Z.; Chen, C.; Yao, G.; Wan, X.; Bao, S.; Ding, J.; Wang, L.; Jiang, H. MPEK: A multitask deep learning framework based on pretrained language models for enzymatic reaction kinetic parameters prediction. Brief. Bioinform. 2024, 25, bbae387. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).