Single-Cell Transcriptomics in Spinal Cord Studies: Progress and Perspectives
Abstract
1. Introduction
2. Single-Cell Transcriptomics: Technologies and Methodologies
2.1. Principles and Evolution of Single-Cell RNA Sequencing Technologies
2.2. Comparison of Major scRNA-Seq Platforms and Their Features
2.3. Technology Selection Strategies and Application Cases in Spinal Cord Research
3. Applications of Single-Cell Transcriptomics in Spinal Cord Research
3.1. Decoding Spinal Cord Developmental Biology
3.2. Investigating Spinal Cord Injury and Neurodegeneration
3.3. Spatial Transcriptomics and Multi-Omics Integration
4. Current Challenges and Limitations
4.1. Technical Challenges
4.2. Computational and Interpretive Bottlenecks
4.3. Biological Complexity and Clinical Translation
5. Future Directions and Perspectives
5.1. Toward a Comprehensive Spinal Cord Cell Atlas
5.2. AI and Machine Learning in Single-Cell Analysis
5.3. Personalized Medicine and Regenerative Therapies
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| scRNA-seq | Single-cell RNA sequencing |
| CNS | Central Nervous System |
| SCI | Spinal Cord Injury |
| SMA | Spinal Muscular Atrophy |
| QC | Quality Control |
| AI | Artificial Intelligence |
| ML | Machine Learning |
| PBMCs | Peripheral Blood Mononuclear Cells |
| SMN | Survival Motor Neuron |
| ALS | Amyotrophic Lateral Sclerosis |
| GNNs | Graph Neural Networks |
| snRNA-seq | Single-nucleus RNA sequencing |
| MSI | mass spectrometry imaging |
References
- Harrow-Mortelliti, M.; Reddy, V.; Jimsheleishvili, G. Physiology, Spinal Cord. [Updated 2023 Mar 17]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK544267/ (accessed on 28 April 2025).
- Bahney, J.; von Bartheld, C.S. The Cellular Composition and Glia-Neuron Ratio in the Spinal Cord of a Human and a Nonhuman Primate: Comparison with Other Species and Brain Regions. Anat. Rec. 2018, 301, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Liau, E.S.; Jin, S.; Chen, Y.C.; Liu, W.S.; Calon, M.; Nedelec, S.; Nie, Q.; Chen, J.A. Single-cell transcriptomic analysis reveals diversity within mammalian spinal motor neurons. Nat. Commun. 2023, 14, 46. [Google Scholar] [CrossRef]
- Blum, J.A.; Klemm, S.; Shadrach, J.L.; Guttenplan, K.A.; Nakayama, L.; Kathiria, A.; Hoang, P.T.; Gautier, O.; Kaltschmidt, J.A.; Greenleaf, W.J.; et al. Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat. Neurosci. 2021, 24, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Zhou, R.; Chen, Z.; Gao, S.; Zhou, F. The Glial Cells Respond to Spinal Cord Injury. Front. Neurol. 2022, 13, 844497. [Google Scholar] [CrossRef] [PubMed]
- Jäkel, S.; Dimou, L. Glial Cells and Their Function in the Adult Brain: A Journey through the History of Their Ablation. Front. Cell. Neurosci. 2017, 11, 24. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K. Glial Cells Shape Pathology and Repair After Spinal Cord Injury. Neurother. J. Am. Soc. Exp. Neurother. 2018, 15, 554–577. [Google Scholar] [CrossRef]
- Wang, N.; Zheng, J.; Chen, Z.; Liu, Y.; Dura, B.; Kwak, M.; Xavier-Ferrucio, J.; Lu, Y.C.; Zhang, M.; Roden, C.; et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 2019, 10, 95. [Google Scholar] [CrossRef]
- Rosenberg, A.B.; Roco, C.M.; Muscat, R.A.; Kuchina, A.; Sample, P.; Yao, Z.; Graybuck, L.T.; Peeler, D.J.; Mukherjee, S.; Chen, W.; et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 2018, 360, 176–182. [Google Scholar] [CrossRef]
- Chuang, H.C.; Li, R.; Huang, H.; Liu, S.W.; Wan, C.; Chaudhuri, S.; Yue, L.; Wong, T.; Dominical, V.; Yen, R.; et al. Single-cell sequencing of full-length transcripts and T-cell receptors with automated high-throughput Smart-seq3. BMC Genom. 2024, 25, 1127. [Google Scholar] [CrossRef]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, Y.; Wang, Z.; Shi, Q. Recent advances in high-throughput single-cell transcriptomics and spatial transcriptomics. Lab Chip 2022, 22, 4774–4791. [Google Scholar] [CrossRef] [PubMed]
- Wirz, J.; Fessler, L.I.; Gehring, W.J. Localization of the Antennapedia protein in Drosophila embryos and imaginal discs. EMBO J. 1986, 5, 3327–3334. [Google Scholar] [CrossRef] [PubMed]
- Ståhl, P.L.; Salmén, F.; Vickovic, S.; Lundmark, A.; Navarro, J.F.; Magnusson, J.; Giacomello, S.; Asp, M.; Westholm, J.O.; Huss, M.; et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 2016, 353, 78–82. [Google Scholar] [CrossRef]
- Molla Desta, G.; Birhanu, A.G. Advancements in single-cell RNA sequencing and spatial transcriptomics: Transforming biomedical research. Acta Biochim. Pol. 2025, 72, 13922. [Google Scholar] [CrossRef]
- Aldridge, S.; Teichmann, S.A. Single cell transcriptomics comes of age. Nat. Commun. 2020, 11, 4307. [Google Scholar] [CrossRef] [PubMed]
- Delile, J.; Rayon, T.; Melchionda, M.; Edwards, A.; Briscoe, J.; Sagner, A. Single cell transcriptomics reveals spatial and temporal dynamics of gene expression in the developing mouse spinal cord. Development 2019, 146, dev173807. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, Y.; Wei, Y.; Chen, H.; Wu, Y.; Wu, L.; Li, J.; Ren, Q.; Miao, C.; Zhu, T.; et al. Spatial transcriptomics and single-nucleus RNA sequencing reveal a transcriptomic atlas of adult human spinal cord. eLife 2024, 12, RP92046. [Google Scholar] [CrossRef]
- Sathyamurthy, A.; Johnson, K.R.; Matson, K.J.E.; Dobrott, C.I.; Li, L.; Ryba, A.R.; Bergman, T.B.; Kelly, M.C.; Kelley, M.W.; Levine, A.J. Massively Parallel Single Nucleus Transcriptional Profiling Defines Spinal Cord Neurons and Their Activity during Behavior. Cell Rep. 2018, 22, 2216–2225. [Google Scholar] [CrossRef]
- Jovic, D.; Liang, X.; Zeng, H.; Lin, L.; Xu, F.; Luo, Y. Single-cell RNA sequencing technologies and applications: A brief overview. Clin. Transl. Med. 2022, 12, e694. [Google Scholar] [CrossRef]
- Ramsköld, D.; Luo, S.; Wang, Y.C.; Li, R.; Deng, Q.; Faridani, O.R.; Daniels, G.A.; Khrebtukova, I.; Loring, J.F.; Laurent, L.C.; et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012, 30, 777–782. [Google Scholar] [CrossRef]
- Wu, X.; Yang, B.; Udo-Inyang, I.; Ji, S.; Ozog, D.; Zhou, L.; Mi, Q.S. Research Techniques Made Simple: Single-Cell RNA Sequencing and its Applications in Dermatology. J. Investig. Dermatol. 2018, 138, 1004–1009. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M.; et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, C.Y. From bulk, single-cell to spatial RNA sequencing. Int. J. Oral Sci. 2021, 13, 36. [Google Scholar] [CrossRef]
- Zheng, G.X.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef] [PubMed]
- Anaparthy, N.; Ho, Y.J.; Martelotto, L.; Hammell, M.; Hicks, J. Single-Cell Applications of Next-Generation Sequencing. Cold Spring Harb. Perspect. Med. 2019, 9, a026898. [Google Scholar] [CrossRef]
- Wang, X.; He, Y.; Zhang, Q.; Ren, X.; Zhang, Z. Direct Comparative Analyses of 10X Genomics Chromium and Smart-seq2. Genom. Proteom. Bioinform. 2021, 19, 253–266. [Google Scholar] [CrossRef]
- Salcher, S.; Heidegger, I.; Untergasser, G.; Fotakis, G.; Scheiber, A.; Martowicz, A.; Noureen, A.; Krogsdam, A.; Schatz, C.; Schäfer, G.; et al. Comparative analysis of 10X Chromium vs. BD Rhapsody whole transcriptome single-cell sequencing technologies in complex human tissues. Heliyon 2024, 10, e28358. [Google Scholar] [CrossRef]
- Picelli, S.; Björklund, Å.K.; Faridani, O.R.; Sagasser, S.; Winberg, G.; Sandberg, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 2013, 10, 1096–1098. [Google Scholar] [CrossRef]
- Hagemann-Jensen, M.; Ziegenhain, C.; Chen, P.; Ramsköld, D.; Hendriks, G.J.; Larsson, A.J.M.; Faridani, O.R.; Sandberg, R. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 2020, 38, 708–714. [Google Scholar] [CrossRef]
- Klein, A.M.; Mazutis, L.; Akartuna, I.; Tallapragada, N.; Veres, A.; Li, V.; Peshkin, L.; Weitz, D.A.; Kirschner, M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015, 161, 1187–1201. [Google Scholar] [CrossRef]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643.e4. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Liao, S.; Cheng, M.; Ma, K.; Wu, L.; Lai, Y.; Qiu, X.; Yang, J.; Xu, J.; Hao, S.; et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 2022, 185, 1777–1792.e21. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.; Thom, N.; Shadrach, J.L.; Chen, X.; Onesto, M.M.; Amin, N.D.; Yoon, S.J.; Li, L.; Greenleaf, W.J.; Müller, F.; et al. Single-cell transcriptomic landscape of the developing human spinal cord. Nat. Neurosci. 2023, 26, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Matson, K.J.E.; Russ, D.E.; Kathe, C.; Hua, I.; Maric, D.; Ding, Y.; Krynitsky, J.; Pursley, R.; Sathyamurthy, A.; Squair, J.W.; et al. Single cell atlas of spinal cord injury in mice reveals a pro-regenerative signature in spinocerebellar neurons. Nat. Commun. 2022, 13, 5628. [Google Scholar] [CrossRef]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Svensson, V.; Vento-Tormo, R.; Teichmann, S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc. 2018, 13, 599–604. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 1–14. [Google Scholar] [CrossRef]
- Ahuja, C.S.; Wilson, J.R.; Nori, S.; Kotter, M.R.N.; Druschel, C.; Curt, A.; Fehlings, M.G. Traumatic spinal cord injury. Nature reviews. Dis. Primers 2017, 3, 17018. [Google Scholar] [CrossRef] [PubMed]
- Wahane, S.; Zhou, X.; Zhou, X.; Guo, L.; Friedl, M.S.; Kluge, M.; Ramakrishnan, A.; Shen, L.; Friedel, C.C.; Zhang, B.; et al. Diversified transcriptional responses of myeloid and glial cells in spinal cord injury shaped by HDAC3 activity. Sci. Adv. 2021, 7, eabd8811. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wu, Z.; Zhou, L.; Shao, J.; Hu, X.; Xu, W.; Ren, Y.; Zhu, X.; Ge, W.; Zhang, K.; et al. Temporal and spatial cellular and molecular pathological alterations with single-cell resolution in the adult spinal cord after injury. Signal Transduct. Target. Ther. 2022, 7, 65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wu, X.; Fan, Y.; Zhang, H.; Yin, M.; Xue, X.; Yin, Y.; Jin, C.; Quan, R.; Jiang, P.; et al. Characterizing progenitor cells in developing and injured spinal cord: Insights from single-nucleus transcriptomics and lineage tracing. Proc. Natl. Acad. Sci. USA 2025, 122, e2413140122. [Google Scholar] [CrossRef]
- Hildebrand, C.; Remahl, S.; Persson, H.; Bjartmar, C. Myelinated nerve fibres in the CNS. Prog. Neurobiol. 1993, 40, 319–384. [Google Scholar] [CrossRef]
- Wu, Z.; Li, G.; Wang, S.; Zhang, N.; Li, X.; Zhang, F.; Niu, J.; Wang, N.; Zu, J.; Wang, Y. Single-cell analysis of spinal cord injury reveals functional heterogeneity of oligodendrocyte lineage cells. Gene 2023, 886, 147713. [Google Scholar] [CrossRef]
- Saraswathy, V.M.; Zhou, L.; Mokalled, M.H. Single-cell analysis of innate spinal cord regeneration identifies intersecting modes of neuronal repair. Nat. Commun. 2024, 15, 6808. [Google Scholar] [CrossRef]
- Gillespie, E.R.; Grice, L.F.; Courtney, I.G.; Lao, H.W.; Jung, W.; Ramkomuth, S.; Xie, J.; Brown, D.A.; Walsham, J.; Radford, K.J.; et al. Single-cell RNA sequencing reveals peripheral blood leukocyte responses to spinal cord injury in mice with humanised immune systems. J. Neuroinflamm. 2024, 21, 63. [Google Scholar] [CrossRef]
- Yao, C.; Cao, Y.; Wang, D.; Lv, Y.; Liu, Y.; Gu, X.; Wang, Y.; Wang, X.; Yu, B. Single-cell sequencing reveals microglia induced angiogenesis by specific subsets of endothelial cells following spinal cord injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22393. [Google Scholar] [CrossRef]
- Chen, B.; Hasan, M.M.; Zhang, H.; Zhai, Q.; Waliullah, A.S.M.; Ping, Y.; Zhang, C.; Oyama, S.; Mimi, M.A.; Tomochika, Y.; et al. UBL3 Interacts with Alpha-Synuclein in Cells and the Interaction Is Downregulated by the EGFR Pathway Inhibitor Osimertinib. Biomedicines 2023, 11, 1685. [Google Scholar] [CrossRef]
- Lefebvre, S.; Bürglen, L.; Reboullet, S.; Clermont, O.; Burlet, P.; Viollet, L.; Benichou, B.; Cruaud, C.; Millasseau, P.; Zeviani, M. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995, 80, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Lobsiger, C.S.; Cleveland, D.W. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat. Neurosci. 2007, 10, 1355–1360. [Google Scholar] [CrossRef]
- Sun, J.; Qiu, J.; Yang, Q.; Ju, Q.; Qu, R.; Wang, X.; Wu, L.; Xing, L. Single-cell RNA sequencing reveals dysregulation of spinal cord cell types in a severe spinal muscular atrophy mouse model. PLoS Genet. 2022, 18, e1010392. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Peng, Y.; Jie, J.; Yang, Y.; Yang, P. The immune microenvironment and tissue engineering strategies for spinal cord regeneration. Front. Cell. Neurosci. 2022, 16, 969002. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.P.; Chen, S.Y.; Pang, Q.M.; Zhang, M.; Wu, X.C.; Wan, X.; Wan, W.H.; Ao, J.; Zhang, T. Advances in the research of the role of macrophage/microglia polarization-mediated inflammatory response in spinal cord injury. Front. Immunol. 2022, 13, 1014013. [Google Scholar] [CrossRef]
- Zhang, Q.; Yu, B.; Zhang, Y.; Tian, Y.; Yang, S.; Chen, Y.; Wu, H. Combination of single-cell and bulk RNA seq reveals the immune infiltration landscape and targeted therapeutic drugs in spinal cord injury. Front. Immunol. 2023, 14, 1068359. [Google Scholar] [CrossRef]
- Mimi, M.A.; Hasan, M.M.; Takanashi, Y.; Waliullah, A.S.M.; Mamun, M.A.; Chi, Z.; Kahyo, T.; Aramaki, S.; Takatsuka, D.; Koizumi, K.; et al. UBL3 overexpression enhances EV-mediated Achilles protein secretion in conditioned media of MDA-MB-231 cells. Biochem. Biophys. Res. Commun. 2024, 738, 150559. [Google Scholar] [CrossRef]
- Russ, D.E.; Cross, R.B.P.; Li, L.; Koch, S.C.; Matson, K.J.E.; Yadav, A.; Alkaslasi, M.R.; Lee, D.I.; Le Pichon, C.E.; Menon, V.; et al. A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat. Commun. 2021, 12, 5722. [Google Scholar] [CrossRef]
- Han, B.; Zhou, S.; Zhang, Y.; Chen, S.; Xi, W.; Liu, C.; Zhou, X.; Yuan, M.; Yu, X.; Li, L.; et al. Integrating spatial and single-cell transcriptomics to characterize the molecular and cellular architecture of the ischemic mouse brain. Sci. Transl. Med. 2024, 16, eadg1323. [Google Scholar] [CrossRef]
- Hasan, M.M.; Eto, F.; Mamun, M.A.; Sato, S.; Islam, A.; Waliullah, A.S.M.; Chi, D.H.; Takahashi, Y.; Kahyo, T.; Naito, Y.; et al. Desorption ionization using through-hole alumina membrane offers higher reproducibility than 2,5-dihydroxybenzoic acid, a widely used matrix in Fourier transform ion cyclotron resonance mass spectrometry imaging analysis. Rapid Commun. Mass Spectrom. RCM 2021, 35, e9076. [Google Scholar] [CrossRef]
- Chi, D.H.; Kahyo, T.; Islam, A.; Hasan, M.M.; Waliullah, A.S.M.; Mamun, M.A.; Nakajima, M.; Ikoma, T.; Akita, K.; Maekawa, Y.; et al. NAD+ Levels Are Augmented in Aortic Tissue of ApoE−/− Mice by Dietary Omega-3 Fatty Acids. Arterioscler. Thromb. Vasc. Biol. 2022, 42, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Mamun, M.A.; Islam, A.; Hasan, M.M.; Waliullah, A.S.M.; Tamannaa, Z.; Huu Chi, D.; Sato, T.; Kahyo, T.; Kikushima, K.; Takahashi, Y.; et al. The human vermilion surface contains a rich amount of cholesterol sulfate than the skin. J. Dermatol. Sci. 2021, 103, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.; Mimi, A.; Kikushima, K.; Kahyo, T.; Setou, M. Mass spectrometry imaging for glycosphingolipids. In Glycoscience Protocols (GlycoPODv2); Japan Consortium for Glycobiology and Glycotechnology: Saitama, Japan, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK593865/ (accessed on 28 April 2025).
- Khan, Y.S.; Lui, F. Neuroanatomy, Spinal Cord. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK559056/ (accessed on 28 April 2025).
- Nabi, M.; Mamun, A.; Islam, A.; Hasan, M.; Waliullah, A.S.M.; Tamannaa, Z.; Sato, T.; Kahyo, T.; Setou, M. Mass spectrometry in the lipid study of cancer. Expert Rev. Proteom. 2021, 18, 201–219. [Google Scholar] [CrossRef]
- Peng, R.; Zhang, L.; Xie, Y.; Guo, S.; Cao, X.; Yang, M. Spatial multi-omics analysis of the microenvironment in traumatic spinal cord injury: A narrative review. Front. Immunol. 2024, 15, 1432841. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, L.; Dong, H.; Yang, M.; Ding, W.; Zhou, X.; Lu, T.; Liu, Z.; Zhou, X.; Wang, M.; et al. Decoding the spatiotemporal regulation of transcription factors during human spinal cord development. Cell Res. 2024, 34, 193–213. [Google Scholar] [CrossRef]
- Chen, K.H.; Boettiger, A.N.; Moffitt, J.R.; Wang, S.; Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348, aaa6090. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. Method of the Year: Spatially resolved transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef]
- Yang, P.; Jin, K.; Yao, Y.; Jin, L.; Shao, X.; Li, C.; Lu, X.; Fan, X. Spatial integration of multi-omics single-cell data with SIMO. Nat. Commun. 2025, 16, 1265. [Google Scholar] [CrossRef]
- Biancalani, T.; Scalia, G.; Buffoni, L.; Avasthi, R.; Lu, Z.; Sanger, A.; Tokcan, N.; Vanderburg, C.R.; Segerstolpe, Å.; Zhang, M.; et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 2021, 18, 1352–1362. [Google Scholar] [CrossRef]
- Miao, Z.; Humphreys, B.D.; McMahon, A.P.; Kim, J. Multi-omics integration in the age of million single-cell data. Nat. Rev. Nephrol. 2021, 17, 710–724. [Google Scholar] [CrossRef]
- Gao, H.; Zhang, B.; Liu, L.; Li, S.; Gao, X.; Yu, B. A universal framework for single-cell multi-omics data integration with graph convolutional networks. Brief. Bioinform. 2023, 24, bbad081. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hu, Z.; Yu, T.; Wang, Y.; Wang, R.; Wei, Y.; Shu, J.; Ma, J.; Li, Y. Con-AAE: Contrastive cycle adversarial autoencoders for single-cell multi-omics alignment and integration. Bioinformatics 2023, 39, btad162. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.J.; Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 2022, 40, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Fu, J.; Lu, Z.; Tu, J. Deep learning in integrating spatial transcriptomics with other modalities. Brief. Bioinform. 2024, 26, bbae719. [Google Scholar] [CrossRef]
- Yadav, A.; Matson, K.J.E.; Li, L.; Hua, I.; Petrescu, J.; Kang, K.; Alkaslasi, M.R.; Lee, D.I.; Hasan, S.; Galuta, A.; et al. A cellular taxonomy of the adult human spinal cord. Neuron 2023, 111, 328–344.e7. [Google Scholar] [CrossRef]
- Taylor, R.; Nikolaou, N. RNA in axons, dendrites, synapses and beyond. Front. Mol. Neurosci. 2024, 17, 1397378. [Google Scholar] [CrossRef]
- Ament, S.A.; Poulopoulos, A. The brain’s dark transcriptome: Sequencing RNA in distal compartments of neurons and glia. Curr. Opin. Neurobiol. 2023, 81, 102725. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Gautier, M.; Teo, A.Y.Y.; Kathe, C.; Hutson, T.H.; Laskaratos, A.; de Coucy, A.; Regazzi, N.; Aureli, V.; James, N.D.; et al. Single-cell and spatial atlases of spinal cord injury in the Tabulae Paralytica. Nature 2024, 631, 150–163. [Google Scholar] [CrossRef]
- Milich, L.M.; Ryan, C.B.; Lee, J.K. The origin, fate, and contribution of macrophages to spinal cord injury pathology. Acta Neuropathol. 2019, 137, 785–797. [Google Scholar] [CrossRef]
- Jiang, R.; Sun, T.; Song, D.; Li, J.J. Statistics or biology: The zero-inflation controversy about scRNA-seq data. Genome Biol. 2022, 23, 31. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Silberstein, L.; Scadden, D.T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 2014, 11, 740–742. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hu, S.K.; Campbell, V.; Tatters, A.O.; Heidelberg, K.B.; Caron, D.A. Single-cell transcriptomics of small microbial eukaryotes: Limitations and potential. ISME J. 2017, 11, 1282–1285. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Nichterwitz, S.; Chen, G.; Aguila Benitez, J.; Yilmaz, M.; Storvall, H.; Cao, M.; Sandberg, R.; Deng, Q.; Hedlund, E. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 2016, 7, 12139. [Google Scholar] [CrossRef]
- Tran, H.T.N.; Ang, K.S.; Chevrier, M.; Zhang, X.; Lee, N.Y.S.; Goh, M.; Chen, J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020, 21, 12. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current best practices in single-cell RNA-seq analysis: A tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef]
- Adil, A.; Kumar, V.; Jan, A.T.; Asger, M. Single-Cell Transcriptomics: Current Methods and Challenges in Data Acquisition and Analysis. Front. Neurosci. 2021, 15, 591122. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef]
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef]
- Li, W.V.; Li, J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018, 9, 997. [Google Scholar] [CrossRef]
- Huang, M.; Wang, J.; Torre, E.; Dueck, H.; Shaffer, S.; Bonasio, R.; Murray, J.I.; Raj, A.; Li, M.; Zhang, N.R. SAVER: Gene expression recovery for single-cell RNA sequencing. Nat. Methods 2018, 15, 539–542. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, D.; Sharma, R.; Nainys, J.; Yim, K.; Kathail, P.; Carr, A.J.; Burdziak, C.; Moon, K.R.; Chaffer, C.L.; Pattabiraman, D.; et al. Recovering Gene Interactions from Single-Cell Data Using Data Diffusion. Cell 2018, 174, 716–729.e27. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P. Embracing the dropouts in single-cell RNA-seq analysis. Nat. Commun. 2020, 11, 1169. [Google Scholar] [CrossRef]
- Rayon, T.; Maizels, R.J.; Barrington, C.; Briscoe, J. Single-cell transcriptome profiling of the human developing spinal cord reveals a conserved genetic programme with human-specific features. Development 2021, 148, dev199711. [Google Scholar] [CrossRef] [PubMed]
- Argelaguet, R.; Velten, B.; Arnol, D.; Dietrich, S.; Zenz, T.; Marioni, J.C.; Buettner, F.; Huber, W.; Stegle, O. Multi-Omics Factor Analysis-a framework for unsupervised integration of multi-omics data sets. Mol. Syst. Biol. 2018, 14, e8124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M.; 3rd Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902.e21. [Google Scholar] [CrossRef]
- Sofroniew, M.V.; Vinters, H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010, 119, 7–35. [Google Scholar] [CrossRef]
- Peirs, C.; Seal, R.P. Neural circuits for pain: Recent advances and current views. Science 2016, 354, 578–584. [Google Scholar] [CrossRef]
- Fan, Y.; Wu, X.; Han, S.; Zhang, Q.; Sun, Z.; Chen, B.; Xue, X.; Zhang, H.; Chen, Z.; Yin, M.; et al. Single-cell analysis reveals region-heterogeneous responses in rhesus monkey spinal cord with complete injury. Nat. Commun. 2023, 14, 4796. [Google Scholar] [CrossRef]
- Zhang, Q.; Wu, X.; Fan, Y.; Jiang, P.; Zhao, Y.; Yang, Y.; Han, S.; Xu, B.; Chen, B.; Han, J.; et al. Single-cell analysis reveals dynamic changes of neural cells in developing human spinal cord. EMBO Rep. 2021, 22, e52728. [Google Scholar] [CrossRef] [PubMed]
- Kushnarev, M.; Pirvulescu, I.P.; Candido, K.D.; Knezevic, N.N. Neuropathic pain: Preclinical and early clinical progress with voltage-gated sodium channel blockers. Expert Opin. Investig. Drugs 2020, 29, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Awuah, W.A.; Ahluwalia, A.; Ghosh, S.; Roy, S.; Tan, J.K.; Adebusoye, F.T.; Ferreira, T.; Bharadwaj, H.R.; Shet, V.; Kundu, M.; et al. The molecular landscape of neurological disorders: Insights from single-cell RNA sequencing in neurology and neurosurgery. Eur. J. Med. Res. 2023, 28, 529. [Google Scholar] [CrossRef] [PubMed]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.R.; Neff, N.F.; Kalisky, T.; Dalerba, P.; Treutlein, B.; Rothenberg, M.E.; Mburu, F.M.; Mantalas, G.L.; Sim, S.; Clarke, M.F.; et al. Quantitative assessment of single-cell RNA-sequencing methods. Nat. Methods 2014, 11, 41–46. [Google Scholar] [CrossRef]
- Moses, L.; Pachter, L. Museum of spatial transcriptomics. Nat. Methods 2022, 19, 534–546. [Google Scholar] [CrossRef]
- BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 2021, 598, 86–102. [Google Scholar] [CrossRef]
- Ma, Q.; Xu, D. Deep learning shapes single-cell data analysis. Nat. Rev. Mol. Cell Biol. 2022, 23, 303–304. [Google Scholar] [CrossRef]
- Wang, J.; Ma, A.; Chang, Y.; Gong, J.; Jiang, Y.; Qi, R.; Wang, C.; Fu, H.; Ma, Q.; Xu, D. Author Correction: scGNN is a novel graph neural network framework for single-cell RNA-Seq analyses. Nat. Commun. 2022, 13, 2554. [Google Scholar] [CrossRef]
- Xi, N.M.; Li, J.J. Exploring the optimization of autoencoder design for imputing single-cell RNA sequencing data. Comput. Struct. Biotechnol. J. 2023, 21, 4079–4095. [Google Scholar] [CrossRef]
- Cao, X.; Xing, L.; Majd, E.; He, H.; Gu, J.; Zhang, X. A Systematic Evaluation of Supervised Machine Learning Algorithms for Cell Phenotype Classification Using Single-Cell RNA Sequencing Data. Front. Genet. 2022, 13, 836798. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Yang, H.; Zhuang, X.; Liao, J.; Yang, P.; Cheng, J.; Lu, X.; Chen, H.; Fan, X. scDeepSort: A pre-trained cell-type annotation method for single-cell transcriptomics using deep learning with a weighted graph neural network. Nucleic Acids Res. 2021, 49, e122. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Pellegrini, M. ACTINN: Automated identification of cell types in single cell RNA sequencing. Bioinformatics 2020, 36, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Wang, W.; Wang, F.; Fang, Y.; Tang, D.; Huang, J.; Lu, H.; Yao, J. scBERT as a large-scale pretrained deep language model for cell type annotation of single-cell RNA-seq data. Nat. Mach. Intell. 2022, 4, 852–866. [Google Scholar] [CrossRef]
- Wang, Y.R.; Du, P.F. WCSGNet: A graph neural network approach using weighted cell-specific networks for cell-type annotation in scRNA-seq. Front. Genet. 2025, 16, 1553352. [Google Scholar] [CrossRef]
- Dobrott, C.I.; Sathyamurthy, A.; Levine, A.J. Decoding Cell Type Diversity Within the Spinal Cord. Curr. Opin. Physiol. 2019, 8, 1–6. [Google Scholar] [CrossRef]
- Liu, L.; Huang, Y.; Zheng, Y.; Liao, Y.; Ma, S.; Wang, Q. ScnML models single-cell transcriptome to predict spinal cord neuronal cell status. Front. Genet. 2024, 15, 1413484. [Google Scholar] [CrossRef]
- Fang, M.; Gorin, G.; Pachter, L. Trajectory inference from single-cell genomics data with a process time model. PLoS Comput. Biol. 2025, 21, e1012752. [Google Scholar] [CrossRef] [PubMed]
- A graph neural network that combines scRNA-seq and protein-protein interaction data. Nat. Methods 2025, 22, 660–661. [CrossRef]
- Jaja, B.N.R.; Badhiwala, J.; Guest, J.; Harrop, J.; Shaffrey, C.; Boakye, M.; Kurpad, S.; Grossman, R.; Toups, E.; Geisler, F.; et al. Trajectory-Based Classification of Recovery in Sensorimotor Complete Traumatic Cervical Spinal Cord Injury. Neurology 2021, 96, e2736–e2748. [Google Scholar] [CrossRef]
- Patel, M.; Magre, N.; Motwani, H.; Brown, N.B. Advances in machine learning, statistical methods, and AI for single-cell RNA annotation using raw count matrices in scRNA-seq data. arXiv 2024, arXiv:2406.05258. [Google Scholar] [CrossRef]
- Shen, X.; Li, X. Deep-learning methods for unveiling large-scale single-cell transcriptomes. Cancer Biol. Med. 2024, 20, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhu, S.; Yu, B.; Yao, C. Single-cell RNA sequencing for traumatic spinal cord injury. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2022, 36, e22656. [Google Scholar] [CrossRef] [PubMed]
- Shu, M.; Xue, X.; Nie, H.; Wu, X.; Sun, M.; Qiao, L.; Li, X.; Xu, B.; Xiao, Z.; Zhao, Y.; et al. Single-cell RNA sequencing reveals Nestin+ active neural stem cells outside the central canal after spinal cord injury. Sci. China Life Sci. 2022, 65, 295–308. [Google Scholar] [CrossRef]
- Li, D.; Ma, Y.; Huang, W.; Li, X.; Liu, H.; Xiong, C.; Zhao, Q.; Wang, B.; Lai, X.; Huang, S.; et al. Modeling human spine-spinal cord organogenesis by hPSC-derived neuromesodermal progenitors. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chelyshev, Y.A.; Ermolin, I.L. RNA Sequencing and Spatial Transcriptomics in Traumatic Spinal Cord Injury (Review). Sovrem. Tekhnologii Meditsine 2023, 15, 75–86. [Google Scholar] [CrossRef]
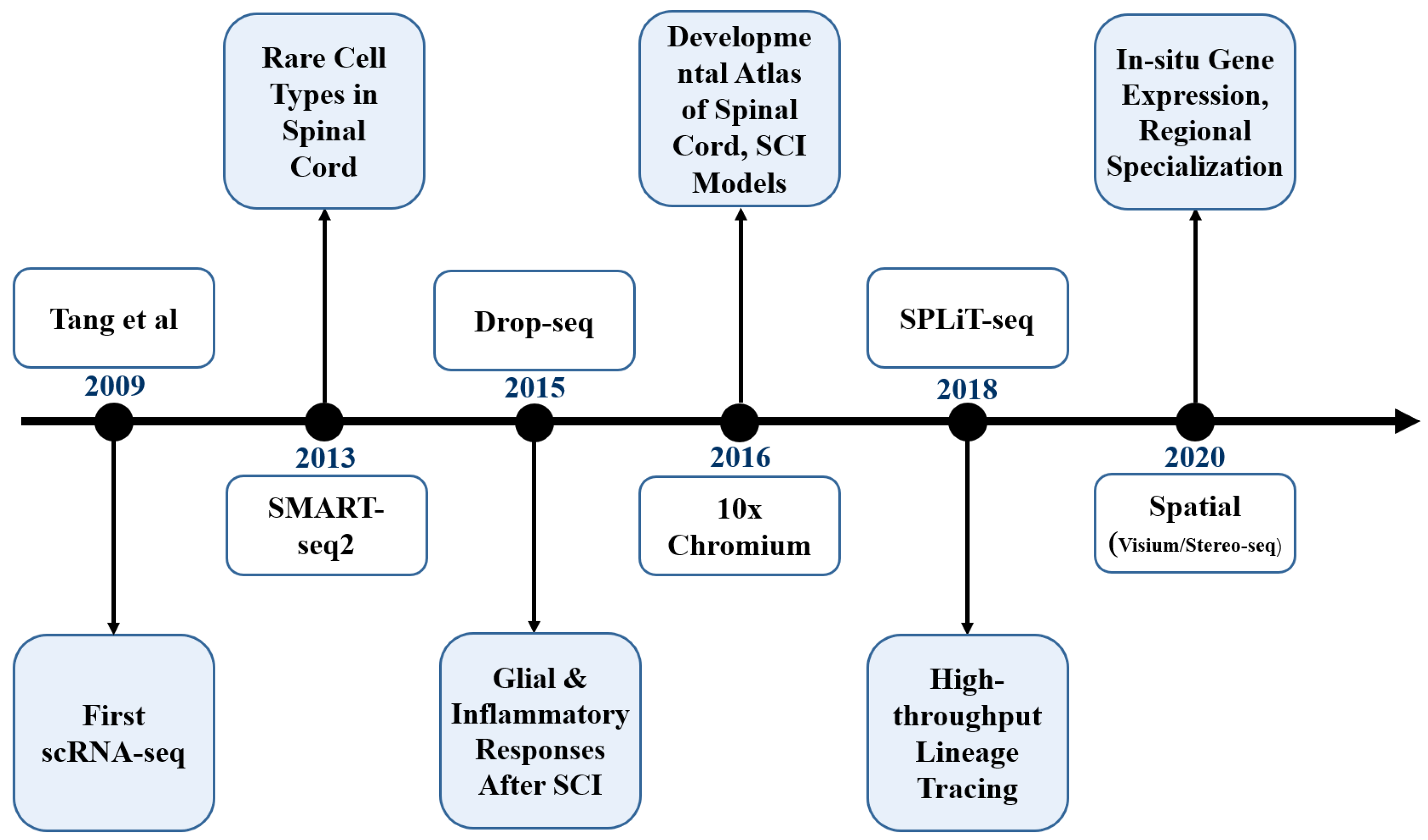
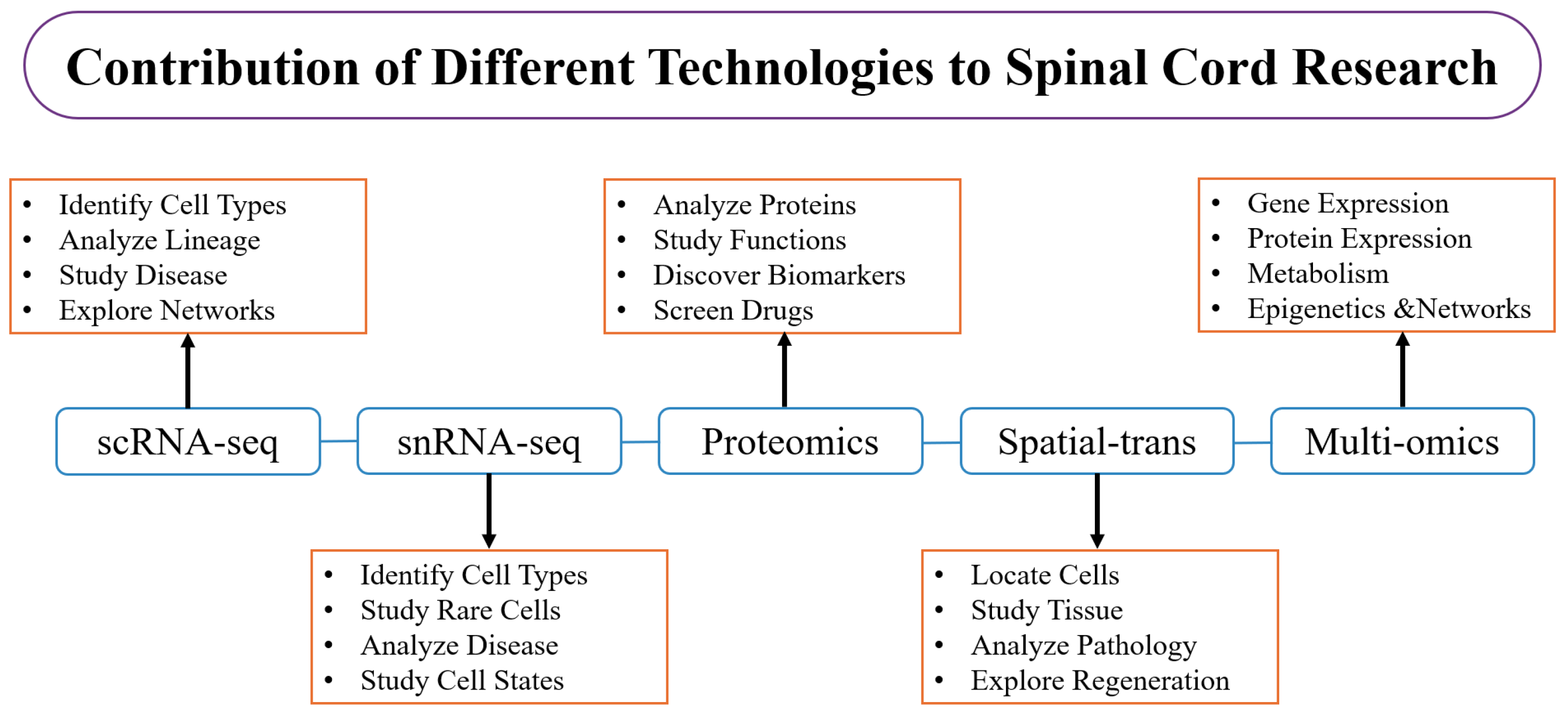
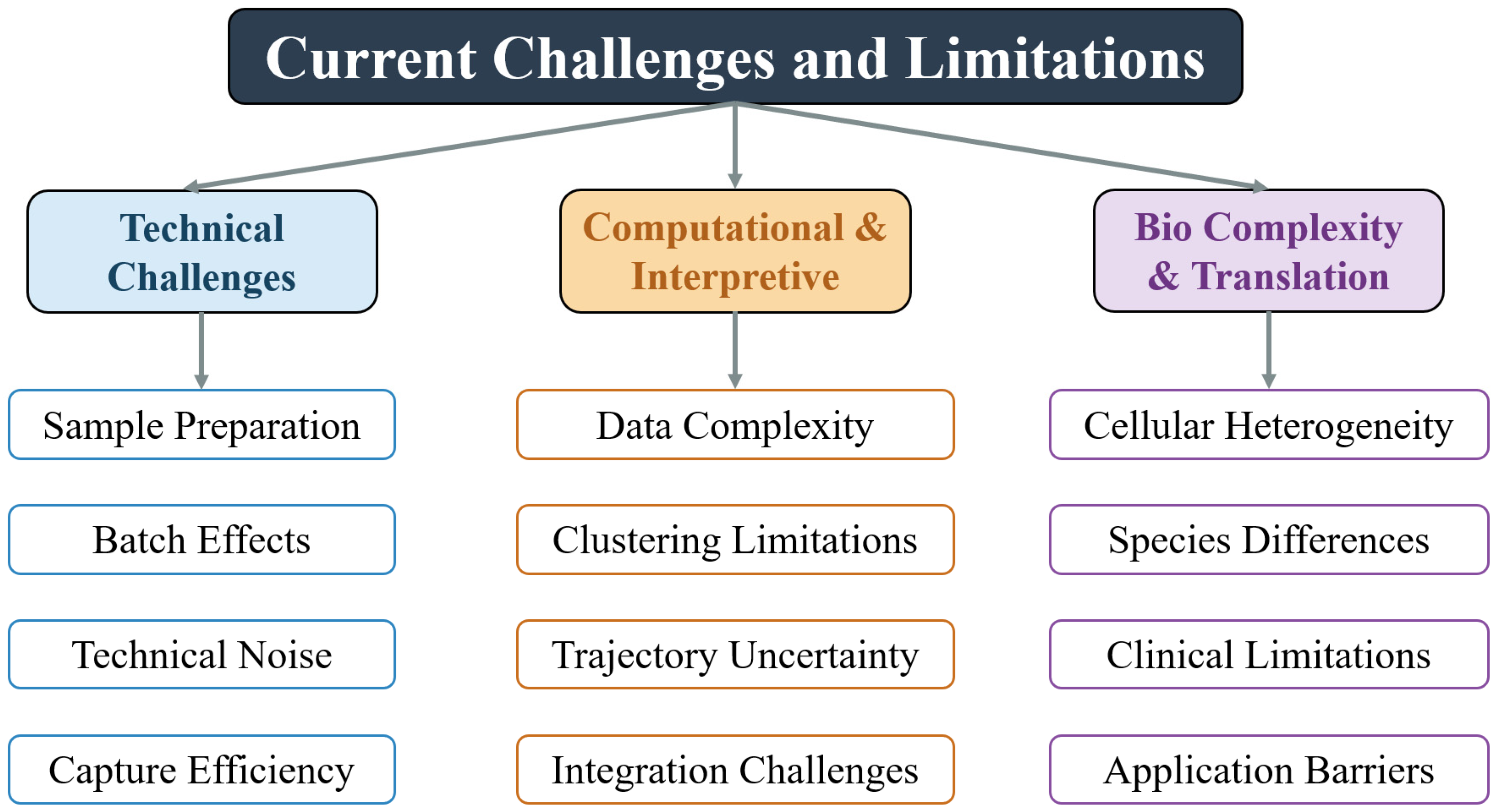
| Sequencing Method | Key Features | Advantages | Limitations | Applications in Spinal Cord Research | Temporal Evolution |
|---|---|---|---|---|---|
| SMART-seq2/SMART-seq3 | Plate-based; full-length transcript coverage (SMART-seq3 improves sensitivity) | High sensitivity; isoform detection; full-length transcripts | Low throughput; expensive; time-consuming | Detecting rare cell types or novel isoforms | SMART-seq2 (~2013); SMART-seq3 (~2019) |
| 10x Genomics Chromium | Droplet-based; barcode encapsulation of cells | High throughput; scalable; relatively low cost | Partial transcript coverage; dropout effect | Developmental mapping; SCI response; human/fetal profiling | Mainstream since ~2016 |
| Drop-seq | Droplet-based; barcoded beads in droplets | Cost-effective; scalable; high efficiency | Lower sensitivity than SMART-seq | Identifying glial/inflammatory responses | Introduced ~2015 |
| inDrops | Improved droplet-based; enhanced barcoding and chemistry | Low background noise; flexible; improved capture | Still limited in sensitivity; complex protocol | Flexible transcriptomic profiling with high consistency | Developed post-2015 |
| SPLiT-seq | Combinatorial barcoding; no physical isolation required | Ultra-high throughput; low equipment requirement | Sparse gene detection; complex processing | Mapping neuron subtypes in mouse spinal cord development | Introduced ~2018 |
| 10x Visium/BGI Stereo-seq | Spatial transcriptomics; spatial gene expression in situ | High spatial resolution; large capture area | Requires tissue sectioning; limited to spatial resolution | Mapping spatial gene expression in human spinal cord | Emerging since ~2020 |
| Study | Species | Developmental Stage | Platform | Key Findings |
|---|---|---|---|---|
| Delile et al. [17] | Mouse | E9.5–E13.5 | 10x Genomics Chromium | Spatial/temporal dynamics of neural progenitors; novel markers in dorsoventral domains |
| Andersen et al. [34] | Human | ~22 weeks gestation | 10x Genomics Chromium | Glial heterogeneity, astrocyte regionalization, disease gene mapping to specific cell types |
| Sathyamurthy et al. [19] | Mouse | Adult | Drop-Seq | 43 neuronal subtypes, region-specific distribution, spinal neuron molecular map |
| Blum et al. [4] | Mouse | Adult | 10x Genomics Chromium | Motor neuron heterogeneity, transcriptional profiles linked to axonal targeting and function |
| Cao et al. [36] | Mouse | Organogenesis (multi-stage) | SPLiT-seq | Spinal progenitor transcriptional transitions, Hox genes, Shh pathway in developmental regulation |
| Zhang et al. [18] | Human | Adult | 10x Genomics Chromium | 21 neuronal subtypes, spatial distribution, human-mouse comparison, sex-specific transcription |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maihemuti, M.; Mimi, M.A.; Sohag, S.M.; Hasan, M.M. Single-Cell Transcriptomics in Spinal Cord Studies: Progress and Perspectives. BioChem 2025, 5, 16. https://doi.org/10.3390/biochem5020016
Maihemuti M, Mimi MA, Sohag SM, Hasan MM. Single-Cell Transcriptomics in Spinal Cord Studies: Progress and Perspectives. BioChem. 2025; 5(2):16. https://doi.org/10.3390/biochem5020016
Chicago/Turabian StyleMaihemuti, Maiweilan, Mst. Afsana Mimi, S. M. Sohag, and Md. Mahmudul Hasan. 2025. "Single-Cell Transcriptomics in Spinal Cord Studies: Progress and Perspectives" BioChem 5, no. 2: 16. https://doi.org/10.3390/biochem5020016
APA StyleMaihemuti, M., Mimi, M. A., Sohag, S. M., & Hasan, M. M. (2025). Single-Cell Transcriptomics in Spinal Cord Studies: Progress and Perspectives. BioChem, 5(2), 16. https://doi.org/10.3390/biochem5020016








