Qualitative Shotgun Proteomics Strategy for Protein Expression Profiling of Fish Otoliths
Abstract
1. Introduction
2. Materials and Methods
2.1. Otoliths
2.2. Chemicals and Standards
2.3. Otolith Protein Extraction
2.4. Shotgun Proteomics by LC-ESI-MS/MS
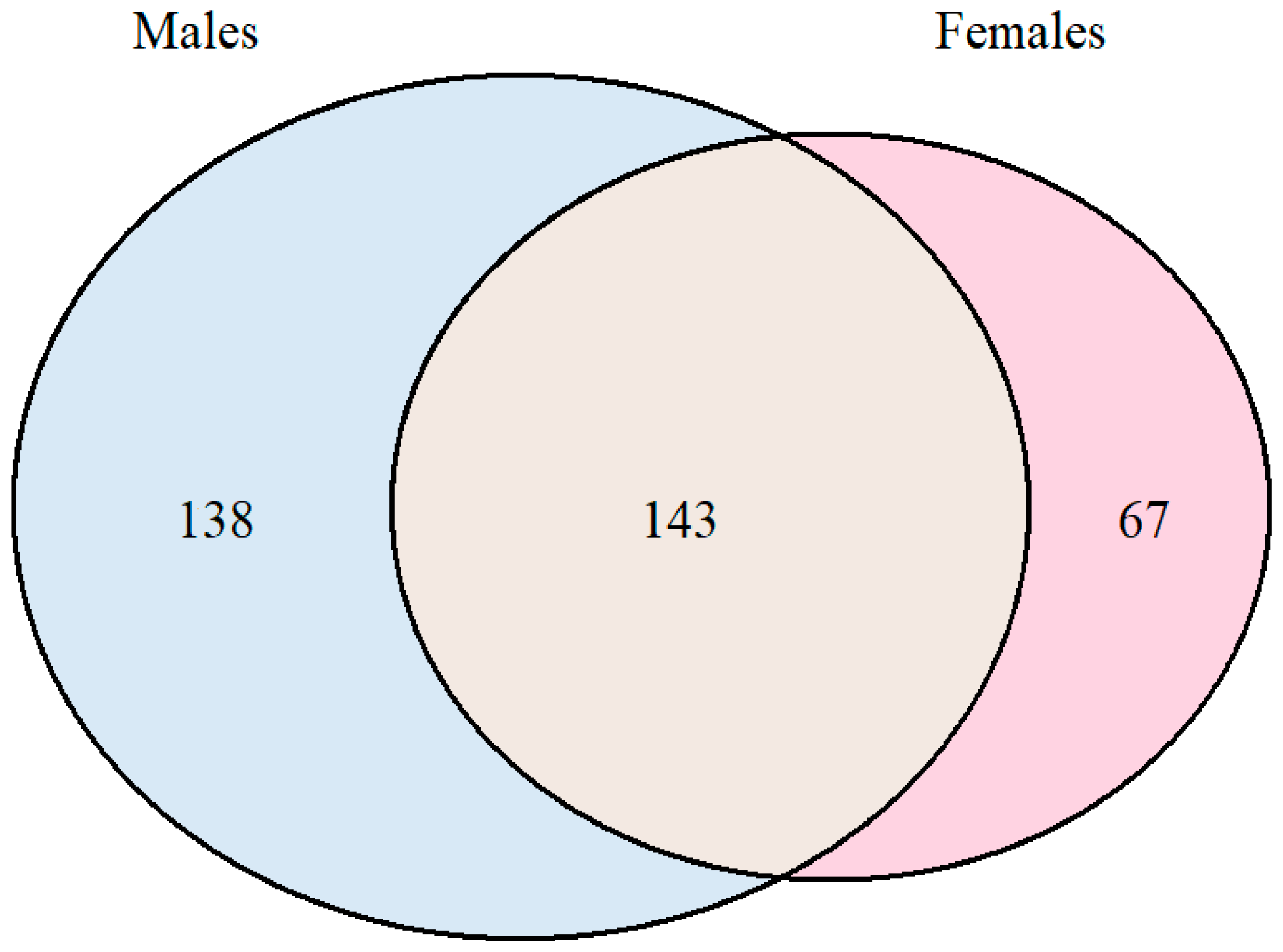
3. Results and Discussion
4. Proteins Linked to Biochemical Processes
4.1. Protein Synthesis
4.2. Biochemical Reaction Processes
5. Proteins Linked to Physiological Processes
5.1. Brain Function
5.2. Mediated Transport Processes
5.3. Mitochondrial Processes
5.4. Cytoskeleton and Extracellular Matrices
5.5. Adhesion and Binding Processes
5.6. Cellular Signaling Processes
5.7. Cellular Immunological Processes
5.8. Physiological Nuclear Processes
5.9. Ion Transport Processes
5.10. Sexual Differentiation
6. Identification of Known Otolith Proteins
7. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popper, A.N.; Hawkins, A.D. The importance of particle motion to fishes and invertebrates. J. Acoust. Soc. Am. 2018, 143, 470. [Google Scholar] [CrossRef]
- Lowenstein, O. 7 The Labyrinth. In Fish Physiology; Hoar, W.S., Randall, D.J., Eds.; Academic Press: Cambridge, MA, USA, 1971; Volume 5, pp. 207–240. [Google Scholar]
- Manning, F.B. Hearing in the goldfish in relation to the structure of its ear. J. Exp. Zool. 1924, 41, 5–20. [Google Scholar] [CrossRef]
- Schnetz, L.; Pfaff, C.; Libowitzky, E.; Johanson, Z.; Stepanek, R.; Kriwet, J. Morphology and evolutionary significance of phosphatic otoliths within the inner ears of cartilaginous fishes (Chondrichthyes). BMC Evol. Biol. 2019, 19, 238. [Google Scholar] [CrossRef]
- Fay, R.R. The Goldfish Ear Codes the Axis of Acoustic Particle Motion in Three Dimensions. Science 1984, 225, 951–954. [Google Scholar] [CrossRef] [PubMed]
- Popper, A.N.; Ramcharitar, J.; Campana, S.E. Why otoliths? Insights from inner ear physiology and fisheries biology. Mar. Freshw. Res. 2005, 56, 497–504. [Google Scholar] [CrossRef]
- Pannella, G. Fish otoliths: Daily growth layers and periodical patterns. Science 1971, 173, 1124–1127. [Google Scholar] [CrossRef] [PubMed]
- Brothers, E.B.; Mathews, C.P.; Lasker, R. Daily growth increments in otoliths from larval and adult fishes. Fish Bull 1976, 74, 1–8. [Google Scholar]
- Allemand, D.; Mayer-Gostan, N.; De Pontual, H.; Boeuf, G.; Payan, P. Fish Otolith Calcification in Relation to Endolymph Chemistry. In Handbook of Biomineralization; Bäuerlein, E., Ed.; WILEY-VCH Verlag GmbH & Co. KgaA: Weinheim, Germany, 2007; pp. 291–308. [Google Scholar]
- Campana, S.E.; Thorrold, S.R. Otoliths, increments, and elements: Key to a comprehensive understanding of fish populations. Can. J. Fish. Aquat. Sci. 2001, 58, 30–38. [Google Scholar] [CrossRef]
- Ricard, D.; Adamack, A.; Burbank, J.; Comeau, P.; Daigle, A.; Debertin, A.; Emond, K.; Loewen, T.; Perreault, A.; Puncher, G.; et al. Canadian Technical Report of Fisheries and Aquatic Sciences. In Proceedings of the Technical Expertise in Stock Assessment (TESA) National Workshop on ‘Best Practices in Age Estimation’, Moncton, NB, Canada, 31 January–2 February 2023. in press. [Google Scholar]
- Campana, S.E. How reliable are growth back-calculations based on otoliths? Can. J. Fish. Aquat. Sci. 1990, 47, 2219–2227. [Google Scholar] [CrossRef]
- Degens, E.T.; Deuser, W.G.; Haedrich, R.L. Molecular structure and composition of fish otoliths. Mar. Biol. 1969, 2, 105–113. [Google Scholar] [CrossRef]
- Campana, S.E. Chemistry and composition of fish otoliths: Pathways, mechanisms and applications. Mar. Ecol. Prog. Ser. 1999, 188, 263–297. [Google Scholar] [CrossRef]
- Payan, P.; De Pontual, H.; Bœuf, G.; Mayer-Gostan, N. Endolymph chemistry and otolith growth in fish. Comptes Rendus Palevol 2004, 3, 535–547. [Google Scholar] [CrossRef]
- Köppl, C.; Wilms, V.; Russell, I.J.; Nothwang, H.G. Evolution of Endolymph Secretion and Endolymphatic Potential Generation in the Vertebrate Inner Ear. Brain Behav. Evol. 2018, 92, 1–31. [Google Scholar] [CrossRef]
- Romanek, C.S.; Gauldie, R.W. A predictive model of otolith growth in fish based on the chemistry of the endolymph. Comp. Biochem. Physiol. Part A Physiol. 1996, 114, 71–79. [Google Scholar] [CrossRef]
- Disspain, M.C.F.; Ulm, S.; Gillanders, B.M. Otoliths in archaeology: Methods, applications and future prospects. J. Archaeol. Sci. Rep. 2016, 6, 623–632. [Google Scholar] [CrossRef]
- Yasuo, M. Diurnal rhythm in otolith formation in the rainbow trout, Salmo gairdneri: Seasonal reversal of the rhythm in relation to plasma calcium concentrations. Comp. Biochem. Physiol. Part A Physiol. 1984, 78, 289–293. [Google Scholar] [CrossRef]
- Thomas, O.R.B.; Swearer, S.E. Otolith Biochemistry—A Review. Rev. Fish. Sci. Aquac. 2019, 27, 458–489. [Google Scholar] [CrossRef]
- Loewen, T.N.; Carriere, B.; Reist, J.D.; Halden, N.M.; Anderson, W.G. Linking physiology and biomineralization processes to ecological inferences on the life history of fishes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2016, 202, 123–140. [Google Scholar] [CrossRef]
- Falini, G.; Albeck, S.; Weiner, S.; Addadi, L. Control of Aragonite or Calcite Polymorphism by Mollusk Shell Macromolecules. Science 1996, 271, 67–69. [Google Scholar] [CrossRef]
- Murayama, E.; Takagi, Y.; Ohira, T.; Davis, J.G.; Greene, M.I.; Nagasawa, H. Fish otolith contains a unique structural protein, otolin-1. Eur. J. Biochem. 2002, 269, 688–696. [Google Scholar] [CrossRef]
- Huang, S.; Qian, S. Advances in otolith-related protein research. Front. Neurosci. 2022, 16, 956200. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, Y.W.; Xu, Y.; Thiessen, K.D.; Kramer, K.L. Mechanisms of otoconia and otolith development. Dev. Dyn. 2015, 244, 239–253. [Google Scholar] [CrossRef]
- Thomas, O.R.B.; Swearer, S.E.; Kapp, E.A.; Peng, P.; Tonkin-Hill, G.Q.; Papenfuss, A.; Roberts, A.; Bernard, P.; Roberts, B.R. The inner ear proteome of fish. FEBS J. 2018, 286, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Mulry, E.; Parham, K. Inner Ear Proteins as Potential Biomarkers. Otol. Neurotol. 2020, 41, 145–152. [Google Scholar] [CrossRef]
- Murayama, E.; Herbomel, P.; Kawakami, A.; Takeda, H.; Nagasawa, H. Otolith matrix proteins OMP-1 and Otolin-1 are necessary for normal otolith growth and their correct anchoring onto the sensory maculae. Mech. Dev. 2005, 122, 791–803. [Google Scholar] [CrossRef]
- Miller, M.B.; Clough, A.M.; Batson, J.N.; Vachet, R.W. Transition metal binding to cod otolith proteins. J. Exp. Mar. Biol. Ecol. 2006, 329, 135–143. [Google Scholar] [CrossRef]
- Fournier, M.L.; Gilmore, J.M.; Martin-Brown, S.A.; Washburn, M.P. Multidimensional separations-based shotgun proteomics. Chem. Rev. 2007, 107, 3654–3686. [Google Scholar] [CrossRef]
- Meyer, J.G. Qualitative and Quantitative Shotgun Proteomics Data Analysis from Data-Dependent Acquisition Mass Spectrometry. Methods Mol. Biol. 2021, 2259, 297–308. [Google Scholar] [CrossRef]
- Chen, H. “VennDiagram: Generate High-Resolution Venn and Euler Plots”. R Package Version 1.7.3. 2022. Available online: https://CRAN.R-project.org/package=VennDiagram (accessed on 1 July 2022).
- Dannevig, E.H. Chemical composition of the zones in cod otoliths. ICES J. Mar. Sci. 1956, 21, 156–159. [Google Scholar] [CrossRef]
- Peters, J.-M.; Cejka, Z.; Harris, J.R.; Kleinschmidt, J.A.; Baumeister, W. Structural Features of the 26 S Proteasome Complex. J. Mol. Biol. 1993, 234, 932–937. [Google Scholar] [CrossRef] [PubMed]
- Vanzi, F.; Vladimirov, S.; Knudsen, C.R.; Goldman, Y.E.; Cooperman, B.S. Protein synthesis by single ribosomes. RNA 2003, 9, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Das, G.; Varshney, U. Peptidyl-tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 2006, 152, 2191–2195. [Google Scholar] [CrossRef]
- Osugi, T.; Ding, Y.; Tanaka, H.; Kuo, C.-H.; Do, E.; Irie, Y.; Miki, N. Involvement of a single-stranded DNA binding protein, ssCRE-BP/Purα, in morphine dependence. FEBS Lett. 1996, 391, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Fey, E.G.; Krochmalnic, G.; Penman, S. The nonchromatin substructures of the nucleus: The ribonucleoprotein (RNP)-containing and RNP-depleted matrices analyzed by sequential fractionation and resinless section electron microscopy. J. Cell Biol. 1986, 102, 1654–1665. [Google Scholar] [CrossRef]
- Kimpler, L.A.; Glosson, N.L.; Downs, D.; Gonyo, P.; May, N.A.; Hudson, A.W. Adaptor Protein Complexes AP-1 and AP-3 Are Required by the HHV-7 Immunoevasin U21 for Rerouting of Class I MHC Molecules to the Lysosomal Compartment. PLoS ONE 2014, 9, e99139. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Du, J.; Cui, K.; Fang, W.; Zhao, Z.; Chen, Q.; Mai, K.; Ai, Q. Acetyl-CoA derived from hepatic mitochondrial fatty acid β-oxidation aggravates inflammation by enhancing p65 acetylation. iScience 2021, 24, 103244. [Google Scholar] [CrossRef]
- Waagbø, R. Water-soluble vitamins in fish ontogeny. Aquac. Res. 2010, 41, 733–744. [Google Scholar] [CrossRef]
- Panepucci, L.; Fernandes, M.N.; Sanches, J.R.; Rantin, F.T. Changes in lactate dehydrogenase and malate dehydrogenase activities during hypoxia and after temperature acclimation in the armored fish, Rhinelepis strigosa (Siluriformes, Loricariidae). Rev. Bras. Biol. 2000, 60, 353–360. [Google Scholar] [CrossRef]
- Valero, Y.; Martínez-Morcillo, F.J.; Esteban, M.Á.; Chaves-Pozo, E.; Cuesta, A. Fish Peroxiredoxins and Their Role in Immunity. Biology 2015, 4, 860–880. [Google Scholar] [CrossRef]
- Rand-Weaver, M.; Noso, T.; Muramoto, K.; Kawauchi, H. Isolation and characterization of somatolactin, a new protein related to growth hormone and prolactin from Atlantic cod (Gadus morhua) pituitary glands. Biochemistry 1991, 30, 1509–1515. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, J.; Ma, A.; Wang, X.; Sun, Z.; Cui, W.; Yuan, C.; Zhu, C. Molecular characterization, expression analysis of 14-3-3 beta/alpha and the effect of RNA interference on ion transporter protein Na+-K+-ATPase, Na+–H+-exchanger and CFTR in turbot (Scophthalmus maximus). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2020, 246–247, 110458. [Google Scholar] [CrossRef] [PubMed]
- Sower, S.A.; Decatur, W.A.; Hausken, K.N.; Marquis, T.J.; Barton, S.L.; Gargan, J.; Freamat, M.; Wilmot, M.; Hollander, L.; Hall, J.A.; et al. Emergence of an Ancestral Glycoprotein Hormone in the Pituitary of the Sea Lamprey, a Basal Vertebrate. Endocrinology 2015, 156, 3026–3037. [Google Scholar] [CrossRef] [PubMed]
- Allah Abdulraheem, B. Aspartate Transaminase (AST) Activity in Selected Tissues & Organs of Clarias Gariepinus Exposed To Different Levels of Paraquat. J. Environ. Anal. Toxicol. 2014, 4, 71–73. [Google Scholar] [CrossRef]
- Blumstein, J.; Faundez, V.; Nakatsu, F.; Saito, T.; Ohno, H.; Kelly, R.B. The Neuronal Form of Adaptor Protein-3 Is Required for Synaptic Vesicle Formation from Endosomes. J. Neurosci. 2001, 21, 8034. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, R.; Jouraku, A.; Nakakura, T.; Kusakabe, T.; Yamamoto, T.; Shinohara, Y.; Miyoshi, H.; Shiotsuki, T. Two Adenine Nucleotide Translocase Paralogues Involved in Cell Proliferation and Spermatogenesis in the Silkworm Bombyx mori. PLoS ONE 2015, 10, e0119429. [Google Scholar] [CrossRef]
- Bieczynski, F.; Painefilú, J.C.; Venturino, A.; Luquet, C.M. Expression and Function of ABC Proteins in Fish Intestine. Front. Physiol. 2021, 12, 791834. [Google Scholar] [CrossRef] [PubMed]
- Katyal, G.; Ebanks, B.; Lucassen, M.; Papetti, C.; Chakrabarti, L. Sequence and structure comparison of ATP synthase F0 subunits 6 and 8 in notothenioid fish. PLoS ONE 2021, 16, e0245822. [Google Scholar] [CrossRef] [PubMed]
- Ni, S.; Zhou, Y.; Chen, Y.; Du, X.; Zhang, S. Identification of ATP synthase α subunit as a new maternal factor capable of protecting zebrafish embryos from bacterial infection. FASEB J. 2019, 33, 12983–13001. [Google Scholar] [CrossRef]
- Naghdi, S.; Hajnóczky, G. VDAC2-specific cellular functions and the underlying structure. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2016, 1863, 2503–2514. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Behera, S.; Swain, S.; Panda, M.K.; Mistri, A.R.; Sahoo, B. Sequence and structural analysis of β- actin protein of fishes, using bioinformatics tools and techniques. Int. J. Biosci. 2014, 4, 249–256. [Google Scholar] [CrossRef]
- Dos Remedios, C.G.; Chhabra, D.; Kekic, M.; Dedova, I.V.; Tsubakihara, M.; Berry, D.A.; Nosworthy, N.J. Actin Binding Proteins: Regulation of Cytoskeletal Microfilaments. Physiol. Rev. 2003, 83, 433–473. [Google Scholar] [CrossRef] [PubMed]
- Tillgren, V.; Ho, J.C.S.; Önnerfjord, P.; Kalamajski, S. The Novel Small Leucine-rich Protein Chondroadherin-like (CHADL) Is Expressed in Cartilage and Modulates Chondrocyte Differentiation. J. Biol. Chem. 2015, 290, 918–925. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, Y.-J.; Ma, D.H.-K.; Ma, K.S.-C.; Wang, T.-K.; Chou, C.-H.; Lin, C.-C.; Huang, M.-C.; Luo, L.-J.; Lai, J.-Y.; Chen, H.-C. Extracellular Matrix Protein Coating of Processed Fish Scales Improves Human Corneal Endothelial Cell Adhesion and Proliferation. Transl. Vis. Sci. Technol. 2019, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Zoeller, J.J.; Pimtong, W.; Corby, H.; Goldoni, S.; Iozzo, A.E.; Owens, R.T.; Ho, S.-Y.; Iozzo, R.V. A Central Role for Decorin during Vertebrate Convergent Extension. J. Biol. Chem. 2009, 284, 11728–11737. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Kasahara, K.; Inagaki, M. Intermediate filaments and IF-associated proteins: From cell architecture to cell proliferation. Proc. Jpn. Acad. Ser. B 2019, 95, 479–493. [Google Scholar] [CrossRef]
- Schaffeld, M.; Markl, J. Fish Keratins. Methods Cell Biol. 2004, 78, 627–671. [Google Scholar] [CrossRef] [PubMed]
- van der Loop, F.T.; Schaart, G.; Timmer, E.D.; Ramaekers, F.C.; van Eys, G.J. Smoothelin, a novel cytoskeletal protein specific for smooth muscle cells. J. Cell Biol. 1996, 134, 401–411. [Google Scholar] [CrossRef]
- Detrich, H.W.; Fitzgerald, T.J.; Dinsmore, J.H.; Marchese-Ragona, S.P. Brain and egg tubulins from antarctic fishes are functionally and structurally distinct. J. Biol. Chem. 1992, 267, 18766–18775. [Google Scholar] [CrossRef] [PubMed]
- Hwang Hyun, J.; Moon Chang, H.; Kim Han, G.; Kim Joo, Y.; Lee Jung, M.; Park Jeong, W.; Chung Dae, K. Identification and Functional Analysis of Salmon Annexin 1 Induced by a Virus Infection in a Fish Cell Line. J. Virol. 2007, 81, 13816–13824. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, S.; Schweitzer, J.; Jahn, O.; Werner, H.B. Molecular evolution of myelin basic protein, an abundant structural myelin component. Glia 2013, 61, 1364–1377. [Google Scholar] [CrossRef]
- Boggs, J.M. Myelin basic protein: A multifunctional protein. Cell. Mol. Life Sci. CMLS 2006, 63, 1945–1961. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Foot, N.; Ang, C.-S.; Gembus, K.M.; Keerthikumar, S.; Adda, C.G.; Mathivanan, S.; Kumar, S. Arrestin-Domain Containing Protein 1 (Arrdc1) Regulates the Protein Cargo and Release of Extracellular Vesicles. Proteomics 2018, 18, 1800266. [Google Scholar] [CrossRef]
- Kang, D.S.; Tian, X.; Benovic, J.L. Role of β-arrestins and arrestin domain-containing proteins in G protein-coupled receptor trafficking. Curr. Opin. Cell Biol. 2014, 27, 63–71. [Google Scholar] [CrossRef]
- Swulius, M.T.; Waxham, M.N. Ca2+/Calmodulin-dependent Protein Kinases. Cell. Mol. Life Sci. 2008, 65, 2637. [Google Scholar] [CrossRef]
- Gupta, M.; Brand, M. Identification and Expression Analysis of Zebrafish Glypicans during Embryonic Development. PLoS ONE 2013, 8, e80824. [Google Scholar] [CrossRef]
- Termini, C.M.; Gillette, J.M. Tetraspanins Function as Regulators of Cellular Signaling. Front. Cell Dev. Biol. 2017, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Tulke, S.; Williams, P.; Hellysaz, A.; Ilegems, E.; Wendel, M.; Broberger, C. Nucleobindin 1 (NUCB1) is a Golgi-resident marker of neurons. Neuroscience 2016, 314, 179–188. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response. Funct. Integr. Genom. 2008, 8, 301–307. [Google Scholar] [CrossRef]
- Evans, P.C.; Ovaa, H.; Hamon, M.; Kilshaw, P.J.; Hamm, S.; Bauer, S.; Ploegh, H.L.; Smith, T.S. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 2004, 378, 727–734. [Google Scholar] [CrossRef]
- Jang, J.H.; Kim, H.; Jung, I.Y.; Cho, J.H. A20 Inhibits LPS-Induced Inflammation by Regulating TRAF6 Polyubiquitination in Rainbow Trout. Int. J. Mol. Sci. 2021, 22, 9801. [Google Scholar] [CrossRef]
- Du Clos, T.W. Pentraxins: Structure, Function, and Role in Inflammation. ISRN Inflamm. 2013, 2013, 379040. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M.; Reeves, R. High-mobility-group chromosomal proteins: Architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol. 1996, 54, 35–100. [Google Scholar] [PubMed]
- DesJarlais, R.; Tummino, P.J. Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 2016, 55, 1584–1599. [Google Scholar] [CrossRef]
- Li, Z.; Langhans, S.A. Transcriptional regulators of Na,K-ATPase subunits. Front. Cell Dev. Biol. 2015, 3, 66. [Google Scholar] [CrossRef] [PubMed]
- Binner, M.I.; Kogan, A.; Panser, K.; Schleiffer, A.; Deneke, V.E.; Pauli, A. The sperm protein Spaca6 is essential for fertilization in zebrafish. bioRxiv 2021, arXiv:2021.2011.2019.469324. [Google Scholar] [CrossRef]
- Yuan, S.; Stratton, C.J.; Bao, J.; Zheng, H.; Bhetwal, B.P.; Yanagimachi, R.; Yan, W. Spata6 is required for normal assembly of the sperm connecting piece and tight head–tail conjunction. Proc. Natl. Acad. Sci. USA 2015, 112, E430–E439. [Google Scholar] [CrossRef]
- Lehti, M.S.; Sironen, A. Formation and function of sperm tail structures in association with sperm motility defects. Biol. Reprod. 2017, 97, 522–536. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.M.; Mansour, A.A.H.; Banoub, J.H. ‘De novo’ sequencing of Atlantic cod vitellogenin tryptic peptides by matrix-assisted laser desorption/ionization quadrupole time-of-flight tandem mass spectrometry: Similarities with haddock vitellogenin. Rapid Commun. Mass Spectrom. 2005, 19, 2454–2460. [Google Scholar] [CrossRef] [PubMed]
- Hara, A.; Hiramatsu, N.; Fujita, T. Vitellogenesis and choriogenesis in fishes. Fish. Sci. 2016, 82, 187–202. [Google Scholar] [CrossRef]
- Hamm, L.L.; Nakhoul, N.; Hering-Smith, K.S. Acid-Base Homeostasis. Clin. J. Am. Soc. Nephrol. 2015, 10, 2232. [Google Scholar] [CrossRef]
- Rhyu, H.-J.; Bae, S.H.; Jung, J.; Hyun, Y.-M. Cochlin-cleaved LCCL is a dual-armed regulator of the innate immune response in the cochlea during inflammation. BMB Rep. 2020, 53, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Girón-Pérez, D.A.; Piedra-Quintero, Z.L.; Santos-Argumedo, L. Class I myosins: Highly versatile proteins with specific functions in the immune system. J. Leukoc. Biol. 2019, 105, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Stooke-Vaughan, G.A.; Obholzer, N.D.; Baxendale, S.; Megason, S.G.; Whitfield, T.T. Otolith tethering in the zebrafish otic vesicle requires Otogelin and α-Tectorin. Development 2015, 142, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Stevenson, A.K.; Yau, P.M.; Kollmar, R. Sparc Protein Is Required for Normal Growth of Zebrafish Otoliths. J. Assoc. Res. Otolaryngol. 2008, 9, 436–451. [Google Scholar] [CrossRef]
- Law, R.H.P.; Zhang, Q.; McGowan, S.; Buckle, A.M.; Silverman, G.A.; Wong, W.; Rosado, C.J.; Langendorf, C.G.; Pike, R.N.; Bird, P.I.; et al. An overview of the serpin superfamily. Genome Biol. 2006, 7, 216. [Google Scholar] [CrossRef][Green Version]
- Ramos-Balderas, J.L.; Carrillo-Rosas, S.; Guzman, A.; Navarro, R.E.; Maldonado, E. The zebrafish mutants for the V-ATPase subunits d, ac45, E, H and c and their variable pigment dilution phenotype. BMC Res. Notes 2013, 6, 39. [Google Scholar] [CrossRef] [PubMed]
| 1. Biochemical Processes | |
|---|---|
| 1.1. Protein Synthesis | |
| A0A8C4ZUE8 | 40S ribosomal protein S12 |
| A0A8C5BT07 | 40S ribosomal protein S18 |
| A0A8C5BHD6 | 40S ribosomal protein S19 |
| A0A8C4ZDL6 | 40S ribosomal protein S28 |
| A0A8C4YW06 | 40S ribosomal protein S3a |
| A0A8C5F524 | 40S ribosomal protein S9 |
| A0A8C5B226 | 40S_SA_C domain-containing protein |
| A0A8C4ZT25 | 60S acidic ribosomal protein P2 |
| A0A8C4Z3G5 | 60S ribosomal protein L27 |
| A0A8C5BNH7 | 60S ribosomal protein L30 |
| A0A8C4Z1D1 | 60S ribosomal protein L31 |
| A0A8C4ZUJ5 | Aminoacyl-tRNA hydrolase |
| A0A8C4ZR55 | Purine-rich element binding protein Ab (PURA) |
| A0A8C5FJ16 | Ribonucloprotein (RNP). |
| 1.2. Biochemical Reaction Processes | |
| A0A8C5BPU1 | Oxoisovalerate dehydrogenase subunit alpha |
| A0A8C5C4N4 | Acetyl-CoA acetyltransferase 1 (ACAT1) |
| A0A8C5ADH8/ A0A8C5FBY3 | Aconitate hydratase |
| A0A8C4ZEH5 | Transketolase b (TKT) |
| A0A8C4YZP8 | Malate dehydrogenase (MDH) |
| A0A8C4Z702 | Peroxiredoxin-1 (PRX1) |
| P21919 | Somatolactin |
| 2. Physiological Processes | |
| 2.1. Brain Function Processes | |
| A0A8C4ZJE1 | 14-3-3 protein beta/alpha-1-like protein |
| A0A8C5A8C5 | Alpha-2-HS-glycoprotein-like |
| A0A8C5BJ01 | Aspartate aminotransferase |
| 2.2. Mediated Transport Processes | |
| A0A8C4Z7S3 | AP-3 complex subunit mu-2 (PA3) |
| A0A8C5F5V1 | Adaptor related protein complex 3 subunit sigma 1 |
| 2.3. Mitochondrial Processes | |
| A0A8C4YYC8 | ADP/ATP translocase |
| A0A8C5AUM7 | ATP-binding cassette sub-family B (MDR/TAP) |
| A0A8C4Z154 | ATP synthase-coupling factor 6 mitochondria |
| A0A8C5AGK0/A0A8C4YZ60 | ATP synthase subunit alpha and beta |
| A0A8C5FKS7 | Voltage-Dependent Anion-selective Channel 2 (VDAC2) |
| 2.4. Cytoskeleton and Extracellular Matrixes | |
| A0A8C5FJI3 | Actin, beta protein |
| A0A8C4ZDZ9 | Actin-related protein 2/3 complex subunit 5 |
| A0A8C4ZHY1 | Chondroadherin-like b (CHADL) |
| A0A8C5FEX0 | Choclin: extracellular matrix (ECM) |
| A0A8C4ZF74 | Decorin or pericellular matrix proteoglycan (SLRP) |
| A0A8C5AFR1 | The Intermediate filament protein ON3-like (IFs) [39] |
| A0A8C5C226/A0A8C4ZFJ9/ A0A8C4YVX1/A0A8C5B373/ A0A8C4YSX2/A0A8C5B602/ A0A8C4ZH54 | Keratins |
| A0A8C5FDF8 | F-actin-capping protein subunit beta complex |
| A0A8C5C7G7 | Smoothelin a |
| A0A8C5BEP0 | Tubulin alpha-1A chain (TUBA 1A) |
| 2.5. Adhesion and Binding Processes | |
| A0A8C5BRQ2 | Annexin |
| A0A8C5AI73 | Secreted acidic cysteine rich glycoprotein (Sparc) |
| A0A8C5B6I8 | Myelin basic protein (MBP) |
| 2.6. Cellular Signaling Processes | |
| A0A8C4ZK56 | Arrestin-domain containing protein 1 (Arrdc1) |
| A0A8C5BSR3 | Calmodulin 2a phosphorylase kinase (CaMKII) |
| A0A8C4Z128 | Glypicans (Gpi) |
| A0A8C5BN57/A0A8C4Z2M4 | Glycoprotein Tetraspanin |
| A0A8C4Z782/A0A8C5B4W1 | Nucleobindin 1 (NUCB1) |
| 2.7. Cellular Immunological-Processes | |
| A0A8C5F947 | A20/AN1 zinc-finger domain-containing protein |
| A0A8C5BVB7 | Pentraxin |
| 2.8. Nuclear Physiological Processes | |
| A0A8C5FTZ2/A0A8C4ZQW9/A0A8C4ZTE5 | High mobility group (HMG) protein |
| A0A8C5B042/A0A8C5FQN1/A0A8C5FAA0 | Histone protein |
| 2.9. Ions Transport Processes | |
| A0A8C5AKI2 | Sodium/potassium-transporting ATPase subunit beta (NKA) |
| A0A8C4Z4K1/A0A8C5ASM5 | Sodium/potassium-transporting ATPase subunit alpha |
| 2.10. Sexual Differentiation Processes | |
| A0A8C5A412 | SPATA6 protein |
| A0A8C5CHW7 | Vitellogenin-2-like protein |
| 3. Identifiecation of Know Otolith Proteins | |
| A0A8C4ZKX9 | Carbonic anhydrase protein (CA) |
| A0A8C5CSC6 | Cochlin |
| A0A8C5AZT6 | Myosin |
| A0A8C5F796 | Otogelin |
| A0A8C5CL43 | Otolin-1 |
| A0A8C5ASM5/A0A8C5AKI2 | Na, K-ATPase proteins |
| A0A8C5AI73 | Secreted acidic cysteine rich glycoprotein (Sparc) |
| A0A8C5BD06/A0A8C5D2V1/A0A8C5AT89 | SERPIN domain-containing protein |
| A0A8C5AFL5/A0A8C4ZVQ3 | α- and β-Tectorins |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rideout, R.M.; Youssef, T.N.; Adamack, A.T.; John, R.; Cohen, A.M.; Fridgen, T.D.; Banoub, J.H. Qualitative Shotgun Proteomics Strategy for Protein Expression Profiling of Fish Otoliths. BioChem 2023, 3, 102-117. https://doi.org/10.3390/biochem3030008
Rideout RM, Youssef TN, Adamack AT, John R, Cohen AM, Fridgen TD, Banoub JH. Qualitative Shotgun Proteomics Strategy for Protein Expression Profiling of Fish Otoliths. BioChem. 2023; 3(3):102-117. https://doi.org/10.3390/biochem3030008
Chicago/Turabian StyleRideout, Rick M., Trevena N. Youssef, Aaron T. Adamack, Rince John, Alejandro M. Cohen, Travis D. Fridgen, and Joseph H. Banoub. 2023. "Qualitative Shotgun Proteomics Strategy for Protein Expression Profiling of Fish Otoliths" BioChem 3, no. 3: 102-117. https://doi.org/10.3390/biochem3030008
APA StyleRideout, R. M., Youssef, T. N., Adamack, A. T., John, R., Cohen, A. M., Fridgen, T. D., & Banoub, J. H. (2023). Qualitative Shotgun Proteomics Strategy for Protein Expression Profiling of Fish Otoliths. BioChem, 3(3), 102-117. https://doi.org/10.3390/biochem3030008







