Abstract
Proper and timely segregation of the cellular genome is a prime requirement of all cell division programs. Mis-segregation of chromosomes and the resulting aneuploidy lead to several clinical consequences. Over the years, shugoshin has emerged as a key protein factor involved in the segregation of genetic material in dividing cells. Deletion or an altered level of shugoshin is reported in several human malignancies; as a result, shugoshin is now emerging as an important tumor-associated gene and a possible target for cancer therapy. Apart from its role in cancer, recent studies have also shown the involvement of shugoshin in several other clinical disorders. In this review, we aim to highlight the clinical relevance of shugoshin.
1. Introduction
Proper cell division is a foremost requirement for reproduction as well as for the survival and continuity of every species. Mis-segregation of the genome during cell division leads to aneuploidy, which is closely associated with numerous medical consequences ranging from tumorigenesis to sterility, mental retardation, spontaneous abortion, and other birth-related defects [1,2,3,4,5,6]. To ensure that the genetic blueprint is duplicated and distributed precisely during cell division, cells employ several mechanisms operating either independently or in coordination with one another. Proper and timely removal of cohesin is an example of one such mechanism. Cohesin, a multiprotein complex, holds sister chromatids together from DNA duplication in S-phase until the onset of anaphase. The premature or untimely loss of cohesion as a result of abrupt separase activity leads to chromosome mis-segregation. Hence, cohesin cleavage by separase is kept under tight cellular control [7,8]. Apart from its prime role of holding sister chromatids together, cohesin is also known for its involvement in diverse cellular processes discussed elsewhere [9]. A detailed account of cohesin and separase falls outside the scope of the present review, and these aspects are summarized elsewhere [8,10]. Apart from the timely cleavage of cohesin, several other mechanisms including DNA damage checkpoint (DDC), spindle assembly checkpoint (SAC), separase activation, and centriole duplication (and maybe more which remain unidentified) ensure that the genetic endowment of the cell or organism, i.e., its genome is duplicated and separated properly. A detailed discussion of all such mechanisms is difficult in the present review, may require a separate volume, and can be found elsewhere [8,11,12,13,14,15,16].
In this review, we will mainly focus on shugoshin, a protein factor required for the protection of centromeric cohesin. The intended purpose of this review is to familiarize the readers with the medical conditions in which shugoshin is implicated. Before discussing the clinical association of shugoshin, we will give a bird’s eye view of shugoshin including its identification, cellular localization, conserved nature, distribution in eukaryotes, and finally, shugoshin as an emerging tumor-associated gene and as a potential target for cancer therapy. The present review does not include a description of shugoshin in the cell cycle (mitosis and meiosis) as those topics have already been discussed in recent reviews [17,18,19,20].
2. Shugoshin Background
Shugoshin (meaning “guardian spirit” in Japanese) is a homo-dimeric phospho-protein belonging to the shugoshin protein family [21,22]. Shugoshin is conserved from single-celled yeast to multicellular mammals including humans. Shugoshin shares several structural features with other members of the shugoshin family, including a basic region at the C-terminus that is essential for centromere binding, chromosome localization, and an N-terminal coiled-coil domain that may regulate its dimerization and interaction with other proteins [23,24,25,26]. Initially, shugoshin was discovered in the fruit fly, D. melanogaster, as a peri-centromeric protein (at the time referred to as MEI-S332) required for the protection of Rec8 (meiotic-specific cohesin subunit) from separase action and its persistence during meiosis-I [27,28,29]. Later, a protein factor with a function equivalent to MEI-S332 was discovered in other eukaryotic species including yeast, insects, vertebrates, and plants [30,31,32,33,34,35].
Based on the sequence (at the gene and protein levels) and structural analysis, it has been observed that all eukaryotic species studied to date possess either one or two genes coding for shugoshin (referred to as SGO1 and SGO2), although several splicing isoforms of shugoshin have been reported in higher eukaryotes [36]. Table 1 shows the number of genes coding for shugoshin in different species. The reason why some species (for example, Saccharomyces cerevisiae) possess only one gene for shugoshin and others two (for example, fission yeast, humans) remains elusive. In human cells, a combined total of 10 splicing isoforms (for SGO1 and SGO2) have been identified (http://www.uniprot.org/uniprot/?query=hugoshin%2C+homo+sapiens&sort=score (accessed on 4 March 2021)). Information related to different isoforms of human SGO1 and SGO2, including the number of amino acid residues and molecular mass, is given in Table 2. The size or number of amino acid residues in shugoshin and molecular mass vary significantly across different eukaryotic species [35] as well as among different isoforms within the same species (for example, Homo sapiens, Table 2). It is important to mention that different shugoshin paralogs are known to exhibit different properties depending on the species under consideration. The expression pattern or profile of shugoshin paralogs may be cell cycle-dependent (i.e., mitosis or meiosis). For example, in fission yeast SGO2 is expressed in both mitosis and meiosis while SGO1 is meiosis-specific [30]. Like fission yeast, mouse SGO2 is required for the completion of meiosis but not for mitosis, suggesting its cell cycle-specific expression [37].

Table 1.
Number of genes coding for shugoshin in different species.

Table 2.
Size and molecular mass of different isoforms of human SGO1 and SGO2.
Shugoshin is present in all the eukaryotic species studied to date, and shugoshin-based protection of centromeric cohesin is conserved across different eukaryotic species. However, the cells of C. elegans use a different strategy that is independent of shugoshin. Unlike other species, chromosome segregation in C. elegans relies on an alternative mechanism that involves LAB-1 (Long Arm of the Bivalent) [38]. This study in C. elegans raised the possibility of shugoshin-independent cohesin protection in other species. Why the cells of C. elegans use this alternative mechanism despite the presence of shugoshin remains an open question. Whether shugoshin-independent protection of centromeric cohesin is exclusive to worm species also remains a matter for future investigation.
3. Cellular Localization
So far, shugoshin has been detected or observed in the nucleus in close association with chromosomes at the kinetochore or centromere, in the spindle pole body (or SPB, the functional equivalent of centrioles of higher eukaryotes), and more recently in the sub-telomeric region of the chromosome [30,39,40,41,42,43]. Cellular localization of shugoshin in yeast and the mammalian cell is shown in Figure 1A,B, respectively. By looking at Figure 1, it can be inferred that the overall cellular localization of shugoshin is similar in yeast and mammalian cells despite a huge evolutionary distance. The similar cellular localization of shugoshin in yeast and mammalian cells shows the importance or suitability of yeast as a model system. It is important to mention that in budding yeast, the nuclear membrane always remains intact (closed mitosis), and the SPB always remains embedded in the nuclear envelope. In mammalian cells, the nuclear envelope is lost completely (open mitosis), and centrioles always remain in the cytoplasm. Therefore, the possibility of a novel shugoshin role and interactors in yeast as well as in mammalian cells cannot be ruled out. Thus, the identification of a novel function or interactors of shugoshin represents both opportunities and challenges to a contemporary biologist working in this direction. We give a comparative localization of human and yeast cells as a great deal of information related to shugoshin is gathered using yeast.
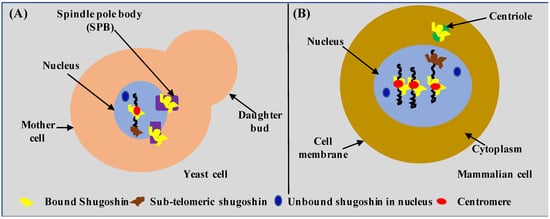
Figure 1.
Cellular localization of shugoshin. Localization of shugoshin in (A) budding yeast and (B) mammalian cell. Note: Diagrams are for demonstration purposes only.
4. Shugoshin as a Tumor-Associated Gene
Being an important player in the cell cycle, cells ensure that shugoshin is present only when and where needed [44]. Cells also maintain an optimum level of shugoshin by regulating its expression and degradation through APC/C (anaphase promoting complex/cyclosome) [45]. A complete absence or altered level (both more and less than the optimum level) of shugoshin may lead to tumorigenesis. Plenty of studies are now available supporting both the oncogenic as well as tumor suppressor nature of shugoshin. Therefore, it would be fair enough to consider shugoshin a tumor-associated gene (Figure 2). The cellular abundance of shugoshin is so critical that even one fold change in its cellular abundance is enough for tumor induction [46]. The role of shugoshin in cancer is confirmed not only from studies in non-human subjects like rats or mice but also from tissue samples collected from actual human cancer patients. In this section of the review, we highlight some of the recent studies in which the complete absence or altered level of shugoshin was linked to cancer.
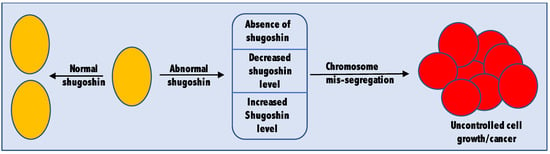
Figure 2.
Shugoshin as a tumor-associated gene. Complete absence as well as increased or decreased level of shugoshin were found to be associated with different types of cancers.
4.1. Shugoshin as a Tumor Suppressor Gene
A decreased level or complete absence of shugoshin has been observed in head and neck cancer [47], nasopharyngeal carcinoma [48], neuroblastoma [49], and prostate cancer [50,51]. Homozygous deletion of SGOL2 has been observed in different types of human tumors including head and neck cancer [52], small-cell lung carcinoma [53], cervical carcinoma [54], and neuroblastoma [55]. It is important to mention that deletion of either allele of shugoshin (i.e., SGOL1 or SGOL2 for shugoshin in humans) can lead to cancer. Among 46 colorectal cancer cases, hSgo1 mRNA expression was decreased in the tumor tissue compared with the corresponding normal tissue [56]. Heterozygous deletion of sgo1+/− leads to systemic chromosome instability in mice [57] and the formation of aberrant crypt foci (ACF) in mice heterozygous for shugoshin-1 [58]. Treatment with the carcinogen azoxymethane (oxide of azomethane, a carcinogenic and neurotoxic chemical compound used in biological research) caused sgo1+/− ME-CIN model mice to develop hepatocellular carcinoma (HCC) within 6 months; in contrast, control mice developed no HCC (p < 0.003) [59].
Although SGO1+/− mice are viable and fertile, they show enhanced colonic tumorigenesis. Enhanced cervical intraepithelial neoplasia (CIN) observed in sgo1-deficient mice leads to an increase in the formation of ACF and an accelerated development of tumors on exposure to azoxymethane [60]. The SGOL1-P1 (one of the splicing forms of shugoshin in humans) transcript containing an exon-skip of exon 3 results in a stop codon occurring within exon 4, whose overexpression in human HCT116 cell line resulted in an increased number of cells with aberrant chromosome alignment, precociously separated chromatids, and delayed mitotic progression, occasionally followed by inaccurate distribution of the chromosomes [61]. Downregulation of shugoshin-1 leads to CIN in colorectal cancer cells [56]. Mice heterozygous for shugoshin-1 showed mild proneness to spontaneous lung and liver cancers. In a recent study using adoptive (T/B cell-based) immunity-deficient RAG1(−/−) (recombination activating gene), sgo1(−/+) double-mutant mice developed lung adenocarcinomas more aggressively compared with sgo1(−/+) or RAG1(−/−) mice, suggesting immune system involvement in CIN-mediated lung carcinogenesis [62]. All these studies clearly showed the tumor suppressor nature of shugoshin.
4.2. Shugoshin as an Oncogene
In the last section, we mentioned some of the studies where shugoshin behaved as a tumor suppressor gene. In this section, we will mention some of the studies which showed the oncogenic behavior of shugoshin. Upregulated expression of shugoshin was observed in 82% of hepatocellular carcinoma (HCC) cases and correlated with elevated alpha-fetoprotein and early disease onset of HCC, while depletion of shugoshin-1 reduced the cell viability of hepatoma cell lines including HuH7, HepG2, Hep3B, and HepaRG due to persistent activation of the spindle assembly checkpoint [63]. Increased expression and level of shugoshin were reported in human leukemia [64] and breast cancers [65,66]. Similarly, overexpression of SGOL1-B1 in a non-small-cell lung carcinoma (NSCLC) cell line induced aberrant chromosome mis-segregation, precociously separated chromatids, and delayed mitotic progression. A higher level of SGO1-B mRNA was related to taxane (diterpenes, compounds originally identified in the plant genus Taxus (yews), used in cancer chemotherapy, e.g., paclitaxel and docetaxel) resistance, while the forced downregulation of SGO1-B increased the sensitivity to taxane [67]. Expression of SGO1C (a non-functional isoform of shugoshin) alone induced aberrant mitosis similar to depletion of SGO1A, promoting premature sister chromatid separation, activation of the spindle assembly checkpoint, and mitotic arrest, suggesting that the expression of SGO1C is tightly regulated to prevent dominant-negative effects of SGO1A and genome instability [68]. In another clinical study, the expression of SGO1 in human prostate tumors was higher than that of adjacent normal tissues and was positively correlated with the poor prognosis of prostate cancer patients [69]. Some of the studies mentioned above clearly showed the oncogenic nature of shugoshin.
Based on the studies mentioned in this section, it can be said that shugoshin can act as an important target for medical intervention in cancer therapy. Not only complete loss of shugoshin but also an altered level of shugoshin can lead to cancer. Whether shugoshin’s association with cancer is due to chromosome mis-segregation or due to derailment of other cellular pathways resulting from a complete absence or altered level of shugoshin remains a topic for future research. Because an altered shugoshin level is associated with various cancers, and chemicals (for example, BPA or Bisphenol A, used as a plasticizer in plastic industries) can potentially alter its expression, it is possible that increased incidences of tumors and associated altered shugoshin levels may be linked and require further research [70]. The identification of chemicals that can modulate the transcription of shugoshin and other tumor-associated genes can be an important field for future research.
Both complete loss or absence and an increased or decreased expression or cellular abundance of shugoshin can lead to cancer. Whether the oncogenic and tumor suppressor natures of shugoshin modulate the same cellular pathways remains unknown.
5. Shugoshin in Other Clinical Disorders
In the last section, we mentioned the tumor-associated nature of shugoshin. However, cancer or tumor is not the only clinical condition where shugoshin is involved. Research over the last several years has also implicated shugoshin in other serious medical conditions. In this section, we would like to draw attention towards some of those clinical disorders. For example, recently it was shown that mutations of SGO2 (frameshift, p.Glu485Lysfs *5) and CLDN14 collectively cause coincidental Perrault syndrome, which is a rare autosomal genetic disorder characterized by sensorineural hearing loss (SNHL) in males and females and ovarian dysfunction in females [71,72]. Some studies in the last few years also showed the involvement of shugoshin in neurological disorders such as late-onset Alzheimer’s disease (LOAD) [73,74]. It is important to mention that not all the diseased conditions are due to chromosome instability, which is induced by impaired shugoshin function of centromeric cohesion. For example, chronic atrial and intestinal dysrhythmia (CAID) is not due to chromosome instability because of impaired shugoshin at the centromere [20,75].
6. Conclusions and Future Directions
Based on results from different scientific groups working on various model systems and tissue samples from cancer patients, it can be clearly said that shugoshin is an important tumor-associated gene. Apart from its role in cancer, shugoshin is also involved in several other medical conditions. It is important to note that shugoshin-associated pathies may also happen even when there is no observed chromosomal instability, and shugoshin may still be involved. Given the way shugoshin is linked to various clinical conditions, it is possible that the list of shugoshin-associated pathies may increase in the future. In such a scenario, the availability of a high-resolution atomic structure of shugoshin will help in the designing or screening of small molecules with applications in cancer therapy and other shugoshin-associated diseases. Aside from that, the availability of the structure will help towards a better understanding of shugoshin’s localization and its interaction with other cellular proteins or complexes. The availability of a high-resolution structure will also be important in better understanding the way shugoshin is involved in different diseases. The diverse and large size of shugoshin poses a big challenge for structure biologists [24,76]. Apart from this, innately disordered regions in most shugoshin proteins make it difficult to get a high-resolution structure. The presence of different splicing isoforms of shugoshin in higher eukaryotes adds another hurdle in structural studies. Furthermore, we are still not aware of the cellular abundance of shugoshin, how it is regulated, and what protein factors and signaling are involved in maintaining the optimum level of shugoshin in a cell. Therefore, future research on shugoshin will be important and rewarding both from the standpoint of basic science and from the perspective of medical science.
Author Contributions
R.K. conceived the idea, started writing the draft, and prepared the figures and tables. M.A. contributed to writing the draft and managed the reference section. All authors have read and agreed to the published version of the manuscript.
Funding
The authors declare no funding to be reported.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We regret for not being able to refer to the work of everyone in the field. We are thankful to the University of California San Francisco for providing space and other necessary facilities which helped us in completing this manuscript.
Conflicts of Interest
The authors declare that no conflict of interest exist.
References
- Rajagopalan, H.; Lengauer, C. Aneuploidy and cancer. Nature 2004, 432, 338–341. [Google Scholar] [CrossRef]
- Hassold, T.; Hall, H.; Hunt, P. The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 2007, 16, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Jallepalli, P.V.; Lengauer, C. Chromosome segregation and cancer: Cutting through the mystery. Nat. Rev. Cancer 2001, 1, 109–117. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instability in colorectal cancers. Nature 1997, 386, 623–627. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K. The incidence, origin, and etiology of aneuploidy. Int. Rev. Cytol. 1996, 167, 263–296. [Google Scholar]
- Sen, S. Aneuploidy and cancer. Curr. Opin. Oncol. 2000, 12, 82–88. [Google Scholar] [CrossRef]
- Uhlmann, F. Secured cutting: Controlling separase at the metaphase to anaphase transition. EMBO Rep. 2001, 2, 487–492. [Google Scholar] [CrossRef]
- Kumar, R. Separase: Function Beyond Cohesion Cleavage and an Emerging Oncogene. J. Cell Biochem. 2017, 118, 1283–1299. [Google Scholar] [CrossRef] [PubMed]
- Mehta, G.D.; Kumar, R.; Srivastava, S.; Ghosh, S.K. Cohesin: Functions beyond sister chromatid cohesion. FEBS Lett. 2013, 587, 2299–2312. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Joglekar, A.P. A Cell Biological Perspective on Past, Present and Future Investigations of the Spindle Assembly Checkpoint. Biology 2016, 5, 44. [Google Scholar] [CrossRef]
- Kamenz, J.; Hauf, S. Time to Split up: Dynamics of Chromosome Separation. Trends Cell Biol. 2017, 27, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Fujita, H.; Yoshino, Y.; Chiba, N. Regulation of the centrosome cycle. Mol. Cell Oncol. 2015, 3, e1075643. [Google Scholar] [CrossRef] [PubMed]
- Simpson-Lavy, K.J.; Oren, Y.S.; Feine, O.; Sajman, J.; Listovsky, T.; Brandeis, M. Fifteen years of APC/cyclosome: A short and impressive biography. Biochem. Soc. Trans. 2010, 38, 78–82. [Google Scholar] [CrossRef][Green Version]
- Agarwal, M.; Mehta, G.; Ghosh, S.K. Role of Ctf3 and COMA subcomplexes in meiosis: Implication in maintaining Cse4 at the centromere and numeric spindle poles. Biochim. Biophys. Acta Mol. Cell Res. 2015, 1853, 671–684. [Google Scholar] [CrossRef][Green Version]
- Agarwal, M.; Jin, H.; McClain, M.; Fan, J.; Koch, B.A.; Jaspersen, S.L.; Yu, H.G. The half-bridge component Kar1 promotes centrosome separation and duplication during budding yeast meiosis. Mol. Biol. Cell 2018, 29, 1798–1810. [Google Scholar] [CrossRef] [PubMed]
- Marston, A.L. Shugoshins: Tension-sensitive pericentromeric adaptors safeguarding chromosome segregation. Mol. Cell Biol. 2015, 35, 634–648. [Google Scholar] [CrossRef]
- Clift, D.; Marston, A.L. The role of shugoshin in meiotic chromosome segregation. Cytogenet. Genome Res. 2011, 133, 234–242. [Google Scholar] [CrossRef]
- Wassmann, K. Sister chromatid segregation in meiosis II: Deprotection through phosphorylation. Cell Cycle 2013, 12, 1352–1359. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, H. Functioning mechanisms of Shugoshin-1 in centromeric cohesion during mitosis. Essays Biochem. 2020, 64, 289–297. [Google Scholar]
- Xu, Z.; Cetin, B.; Anger, M.; Cho, U.S.; Helmhart, W.; Nasmyth, K.; Xu, W. Structure and function of the PP2A-shugoshin inter-action. Mol. Cell 2009, 35, 426–441. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villén, J. Global analysis of phosphorylation and ubiquitylation crosstalk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.T.; Bickel, S.E.; Young, L.M.; Orr-Weaver, T.L. Maintenance of sister-chromatid cohesion at the centromere by the Drosophila MEI-S332 protein. Genes Dev. 1998, 12, 3843–3856. [Google Scholar] [CrossRef]
- Watanabe, Y. Shugoshin: Guardian spirit at the centromere. Curr. Opin. Cell Biol. 2005, 17, 590–595. [Google Scholar] [CrossRef]
- Watanabe, Y.; Kitajima, T.S. Shugoshin protects cohesin complexes at centromeres. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 515–521. [Google Scholar] [CrossRef]
- Watanabe, Y. Temporal and spatial regulation of targeting aurora B to the inner centromere. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 419–423. [Google Scholar] [CrossRef]
- Davis, B.K. A analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. Mol. Gen. Genet. 1971, 113, 251–272. [Google Scholar] [CrossRef] [PubMed]
- Kerrebrock, A.W.; Miyazaki, W.Y.; Birnby, D.; Orr-Weaver, T.L. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics 1992, 130, 827–841. [Google Scholar] [CrossRef]
- Kerrebrock, A.; Moore, D.; Wu, J.; Orr-Weaver, T. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell 1995, 83, 247–256. [Google Scholar] [CrossRef]
- Kitajima, T.S.; Kawashima, S.A.; Watanabe, Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 2004, 427, 510–517. [Google Scholar] [CrossRef]
- Katis, V.L.; Galova, M.; Rabitsch, K.P.; Gregan, J.; Nasmyth, K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004, 14, 560–572. [Google Scholar] [CrossRef] [PubMed]
- Marston, A.L.; Tham, W.H.; Shah, H.; Amon, A. A genome-wide screen identifies genes required for centromeric cohesion. Science 2004, 303, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Rabitsch, K.P.; Gregan, J.; Schleiffer, A.; Javerzat, J.P.; Eisenhaber, F.; Nasmyth, K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis-I and II. Curr. Biol. 2004, 14, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Hamant, O.; Golubovskaya, I.; Meeley, R.; Fiume, E.; Timofejeva, L.; Schleiffer, A.; Nasmyth, K.; Cande, W.Z. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 2005, 15, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Tang, D.; Wang, K.; Shen, Y.; Qin, B.; Miao, C.; Li, M.; Cheng, Z. OsSGO1 maintains synaptonemal complex stabilization in addition to protecting centromeric cohesion during rice meiosis. Plant J. 2011, 67, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Dai, W. Shugoshins function as a guardian for chromosomal stability in nuclear division. Cell Cycle 2012, 11, 2631–2642. [Google Scholar] [CrossRef]
- Llano, E.; Gómez, R.; Gutiérrez-Caballero, C.; Herrán, Y.; Sánchez-Martín, M.; Vázquez-Quiñones, L.E.; Hernández, T.; de Álava, E.; Cuadrado, A.; Barbero, J.; et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008, 22, 2400–2413. [Google Scholar] [CrossRef]
- de Carvalho, C.E.; Zaaijer, S.; Smolikov, S.; Gu, Y.; Schumacher, J.M.; Colaiácovo, M.P. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 2008, 22, 2869–2885. [Google Scholar] [CrossRef]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Kiburz, B.M.; Reynolds, D.B.; Megee, P.C.; Marston, A.L.; Lee, B.H.; Lee, T.I.; Levine, S.S.; Young, R.A.; Amon, A. The core centromere and Sgo1 establish a 50-kb cohesin-protected domain around centromeres during meiosis I. Genes Dev. 2005, 19, 3017–3030. [Google Scholar] [CrossRef][Green Version]
- Mishra, P.K.; Thapa, K.S.; Chen, P.; Wang, S.; Hazbun, T.R.; Basrai, M.A. Budding yeast CENP-ACse4 interacts with the N-terminus of Sgo1 and regulates its association with centromeric chromatin. Cell Cycle 2018, 17, 11–23. [Google Scholar] [CrossRef]
- Tashiro, S.; Handa, T.; Matsuda, A.; Ban, T.; Takigawa, T.; Miyasato, K.; Ishii, K.; Kugou, K.; Ohta, K.; Hiraoka, Y.; et al. Shugoshin forms a specialized chromatin domain at subtelomeres that regulates transcription and replication timing. Nat. Commun. 2016, 7, 10393. [Google Scholar] [CrossRef] [PubMed]
- Kanoh, J. Unexpected roles of a shugoshin protein at subtelomeres. Genes Genet. Syst. 2018, 92, 127–133. [Google Scholar] [CrossRef]
- McGuinness, B.E.; Hirota, T.; Kudo, N.R.; Peters, J.M.; Nasmyth, K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005, 3, e86. [Google Scholar] [CrossRef]
- Karamysheva, Z.; Diaz-Martinez, L.A.; Crow, S.E.; Li, B.; Yu, H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J. Biol. Chem. 2009, 284, 1772–1780. [Google Scholar] [CrossRef]
- Mu, J.; Fan, L.; Liu, D.; Zhu, D. Overexpression of shugoshin1 predicts a poor prognosis for prostate cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Onco Targets Ther. 2019, 12, 1111–1118. [Google Scholar] [CrossRef]
- Coon, S.W.; Savera, A.T.; Zarbo, R.J.; Benninger, M.S.; Chase, G.A.; Rybicki, B.A.; Van Dyke, D.L. Prognostic implications of loss of heterozygosity at 8p21 and 9p21 in head and neck squamous cell carcinoma. Int. J. Cancer 2004, 111, 206–212. [Google Scholar] [CrossRef]
- Shao, J.Y.; Wang, H.Y.; Huang, X.M.; Feng, Q.S.; Huang, P.; Feng, B.J.; Huang, L.X.; Yu, X.J.; Li, J.T.; Hu, L.F.; et al. Genome-wide allele type analysis of sporadic primary nasopharyngeal carcinoma from southern China. Int. J. Oncol. 2000, 17, 1267–1275. [Google Scholar]
- Altura, R.A.; Maris, J.M.; Li, H.; Boyett, J.M.; Brodeur, G.M.; Look, A.T. Novel regions of chromosomal loss in familial neuroblastoma by comparative genomic hybridization. Genes Chromosomes Cancer 1997, 19, 176–184. [Google Scholar] [CrossRef]
- Pallai, R.; Bhaskar, A.; Barnett-Bernodat, N.; Gallo-Ebert, C.; Nickels, J.T., Jr.; Rice, L.M. Cancerous inhibitor of protein phosphatase 2A promotes premature chromosome segregation and aneuploidy in prostate cancer cells through association with shugoshin. Tumor Biol. 2015, 36, 6067–6074. [Google Scholar] [CrossRef]
- Dahiya, R.; McCarville, J.; Hu, W.; Lee, C.; Chui, R.M.; Kaur, G.; Deng, G. Chromosome 3p24–26 and 3p22–12 loss in human prostatic adenocarcinoma. Int. J. Cancer 1997, 71, 20–25. [Google Scholar] [CrossRef]
- Beder, L.B.; Gunduz, M.; Ouchida, M.; Fukushima, K.; Gunduz, E.; Ito, S.; Sakai, A.; Nagai, N.; Nishizaki, K.; Shimizu, K. Genome-wide analyses on loss of heterozygosity in head and neck squamous cell carcinomas. Lab. Investig. 2003, 83, 99–105. [Google Scholar] [CrossRef]
- Kohno, T.; Morishita, K.; Takano, H.; Shapiro, D.N.; Yokota, J. Homozygous deletion at chromosome 2q33 in human small-cell lung carcinoma identified by arbitrarily primed PCR genomic fingerprinting. Oncogene 1994, 9, 103–108. [Google Scholar]
- Rader, J.S.; Kamarasova, T.; Huettner, P.C.; Li, L.; Li, Y.; Gerhard, D.S. Allelotyping of all chromosomal arms in invasive cervical cancer. Oncogene 1996, 13, 2737–2741. [Google Scholar]
- Takita, J.; Yang, H.W.; Chen, Y.Y.; Hanada, R.; Yamamoto, K.; Teitz, T.; Kidd, V.; Hayashi, Y. Allelic imbalance on chromosome 2q and alterations of the caspase 8 gene in neuroblastoma. Oncogene 2001, 20, 4424–4432. [Google Scholar] [CrossRef][Green Version]
- Iwaizumi, M.; Shinmura, K.; Mori, H.; Yamada, H.; Suzuki, M.; Kitayama, Y.; Igarashi, H.; Nakamura, T.; Suzuki, H.; Watanabe, Y.; et al. Human Sgo1 downregulation leads to chromosomal instability in colorectal cancer. Gut 2009, 58, 249–260. [Google Scholar] [CrossRef]
- Rao, C.V.; Sanghera, S.; Zhang, Y.; Biddick, L.; Reddy, A.; Lightfoot, S.; Janakiram, N.B.; Mohammed, A.; Dai, W.; Yamada, H.Y. Systemic Chromosome Instability Resulted in Colonic Transcriptomic Changes in Metabolic, Proliferation, and Stem Cell Regulators in Sgo1-/+ Mice. Cancer Res. 2016, 76, 630–642. [Google Scholar] [CrossRef]
- Rao, C.V.; Sanghera, S.; Zhang, Y.; Biddick, L.; Reddy, A.; Lightfoot, S.; Dai, W.; Yamada, H.Y. Antagonizing pathways leading to differential dynamics in colon carcinogenesis in Shugoshin1 (Sgo1)-haploinsufficient chromosome instability model. Mol. Carcinog. 2016, 55, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.Y.; Zhang, Y.; Reddy, A.; Mohammed, A.; Lightfoot, S.; Dai, W.; Rao, C.V. Tumor-promoting/progressing role of additional chromosome instability in hepatic carcinogenesis in Sgo1 (Shugoshin 1) haploinsufficient mice. Carcinogenesis 2015, 36, 429–440. [Google Scholar] [CrossRef]
- Yamada, H.Y.; Yao, Y.; Wang, X.; Zhang, Y.; Huang, Y.; Dai, W.; Rao, C.V. Haploinsufficiency of SGO1 results in deregulated centrosome dynamics, enhanced chromosomal instability and colon tumorigenesis. Cell Cycle 2012, 11, 479–488. [Google Scholar] [CrossRef]
- Kahyo, T.; Iwaizumi, M.; Shinmura, K.; Matsuura, S.; Nakamura, T.; Watanabe, Y.; Yamada, H.; Sugimura, H. A novel tumor-derived SGOL1 variant causes abnormal mitosis and unstable chromatid cohesion. Oncogene 2011, 30, 4453–4463. [Google Scholar] [CrossRef]
- Yamada, H.Y.; Kumar, G.; Zhang, Y.; Rubin, E.; Lightfoot, S.; Dai, W.; Rao, C.V. Systemic chromosome instability in Shugosh-in-1 mice resulted in compromised glutathione pathway, activation of Wnt signaling and defects in immune system in the lung. Oncogenesis 2016, 5, e256. [Google Scholar] [CrossRef]
- Wang, L.H.; Yen, C.J.; Li, T.N.; Elowe, S.; Wang, W.C.; Wang, L.H. Sgo1 is a potential therapeutic target for hepatocellular carcinoma. Oncotarget 2015, 6, 2023–2033. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ikezoe, T.; Nishioka, C.; Yokoyama, A. A novel treatment strategy targeting shugoshin 1 in hematological malignancies. Leuk. Res. 2013, 37, 76–82. [Google Scholar] [CrossRef]
- Scanlan, M.J.; Gout, I.; Gordon, C.M.; Williamson, B.; Stocker Et Gure, A.O.; Jager, D.; Chen, Y.T.; Mackay, A.; O’Hare, M.J.; Old, L.J. Humoral immunity to human breast cancer: Antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 2001, 1, 4. [Google Scholar] [PubMed]
- Yang, Q.; Yoshimura, G.; Nakamura, M.; Nakamura, Y.; Shan, L.; Suzuma, T.; Tamaki, T.; Umemura, T.; Mori, I.; Kakudo, K. Allelic loss of chromosome 3p24 correlates with tumor progression rather than with retinoic acid receptor beta2 expression in breast carcinoma. Breast Cancer Res. Treat. 2011, 70, 39–45. [Google Scholar] [CrossRef]
- Matsuura, S.; Kahyo, T.; Shinmura, K.; Iwaizumi, M.; Yamada, H.; Funai, K.; Kobayashi, J.; Tanahashi, M.; Niwa, H.; Ogawa, H.; et al. SGOL1 variant B induces abnormal mitosis and resistance to taxane in non-small cell lung cancers. Sci. Rep. 2013, 3, 3012. [Google Scholar] [CrossRef]
- Wong, W.K.; Kelly, T.; Li, J.; Ma, H.T.; Poon, R.Y. SGO1C is a non-functional isoform of Shugoshin and can disrupt sister chromatid cohesion by interacting with PP2A-B56. Cell Cycle 2015, 14, 3965–3977. [Google Scholar] [CrossRef]
- Chen, Q.; Wan, X.; Chen, Y.; Liu, C.; Gu, M.; Wang, Z. SGO1 induces proliferation and metastasis of prostate cancer through AKT-mediated signaling pathway. Am. J. Cancer Res. 2019, 9, 2693–2705. [Google Scholar] [PubMed]
- Ribeiro-Varandas, E.; Viegas, W.; Sofia Pereira, H.; Delgado, M. Bisphenol A at concentrations found in human serum induces aneugenic effects in endothelial cells. Mutat. Res. 2013, 751, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Domínguez-Ruiz, M.; García-Martínez, A.; Corral-Juan, M.; Pérez-Álvarez, Á.I.; Plasencia, A.M.; Villamar, M.; Moreno-Pelayo, M.A.; Matilla-Dueñas, A.; Menéndez-González, M.; Del Castillo, I. Perrault syndrome with neurological features in a compound heterozygote for two TWNK mutations: Overlap of TWNK-related recessive disorders. J. Transl. Med. 2019, 17, 290. [Google Scholar] [CrossRef]
- Faridi, R.; Rehman, A.U.; Morell, R.J.; Friedman, P.L.; Demain, L.; Zahra, S.; Khan, A.A.; Tohlob, D.; Assir, M.Z.; Beaman Khan, S.N.; et al. Mutations of SGO2 and CLDN14 collectively cause coincidental Perrault syndrome. Clin. Genet. 2017, 91, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Farooqui, M.; Zhang, Y.; Asch, A.S.; Yamada, H.Y. Spontaneous development of Alzheimer’s disease-associated brain pathology in a Shugoshin-1 mouse cohesinopathy model. Aging Cell 2018, 17, e12797. [Google Scholar] [CrossRef]
- Rao, C.V.; Farooqui, M.; Madhavaram, A.; Zhang, Y.; Asch, A.S.; Yamada, H.Y. GSK3-ARC/Arg3.1 and GSK3-Wnt signaling axes trigger amyloid-β accumulation and neuroinflammation in middle-aged Shugoshin 1 mice. Aging Cell 2020, 19, e13221. [Google Scholar] [CrossRef]
- Chetaille, P.; Preuss, C.; Burkhard, S.; Côté, J.-M.; Houde, C.; Castilloux, J.; Piché, J.; Gosset, N.; Leclerc, S.; Wünnemann, F.; et al. Mutations in SGOL1 cause a novel cohesinopathy affecting heart and gut rhythm. Nat. Genet. 2014, 46, 1245–1249. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dai, W. Shugoshin, a guardian for sister chromatid segregation. Exp. Cell Res. 2005, 310, 1–9. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).