The Hsp60 Protein of Helicobacter Pylori Exhibits Chaperone and ATPase Activities at Elevated Temperatures
Abstract
:1. Introduction
2. Materials and Methods
3. Results and Discussion
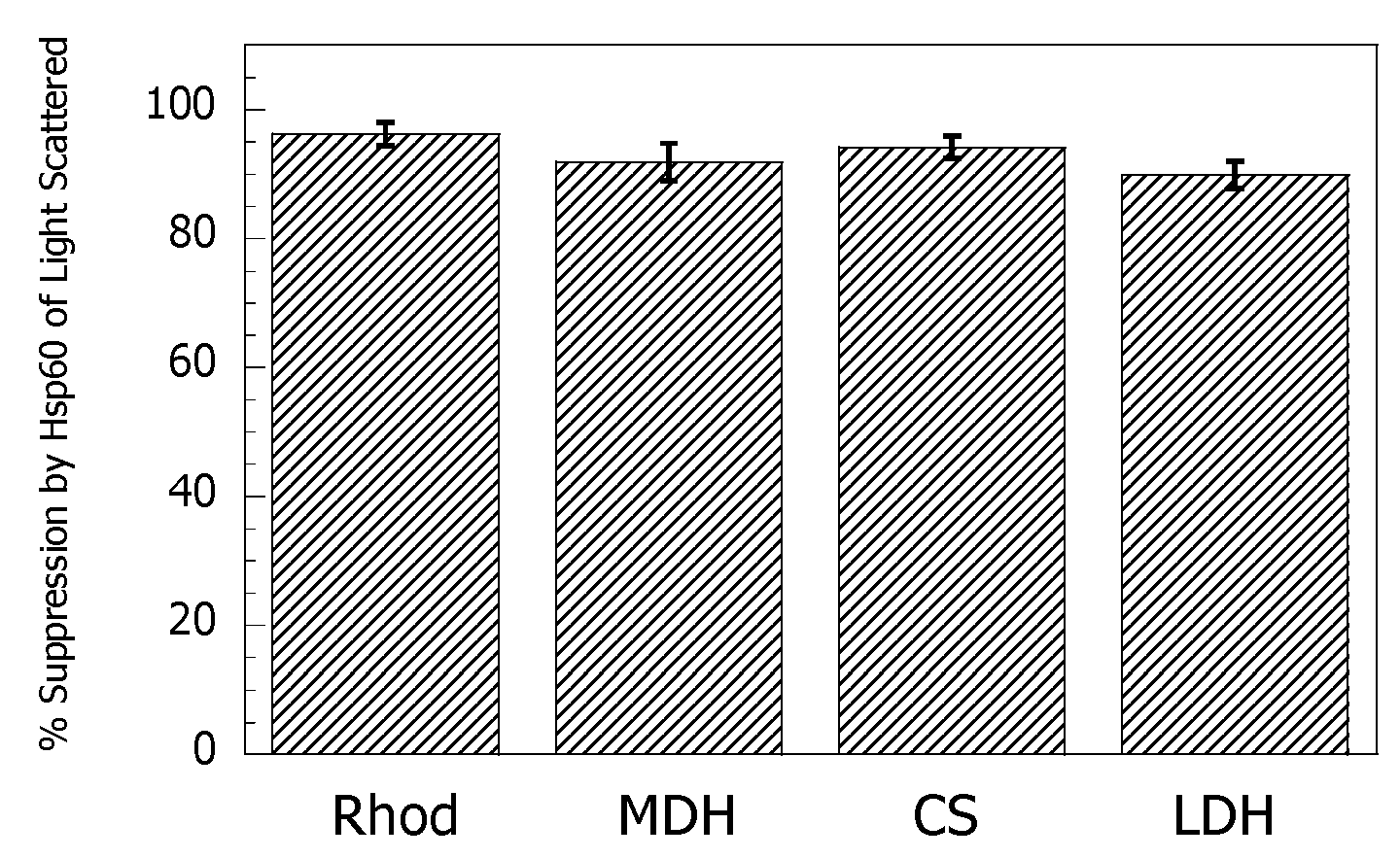
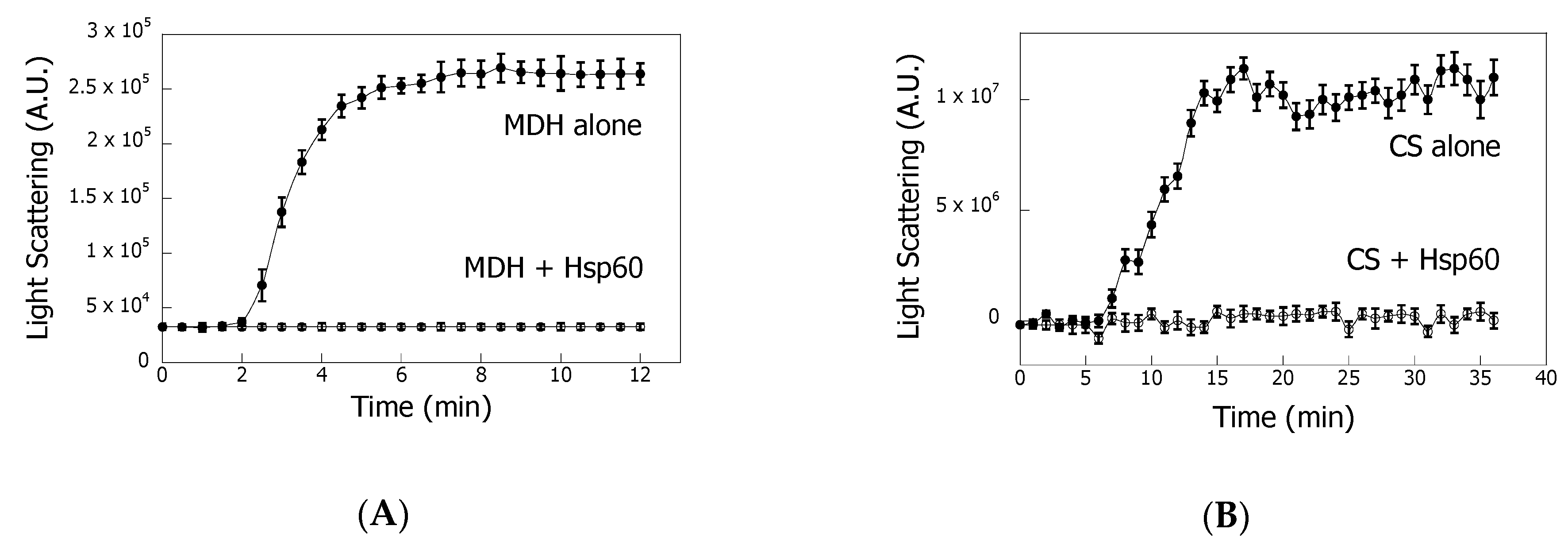
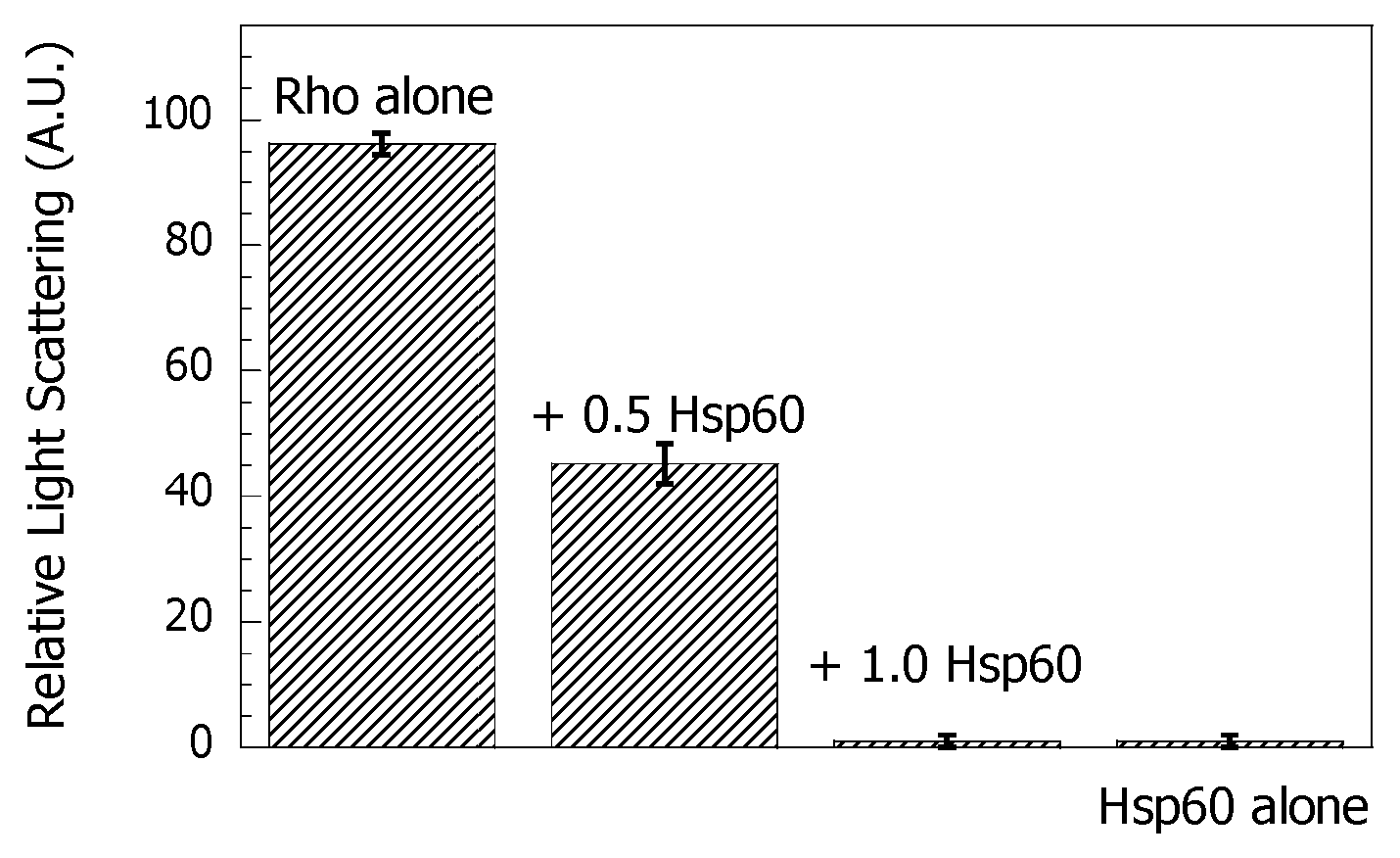
3.1. Hsp60 Reduces Protein Thermal Aggregation
3.2. ATPase Activity of Hsp60
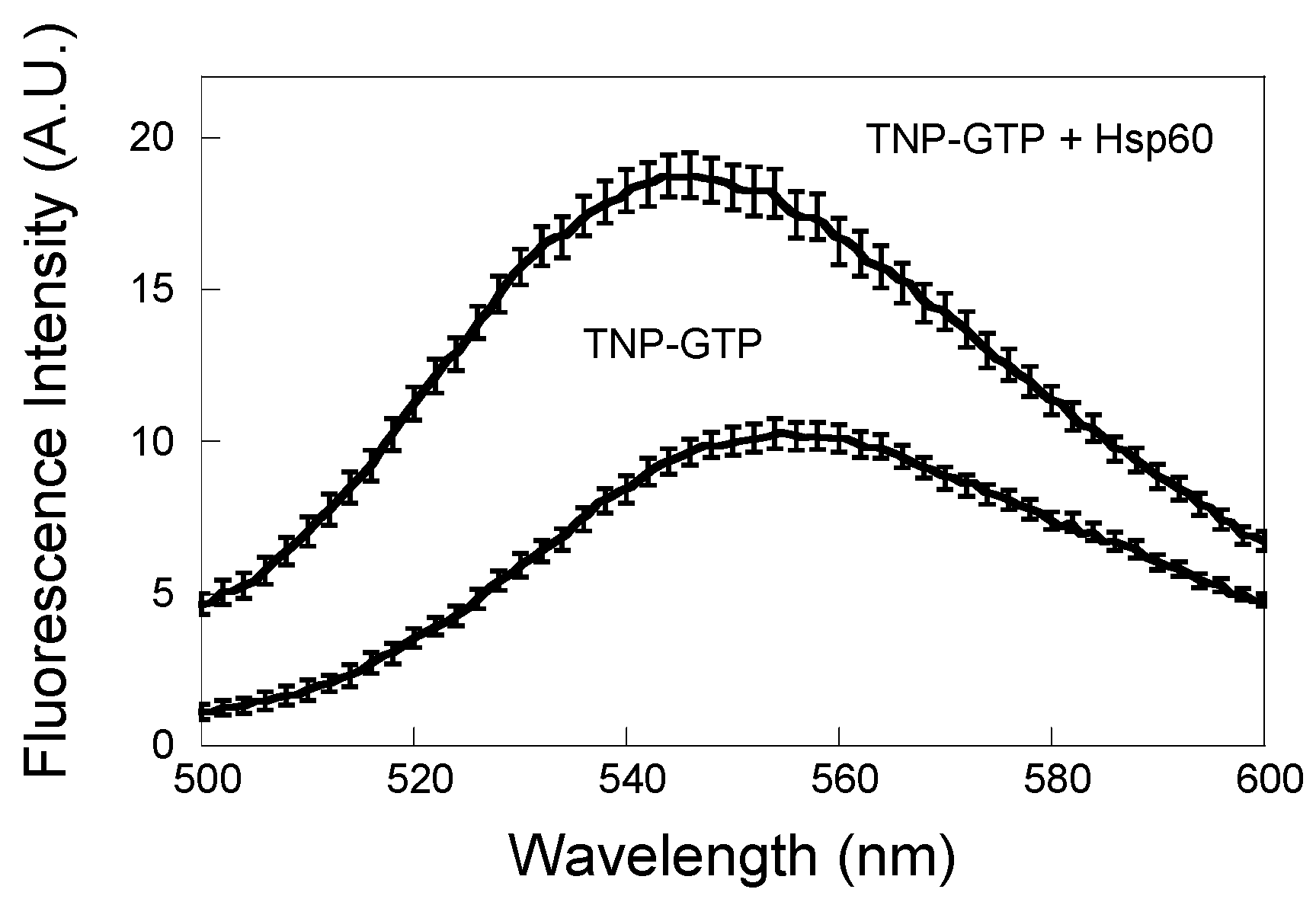
3.3. Binding of GTP to Hsp60
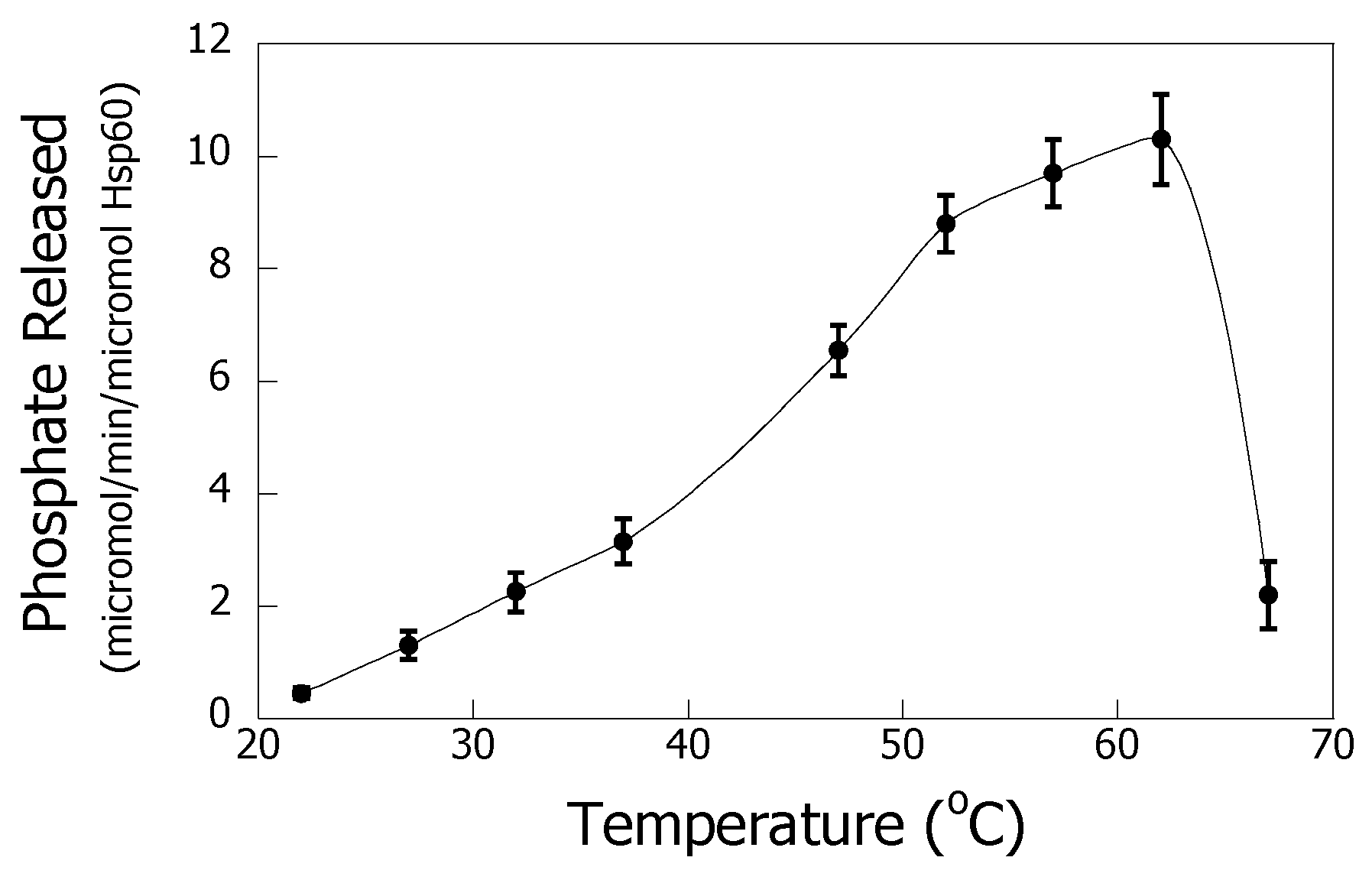
3.4. Temperature Dependence of the ATPase Activity of Hsp60
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Covacci, A.; Telford, G.; Del Giudice, J.; Parsonnet, J.; Rappuoli, R. Helicobacter pylori virulence and genetic geography. Science 1999, 284, 1328–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusters, J.G.; van Vliet, A.H.; Kuipers, E.J. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.C.; Isomoto, H.; Moriyama, M.; Fujioka, T.; Machado, J.C.; Yamaoka, Y. Helicobacter and Gastric Malignancies. Helicobacter 2008, 13, 28–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slonczewski, J.L.; McGee, D.J.; Phillips, J.; Kirkpatrick, C.; Mobley, H.L.T. pH-Dependent Protein Profiles of Helicobacter pylori Analyzed by Two-Dimensional Gels. Helicobacter 2000, 5, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Sachs, G.; Weeks, D.L.; Wen, Y.; Marcus, E.A.; Scott, D.R. Acid acclimation by Helicobacter pylori. Physiology 2005, 20, 429–438. [Google Scholar] [CrossRef]
- Jungblut, P.R.; Bumann, D.; Haas, G.; Zimny-Arndt, U.; Holland, P.; Lamer, S.; Siejak, F.; Aebischer, A.; Meyer, T.F. Comparative proteome analysis of Helicobacter pylori. Mol. Microbiol. 2000, 36, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Macchia, G.; Massone, A.; Burroni, D.; Covacci, A.; Censini, S.; Rappuoli, R. The Hsp60 protein of Helicobacter pylori: Structure and immune response in patients with gastroduodenal diseases. Mol. Microbiol. 1993, 9, 645–652. [Google Scholar] [CrossRef]
- Young, J.C.; Agashe, V.R.; Siegers, K.; Hart, F.U. Pathways of chaperone-mediated protein folding in the cytosol. Nature Reviews Mol. Cell Biol. 2004, 5, 781–791. [Google Scholar] [CrossRef]
- Vabulas, R.M.; Raychaudhuri, S.; Hayer-Hartl, M.; Hartl, F.U. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2010, 2, a004390. [Google Scholar] [CrossRef]
- Voth, W.; Jakob, U. Stress-activated chaperones: A first line of defense. Trends Biochem. Sci. 2017, 42, 899–913. [Google Scholar] [CrossRef]
- Muga, A.; Moro, F. Thermal Adaptation of Heat Shock Proteins. Curr. Protein Pept. Sci. 2008, 9, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Kumsta, C.; Jakob, U. Redox-Regulated Chaperones. Biochemistry 2009, 48, 4666–4676. [Google Scholar] [CrossRef] [Green Version]
- Hong, W.; Wu, Y.E.; Fu, X.; Chang, Z. Chaperone-dependent mechanisms for acid resistance in enteric bacteria. Trends Microbiol. 2012, 20, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Suerbaum, S.; Thiberge, J.M.; Kansau, I.; Ferrero, R.L.; Labigne, A. Helicobacter pylori hspA-hspB heat-shock gene cluster: Nucleotide sequence, expression, putative function and immunogenicity. Mol. Microbiol. 1994, 14, 959–974. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.A.; Weinberger, K.K.; Swan, M.J. The Hsp60 protein of helicobacter pylori displays chaperone activity under acidic conditions. Biochem. Biophys. Rep. 2017, 9, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, J.A.; Lorimer, G.H.; Horowitz, P.M. Chaperonin cpn60 from Escherichia coli protects the mitochondrial enzyme rhodanese against heat inactivation and supports folding at elevated temperatures. J. Biol. Chem. 1992, 267, 17631–17634. [Google Scholar] [CrossRef]
- Hartman, D.J.; Surin, B.P.; Dixon, N.E.; Hoogenraad, N.J.; Høj, P.B. Substoichiometric amounts of the molecular chaperones GroEL and GroES prevent thermal denaturation and aggregation of mammalian mitochondrial malate dehydrogenase in vitro. Proc. Natl. Acad. Sci. USA 1993, 90, 2276–2280. [Google Scholar] [CrossRef] [Green Version]
- Jakob, U.; Lilie, H.; Meyer, I.; Buchner, J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase: Implications for heat shock in vivo. J. Biol. Chem. 1995, 270, 7288–7294. [Google Scholar] [CrossRef] [Green Version]
- Delmas, F.; Pierre, F.; Coucheney, F.; Divies, C.; Guzzo, J. Biochemical and physiological studies of the small heat shock protein Lo18 from the lactic acid bacterium Oenococcus oeni. J. Mol. Microbiol. Biotechnol. 2001, 3, 601–610. [Google Scholar]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Brocchieri, L.; Karlin, S. Conservation among HSP60 sequences in relation to structure, function, and evolution. Protein Sci. 2000, 9, 476–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.Y.; Huang, Y.S.; Li, C.H.; Hsieh, Y.T.; Tsai, N.M.; He, P.J.; Hsu, W.T.; Yeh, Y.C.; Chiang, F.H.; Wu, M.S.; et al. Characterizing the polymeric status of Helicobacter pylori heat shock protein 60. Biochem. Biophys. Res. Commun. 2009, 388, 283–289. [Google Scholar] [CrossRef]
- Qamra, R.; Srinivas, V.; Mande, S.C. Mycobacterium tuberculosis GroEL homologues unusually exist as lower oligomers and retain the ability to suppress aggregation of substrate proteins. Mol. Biol. 2004, 342, 605–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braig, K.; Otwinowski, Z.; Hegde, R.; Boisvert, D.C.; Joachimiak, A.; Horwich, A.L.; Sigler, P.B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature 1994, 371, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Gray, T.E.; Fersht, A.R. Cooperativity in ATP hydrolysis by GroEL is increased by GroES. FEBS Lett. 1991, 292, 254–258. [Google Scholar]
- Todd, M.J.; Viitanen, P.V.; Lorimer, G.H. Hydrolysis of adenosine 5′-triphosphate by Escherichia coli GroEL: Effects of GroES and potassium ion. Biochemistry 1993, 32, 8560–8567. [Google Scholar] [CrossRef]
- Hiratsuka, T. Chromophoric and fluorescent analog of GTP, 2′,3′-O-(2,4,6-trinitrocyclohexadienylidene)-GTP, as a spectroscopic probe for the GTP inhibitory site of liver glutamate dehydrogenase. J. Biol. Chem. 1985, 260, 4784–4790. [Google Scholar] [CrossRef]
- Hiratsuka, T. Fluorescent and colored trinitrophenylated analogs of ATP and GTP. Eur. J. Biochem. 2003, 270, 3479–3485. [Google Scholar] [CrossRef] [Green Version]
- Mendoza, J.A.; Warren, T.; Dulin, P. The ATPase activity of chaperonin GroEL is highly stimulated at elevated temperatures. Biochem. Biophys. Res. Commun. 1996, 229, 271–274. [Google Scholar] [CrossRef]
- Melkani, G.C.; Zardeneta, G.; Mendoza, J.A. The ATPase activity of GroEL is supported at high temperatures by divalent cations that stabilize its structure. Biometals 2003, 16, 479–484. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendoza, J.A.; Ignacio, J.L.; Buckley, C.M. The Hsp60 Protein of Helicobacter Pylori Exhibits Chaperone and ATPase Activities at Elevated Temperatures. BioChem 2021, 1, 19-25. https://doi.org/10.3390/biochem1010002
Mendoza JA, Ignacio JL, Buckley CM. The Hsp60 Protein of Helicobacter Pylori Exhibits Chaperone and ATPase Activities at Elevated Temperatures. BioChem. 2021; 1(1):19-25. https://doi.org/10.3390/biochem1010002
Chicago/Turabian StyleMendoza, Jose A., Julian L. Ignacio, and Christopher M. Buckley. 2021. "The Hsp60 Protein of Helicobacter Pylori Exhibits Chaperone and ATPase Activities at Elevated Temperatures" BioChem 1, no. 1: 19-25. https://doi.org/10.3390/biochem1010002
APA StyleMendoza, J. A., Ignacio, J. L., & Buckley, C. M. (2021). The Hsp60 Protein of Helicobacter Pylori Exhibits Chaperone and ATPase Activities at Elevated Temperatures. BioChem, 1(1), 19-25. https://doi.org/10.3390/biochem1010002






