Isoform-Specific Roles and Therapeutic Targeting of RUNX1 in Hematopoiesis and Leukemogenesis
Abstract
1. Introduction
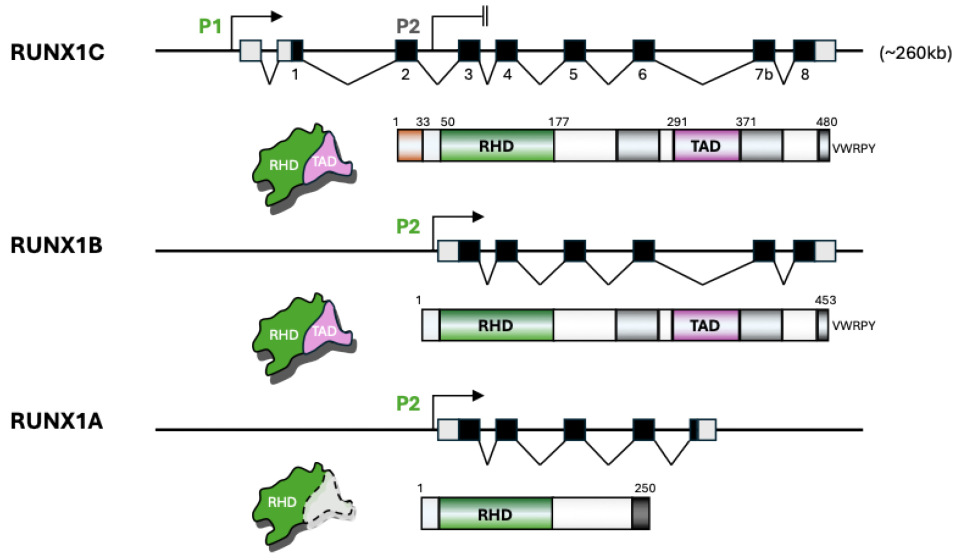
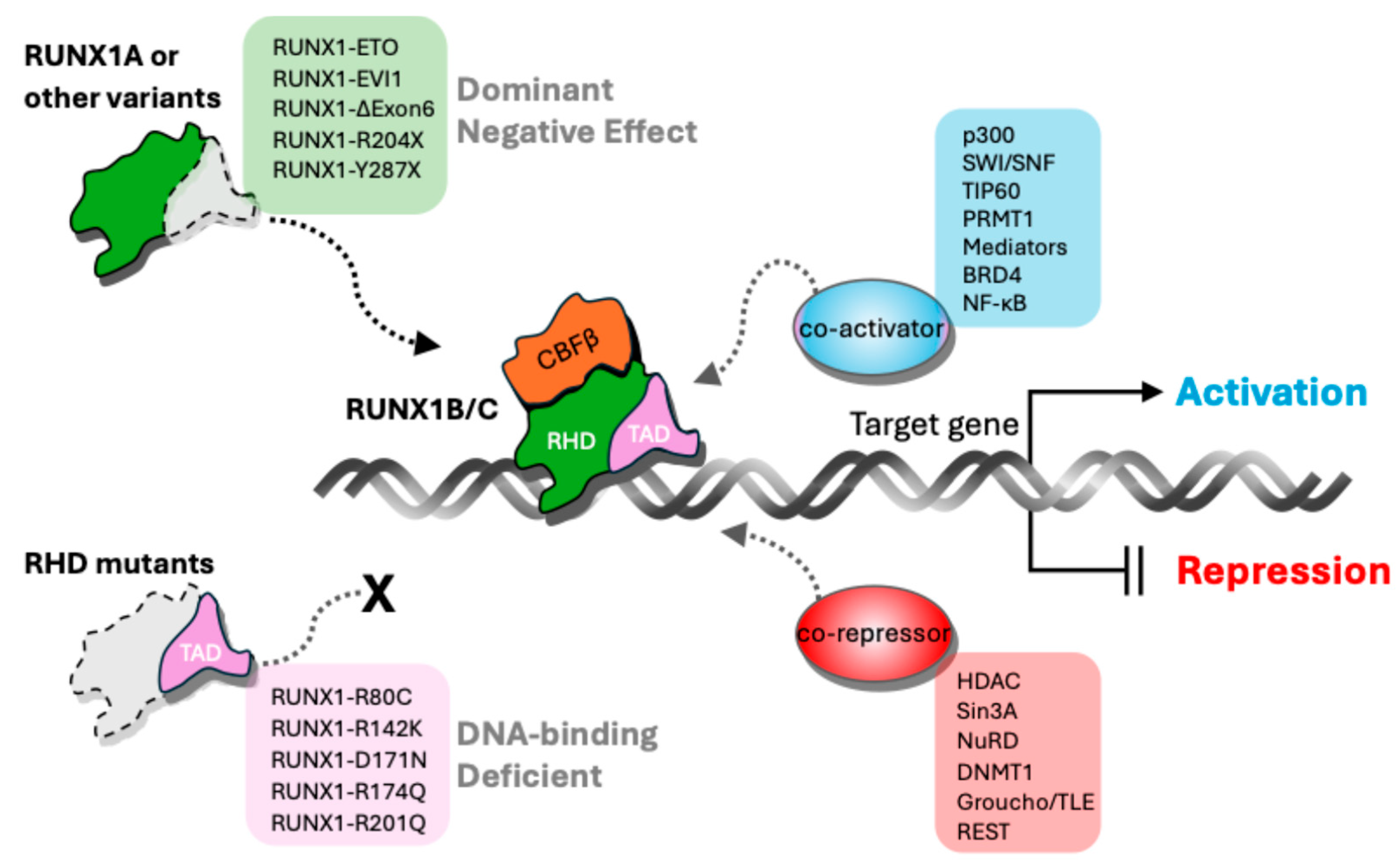
2. Distinctive Features of RUNX1 Isoforms and Variants
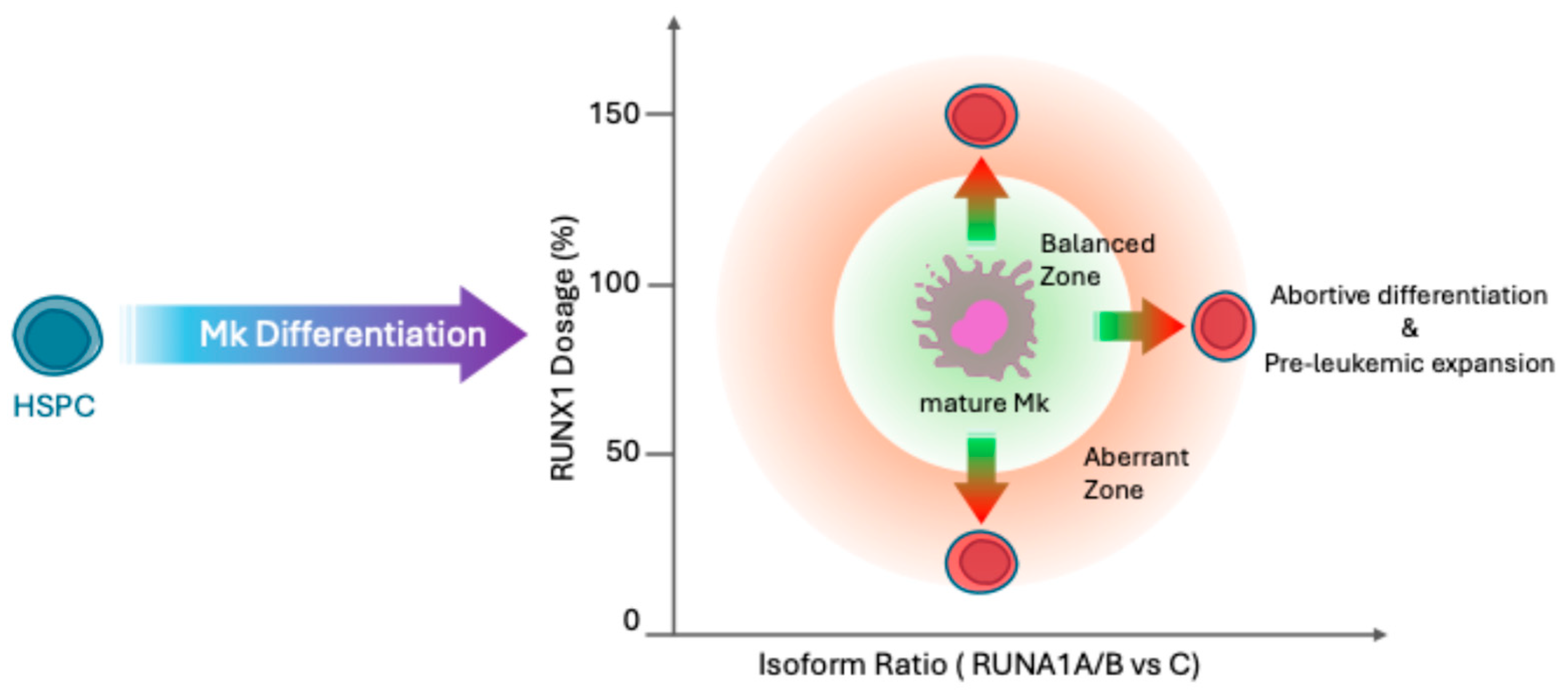
3. Isoform-Specific Roles of RUNX1 in Hematopoiesis and Leukemogenesis
4. Multilayered Regulation of RUNX1 Isoform Expression
5. Therapeutic Strategies Targeting RUNX1 Isoform Regulation
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ichikawa, M.; Asai, T.; Chiba, S.; Kurokawa, M.; Ogawa, S. Runx1/AML-1 ranks as a master regulator of adult hematopoiesis. Cell Cycle 2004, 3, 720–722. [Google Scholar] [CrossRef]
- Bonifer, C.; Levantini, E.; Kouskoff, V.; Lacaud, G. Runx1 Structure and Function in Blood Cell Development. Adv. Exp. Med. Biol. 2017, 962, 65–81. [Google Scholar]
- Sood, R.; Kamikubo, Y.; Liu, P. Role of RUNX1 in hematological malignancies. Blood 2017, 129, 2070–2082. [Google Scholar] [PubMed]
- Okuda, T.; van Deursen, J.; Hiebert, S.W.; Grosveld, G.; Downing, J.R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 1996, 84, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Yokomizo, T.; Zeigler, B.M.; Dzierzak, E.; Speck, N.A. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 2009, 457, 887–891. [Google Scholar] [CrossRef]
- Kuo, M.C.; Liang, D.C.; Huang, C.F.; Shih, Y.S.; Wu, J.H.; Lin, T.L.; Shih, L.Y. RUNX1 mutations are frequent in chronic myelomonocytic leukemia and mutations at the C-terminal region might predict acute myeloid leukemia transformation. Leukemia 2009, 23, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Lam, K.; Zhang, D.E. RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front. Biosci. (Landmark Ed.) 2012, 17, 1120–1139. [Google Scholar] [CrossRef]
- Yokota, A.; Huo, L.; Lan, F.; Wu, J.; Huang, G. The Clinical, Molecular, and Mechanistic Basis of RUNX1 Mutations Identified in Hematological Malignancies. Mol. Cells 2020, 43, 145–152. [Google Scholar]
- Hayashi, Y.; Harada, Y.; Harada, H. Myeloid neoplasms and clonal hematopoiesis from the RUNX1 perspective. Leukemia 2022, 36, 1203–1214. [Google Scholar] [CrossRef]
- Sroczynska, P.; Lancrin, C.; Kouskoff, V.; Lacaud, G. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 2009, 114, 5279–5289. [Google Scholar] [CrossRef]
- Challen, G.A.; Goodell, M.A. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp. Hematol. 2010, 38, 403–416. [Google Scholar] [CrossRef]
- Menegatti, S.; Potts, B.; Garcia-Alegria, E.; Paredes, R.; Lie-A.-Ling, M.; Lacaud, G.; Kouskoff, V. The RUNX1b Isoform Defines Hemogenic Competency in Developing Human Endothelial Cells. Front. Cell Dev. Biol. 2021, 9, 812639. [Google Scholar] [CrossRef]
- Guan, L.; Voora, D.; Myers, R.; Del Carpio-Cano, F.; Rao, A.K. RUNX1 isoforms regulate RUNX1 and target genes differentially in platelets-megakaryocytes: Association with clinical cardiovascular events. J. Thromb. Haemost. 2024, 22, 3581–3598. [Google Scholar] [CrossRef]
- Brady, G.; Elgueta Karstegl, C.; Farrell, P.J. Novel function of the unique N-terminal region of RUNX1c in B cell growth regulation. Nucleic Acids Res. 2013, 41, 1555–1568. [Google Scholar] [CrossRef]
- Komeno, Y.; Yan, M.; Matsuura, S.; Lam, K.; Lo, M.C.; Huang, Y.J.; Tenen, D.G.; Downing, J.R.; Zhang, D.E. Runx1 exon 6-related alternative splicing isoforms differentially regulate hematopoiesis in mice. Blood 2014, 123, 3760–3769. [Google Scholar] [CrossRef] [PubMed]
- Draper, J.E.; Sroczynska, P.; Tsoulaki, O.; Leong, H.S.; Fadlullah, M.Z.; Miller, C.; Kouskoff, V.; Lacaud, G. RUNX1B Expression Is Highly Heterogeneous and Distinguishes Megakaryocytic and Erythroid Lineage Fate in Adult Mouse Hematopoiesis. PLoS Genet. 2016, 12, e1005814. [Google Scholar]
- Davis, A.G.; Einstein, J.M.; Zheng, D.; Jayne, N.D.; Fu, X.D.; Tian, B.; Yeo, G.W.; Zhang, D.E. A CRISPR RNA-binding protein screen reveals regulators of RUNX1 isoform generation. Blood Adv. 2021, 5, 1310–1323. [Google Scholar] [CrossRef] [PubMed]
- Jayne, N.D.; Liang, Z.; Lim, D.H.; Chen, P.B.; Diaz, C.; Arimoto, K.I.; Xia, L.; Liu, M.; Ren, B.; Fu, X.D.; et al. RUNX1 C-terminal mutations impair blood cell differentiation by perturbing specific enhancer-promoter networks. Blood Adv. 2024, 8, 2410–2423. [Google Scholar] [CrossRef]
- Neldeborg, S.; Soerensen, J.F.; Møller, C.T.; Bill, M.; Gao, Z.; Bak, R.O.; Holm, K.; Sorensen, B.; Nyegaard, M.; Luo, Y.; et al. Dual intron-targeted CRISPR-Cas9-mediated disruption of the AML RUNX1-RUNX1T1 fusion gene effectively inhibits proliferation and decreases tumor volume in vitro and in vivo. Leukemia 2023, 37, 1792–1801. [Google Scholar] [CrossRef]
- Khan, N.M.; Wilderman, A.; Kaiser, J.M.; Kamalakar, A.; Goudy, S.L.; Cotney, J.; Drissi, H. Enhanced osteogenic potential of iPSC-derived mesenchymal progenitor cells following genome editing of GWAS variants in the RUNX1 gene. Bone Res. 2024, 12, 70. [Google Scholar] [CrossRef]
- Grinev, V.V.; Barneh, F.; Ilshonak, I.M.; Nakjang, S.; Smink, J.; van Oort, A.; Clough, R.; Seyani, M.; McNeill, H.; Reza, M.; et al. RUNX1/RUNX1T1 mediates alternative splicing and reorganises the transcriptional landscape in leukemia. Nat. Commun. 2021, 12, 520. [Google Scholar] [CrossRef]
- Gialesaki, S.; Bräuer-Hartmann, D.; Issa, H.; Bhayadia, R.; Alejo-Valle, O.; Verboon, L.; Schmell, A.L.; Laszig, S.; Regényi, E.; Schuschel, K.; et al. RUNX1 isoform disequilibrium promotes the development of trisomy 21-associated myeloid leukemia. Blood 2023, 141, 1105–1118. [Google Scholar] [CrossRef]
- Decker, M.; Lammens, T.; Ferster, A.; Erlacher, M.; Yoshimi, A.; Niemeyer, C.M.; Ernst, M.P.T.; Raaijmakers, M.H.G.P.; Duployez, N.; Flaum, A.; et al. Functional classification of RUNX1 variants in familial platelet disorder with associated myeloid malignancies. Leukemia 2021, 35, 3304–3308. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Deuitch, N.; Merguerian, M.; Cunningham, L.; Davis, J.; Bresciani, E.; Diemer, J.; Andrews, E.; Young, A.; Donovan, F.; et al. Genomic landscape of patients with germline RUNX1 variants and familial platelet disorder with myeloid malignancy. Blood Adv. 2024, 8, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Levanon, D.; Glusman, G.; Bangsow, T.; Ben-Asher, E.; Male, D.A.; Avidan, N.; Bangsow, C.; Hattori, M.; Taylor, T.D.; Taudien, S.; et al. Architecture and anatomy of the genomic locus encoding the human leukemia-associated transcription factor RUNX1/AML1. Gene 2001, 262, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.; Hinojosa, M.; Trombly, D.; Morin, V.; Stein, J.; Stein, G.; Javed, A.; Gutierrez, S.E. Transcriptional Auto-Regulation of RUNX1 P1 Promoter. PLoS ONE 2016, 11, e0149119. [Google Scholar] [CrossRef]
- Riddell, A.; McBride, M.; Braun, T.; Nicklin, S.A.; Cameron, E.; Loughrey, C.M.; Martin, T.P. RUNX1: An emerging therapeutic target for cardiovascular disease. Cardiovasc. Res. 2020, 116, 1410–1423. [Google Scholar] [CrossRef]
- Owens, D.D.G.; Anselmi, G.; Oudelaar, A.M.; Downes, D.J.; Cavallo, A.; Harman, J.R.; Schwessinger, R.; Bucakci, A.; Greder, L.; de Ornellas, S.; et al. Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development. Nat. Commun. 2022, 13, 773. [Google Scholar] [CrossRef]
- Miyoshi, H.; Ohira, M.; Shimizu, K.; Mitani, K.; Hirai, H.; Imai, T.; Yokoyama, K.; Soeda, E.; Ohki, M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995, 23, 2762–2769. [Google Scholar] [CrossRef] [PubMed]
- Barutcu, A.R.; Hong, D.; Lajoie, B.R.; McCord, R.P.; van Wijnen, A.J.; Lian, J.B.; Stein, J.L.; Dekker, J.; Imbalzano, A.N.; Stein, G.S. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochim. Biophys. Acta 2016, 1859, 1389–1397. [Google Scholar] [CrossRef]
- Imai, Y.; Kurokawa, M.; Tanaka, K.; Friedman, A.D.; Ogawa, S.; Mitani, K.; Yazaki, Y.; Hirai, H. TLE, the human homolog of Groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem. Biophys. Res. Commun. 1998, 252, 582–589. [Google Scholar] [CrossRef]
- Kellaway, S.G.; Keane, P.; Edginton-White, B.; Regha, K.; Kennett, E.; Bonifer, C. Different mutant RUNX1 oncoproteins program alternate haematopoietic differentiation trajectories. Life Sci. Alliance 2021, 4, e202000864. [Google Scholar] [CrossRef] [PubMed]
- Lutterbach, B.; Hiebert, S.W. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene 2000, 245, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Yzaguirre, A.D.; de Bruijn, M.F.; Speck, N.A. The Role of Runx1 in Embryonic Blood Cell Formation. Adv. Exp. Med. Biol. 2017, 962, 47–64. [Google Scholar]
- Morino-Koga, S.; Yokomizo, T. Deciphering hematopoietic stem cell development: Key signaling pathways and mechanisms. Front. Cell Dev. Biol. 2024, 12, 1510198. [Google Scholar] [CrossRef]
- Draper, J.E.; Sroczynska, P.; Leong, H.S.; Fadlullah, M.Z.H.; Miller, C.; Kouskoff, V.; Lacaud, G. Mouse RUNX1C regulates premegakaryocytic/erythroid output and maintains survival of megakaryocyte progenitors. Blood 2017, 130, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Estevez, B.; Borst, S.; Jarocha, D.; Sudunagunta, V.; Gonzalez, M.; Garifallou, J.; Hakonarson, H.; Gao, P.; Tan, K.; Liu, P.; et al. RUNX-1 haploinsufficiency causes a marked deficiency of megakaryocyte-biased hematopoietic progenitor cells. Blood 2021, 137, 2662–2675. [Google Scholar] [CrossRef]
- Lee, B.C.; Zhou, Y.; Bresciani, E.; Ozkaya, N.; Dulau-Florea, A.; Carrington, B.; Shin, T.H.; Baena, V.; Syed, Z.A.; Hong, S.G.; et al. A RUNX1-FPDMM rhesus macaque model reproduces the human phenotype and predicts challenges to curative gene therapies. Blood 2023, 141, 231–237. [Google Scholar] [CrossRef]
- Sakurai, H.; Harada, Y.; Ogata, Y.; Kagiyama, Y.; Shingai, N.; Doki, N.; Ohashi, K.; Kitamura, T.; Komatsu, N.; Harada, H. Overexpression of RUNX1 short isoform has an important role in the development of myelodysplastic/myeloproliferative neoplasms. Blood Adv. 2017, 1, 1382–1386. [Google Scholar] [CrossRef][Green Version]
- Bender, A.; Boydere, F.; Jayavelu, A.K.; Tibello, A.; König, T.; Aleth, H.; Meyer Zu Hörste, G.; Vogl, T.; Rosenbauer, F. Redistribution of PU.1 partner transcription factor RUNX1 binding secures cell survival during leukemogenesis. EMBO J. 2024, 43, 6291–6309. [Google Scholar] [CrossRef]
- Wilkinson, A.; Ballabio, E.; Geng, H.; North, P.; Tapia, M.; Kerry, J.; Biswas, D.; Roeder, R.; Allis, C.; Melnick, A.; et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013, 3, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Ahn, H.S.; Estevez, B.; Poncz, M. RUNX1-deficient human megakaryocytes demonstrate thrombopoietic and platelet half-life and functional defects. Blood 2023, 141, 260–270. [Google Scholar] [CrossRef]
- Liu, S.; Yang, J.; Sun, G.; Zhang, Y.; Cheng, C.; Xu, J.; Yen, K.; Lu, T. RUNX1 Upregulates CENPE to Promote Leukemic Cell Proliferation. Front. Mol. Biosci. 2021, 8, 692880. [Google Scholar] [CrossRef] [PubMed]
- Bellissimo, D.; Chen, C.; Zhu, Q.; Bagga, S.; Lee, C.; He, B.; Wertheim, G.; Jordan, M.; Tan, K.; Worthen, G.; et al. Runx1 negatively regulates inflammatory cytokine production by neutrophils in response to Toll-like receptor signaling. Blood Adv. 2020, 4, 1145–1158. [Google Scholar] [CrossRef]
- Zezulin, A.; Yen, D.; Ye, D.; Howell, E.; Bresciani, E.; Diemer, J.; Ren, J.; Ahmad, M.; Castilla, L.; Touw, I.; et al. RUNX1 is required in granulocyte-monocyte progenitors to attenuate inflammatory cytokine production by neutrophils. Genes. Dev. 2023, 37, 605–620. [Google Scholar] [CrossRef]
- Mohammadhosseini, M.; Enright, T.; Duvall, A.; Chitsazan, A.; Lin, H.; Ors, A.; Davis, B.; Nikolova, O.; Bresciani, E.; Diemer, J.; et al. Targeting the CD74 signaling axis suppresses inflammation and rescues defective hematopoiesis in RUNX1-familial platelet disorder. Sci. Transl. Med. 2025, 17, eadn9832. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Gao, L.; Teng, L.; Ge, J.; Oo, Z.; Kumar, A.; Gilliland, D.; Mason, P.; Tan, K.; Speck, N. Runx1 Deficiency Decreases Ribosome Biogenesis and Confers Stress Resistance to Hematopoietic Stem and Progenitor Cells. Cell Stem Cell 2015, 17, 165–177. [Google Scholar] [CrossRef]
- Bee, T.; Liddiard, K.; Swiers, G.; Bickley, S.R.; Vink, C.S.; Jarratt, A.; Hughes, J.R.; Medvinsky, A.; de Bruijn, M.F. Alternative Runx1 promoter usage in mouse developmental hematopoiesis. Blood Cells Mol. Dis. 2009, 43, 35–42. [Google Scholar] [CrossRef]
- Bee, T.; Swiers, G.; Muroi, S.; Pozner, A.; Nottingham, W.; Santos, A.C.; Li, P.S.; Taniuchi, I.; de Bruijn, M.F. Nonredundant roles for Runx1 alternative promoters reflect their activity at discrete stages of developmental hematopoiesis. Blood 2010, 115, 3042–3050. [Google Scholar] [CrossRef]
- Ferrell, P.I.; Xi, J.; Ma, C.; Adlakha, M.; Kaufman, D.S. The RUNX1 +24 enhancer and P1 promoter identify a unique subpopulation of hematopoietic progenitor cells derived from human pluripotent stem cells. Stem Cells 2015, 33, 1130–1141. [Google Scholar] [CrossRef][Green Version]
- Navarro-Montero, O.; Ayllon, V.; Lamolda, M.; López-Onieva, L.; Montes, R.; Bueno, C.; Ng, E.; Guerrero-Carreno, X.; Romero, T.; Romero-Moya, D.; et al. RUNX1c Regulates Hematopoietic Differentiation of Human Pluripotent Stem Cells Possibly in Cooperation with Proinflammatory Signaling. Stem Cells 2017, 35, 2253–2266. [Google Scholar] [CrossRef]
- Guo, H.; Ma, O.; Speck, N.A.; Friedman, A.D. Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood 2012, 119, 4408–4418. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Guo, H.; Friedman, A.D. The +37 kb Cebpa Enhancer Is Critical for Cebpa Myeloid Gene Expression and Contains Functional Sites that Bind SCL, GATA2, C/EBPα, PU.1, and Additional Ets Factors. PLoS ONE 2015, 10, e0126385. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohmori, S.; Moriguchi, T.; Noguchi, Y.; Ikeda, M.; Kobayashi, K.; Tomaru, N.; Ishijima, Y.; Ohneda, O.; Yamamoto, M.; Ohneda, K. GATA2 is critical for the maintenance of cellular identity in differentiated mast cells derived from mouse bone marrow. Blood 2015, 125, 3306–3315. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Zhou, L.; Wang, H.; Chen, N.; Jia, L.; Wang, C.; Wang, Y.; Chen, J.; Wen, X.; Niu, C.; et al. Profiling the epigenetic interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer cells by gene in situ cis-activation. Am. J. Cancer Res. 2019, 9, 1635–1649. [Google Scholar]
- Webber, B.R.; Iacovino, M.; Choi, S.H.; Tolar, J.; Kyba, M.; Blazar, B.R. DNA methylation of Runx1 regulatory regions correlates with transition from primitive to definitive hematopoietic potential in vitro and in vivo. Blood 2013, 122, 2978–2986. [Google Scholar] [CrossRef]
- Thomas, A.L.; Marsman, J.; Antony, J.; Schierding, W.; O’Sullivan, J.M.; Horsfield, J.A. Transcriptional Regulation of RUNX1: An Informatics Analysis. Genes 2021, 12, 1175. [Google Scholar] [CrossRef]
- Yi, H.; He, Y.; Zhu, Q.; Fang, L. RUNX Proteins as Epigenetic Modulators in Cancer. Cells 2022, 11, 3687. [Google Scholar] [CrossRef]
- Phillips-Cremins, J.E.; Sauria, M.E.; Sanyal, A.; Gerasimova, T.I.; Lajoie, B.R.; Bell, J.S.; Ong, C.T.; Hookway, T.A.; Guo, C.; Sun, Y.; et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 2013, 153, 1281–1295. [Google Scholar] [CrossRef]
- Pugacheva, E.M.; Kubo, N.; Loukinov, D.; Tajmul, M.; Kang, S.; Kovalchuk, A.L.; Strunnikov, A.V.; Zentner, G.E.; Ren, B.; Lobanenkov, V.V. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. USA 2020, 117, 2020–2031. [Google Scholar] [CrossRef]
- Davidson, I.F.; Barth, R.; Zaczek, M.; van der Torre, J.; Tang, W.; Nagasaka, K.; Janissen, R.; Kerssemakers, J.; Wutz, G.; Dekker, C.; et al. CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nature 2023, 616, 822–827. [Google Scholar] [CrossRef]
- Mercatante, D.R.; Mohler, J.L.; Kole, R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem. 2002, 277, 49374–49382. [Google Scholar] [CrossRef]
- Havens, M.A.; Hastings, M.L. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res. 2016, 44, 6549–6563. [Google Scholar] [CrossRef]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Tijssen, M.R.; Cvejic, A.; Joshi, A.; Hannah, R.L.; Ferreira, R.; Forrai, A.; Bellissimo, D.C.; Oram, S.H.; Smethurst, P.A.; Wilson, N.K.; et al. Genome-wide analysis of simultaneous GATA1/2, RUNX1, FLI1, and SCL binding in megakaryocytes identifies hematopoietic regulators. Dev. Cell 2011, 20, 597–609. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, F.; Barthélémy, A.; Peyrouze, P.; Fenwarth, L.; Preudhomme, C.; Duployez, N.; Cheok, M. Targeting RUNX1 in acute myeloid leukemia: Preclinical innovations and therapeutic implications. Expert. Opin. Ther. Targets 2021, 4, 299–309. [Google Scholar] [CrossRef]
- Mill, C.; Fiskus, W.; DiNardo, C.; Qian, Y.; Raina, K.; Rajapakshe, K.; Perera, D.; Coarfa, C.; Kadia, T.; Khoury, J.; et al. RUNX1-targeted therapy for AML expressing somatic or germline mutation in RUNX1. Blood 2019, 134, 59–73. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Ottis, P.; Crews, C.M. Proteolysis-Targeting Chimeras: Induced Protein Degradation as a Therapeutic Strategy. ACS Chem. Biol. 2017, 12, 892–898. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, K. Isoform-Specific Roles and Therapeutic Targeting of RUNX1 in Hematopoiesis and Leukemogenesis. Hemato 2025, 6, 33. https://doi.org/10.3390/hemato6030033
Kim S, Lee K. Isoform-Specific Roles and Therapeutic Targeting of RUNX1 in Hematopoiesis and Leukemogenesis. Hemato. 2025; 6(3):33. https://doi.org/10.3390/hemato6030033
Chicago/Turabian StyleKim, Seungjun, and Kiwon Lee. 2025. "Isoform-Specific Roles and Therapeutic Targeting of RUNX1 in Hematopoiesis and Leukemogenesis" Hemato 6, no. 3: 33. https://doi.org/10.3390/hemato6030033
APA StyleKim, S., & Lee, K. (2025). Isoform-Specific Roles and Therapeutic Targeting of RUNX1 in Hematopoiesis and Leukemogenesis. Hemato, 6(3), 33. https://doi.org/10.3390/hemato6030033





