Abstract
Myeloid leukemias are a broad group of hematological disorders, characterized by heterogeneous clinical and biological features. In recent years, unprecedented genetic discoveries and clinical–biological correlations have revolutionized the field of myeloid leukemias. The most relevant changes have specifically occurred in acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), chronic myeloid leukemia (CML) and myeloid neoplasms (MNs) with eosinophilia. The recently published International Consensus Classification (ICC) of myeloid neoplasms has addressed these changes, providing an updated framework and revised diagnostic criteria for such entities. This is also the aim of the 5th edition of the WHO classification of hematopoietic tumors, whose preliminary version was published in 2022. Parallel to this, new therapeutic options and novel molecular targets have changed the management of many myeloid entities, including AML and CML. This review aims to address the most relevant updates in the classification and diagnosis of AML, CMML, CML and MNs with eosinophilia. The state of the art of treatment and future therapeutic options for such disorders are also discussed.
1. Introduction
Leukemias are a heterogeneous group of hematologic neoplasms, characterized by clones of circulating myeloid or lymphoid cells. The first description of leukemias as distinct disease entities dates back to the work of J.H. Bennett (1812–1875), R. Virchow (1821–1902) and A.F. Donné (1801–1878) in the mid 1840s [1]. Early classification attempts distinguished two forms of leukemia, namely splenic and lymphatic. The former corresponded to chronic myeloid leukemia (CML), the latter to chronic lymphocytic leukemia/small lymphocytic lymphoma [2]. In 1895, more aggressive disease variants were described and referred to as “acute leukemia” by A. Fränkel (1848–1916) [3]. Finally, in 1913, V. Schilling-Torgau (1883–1960) coined the term “monocytic leukemia” for cases featuring high monocyte counts [4].
Over the twentieth century, the classification of leukemia progressively broadened to include better-defined entities with distinct clinical, morphological, phenotypic and genetic features [5,6]. This integrated approach was endorsed by the 2008 World Health Organization (WHO) classification of hematopoietic neoplasms [7] and by its 2016 revision [8]. Since then, further data have been obtained and two new classifications have recently been published. These are the 2022 International Consensus Classification (ICC) [9] and the 5th edition of the WHO Classification of Hematolymphoid Tumors [10]. Both of them introduced significant changes in the classification and diagnostic criteria of many hematologic neoplasms. Among myeloid leukemias, the most relevant changes regard acute myeloid leukemia (AML), chronic myelomonocytic leukemia (CMML), CML and myeloid neoplasms (MNs) with eosinophilia.
This review aims at presenting the revised classification and diagnostic criteria of such entities, specifically considering the rationale for changes to prior classifications. For each disease, new therapeutic options and future treatment perspectives are also addressed.
2. Updates on the Classification, Diagnosis and Therapy of Acute Myeloid Leukemia (AML)
Over the last few years, the scientific discoveries that occurred in the field of AML have revolutionized our understanding of this neoplasm. High-throughput molecular studies have highlighted new recurrent genetic aberrations, bearing relevant prognostic and therapeutic implications. This, in turn, led to reconsider the classification of AML and to propose new therapeutic approaches.
2.1. New Classification and Updates in the Diagnosis of Acute Myeloid Leukemia (AML)
One of the most relevant challenges in the classification of AML is how to integrate clinical, morphological and genetic data to identify non-ambiguous and clinically relevant disease subtypes. The 1976 French–American–British (FAB) classification relied essentially on blast morphology [11]. Since then, phenotypic and molecular studies have demonstrated that AML is highly heterogeneous and that such diversity cannot be captured by morphology alone. This prompted new classification proposals, integrating morphology with clinical, phenotypic and genetic data [6,7,8,12].
Even this integrated approach, however, poses relevant questions, concerning (i) the type and hierarchy of genetic derangements to include in AML classifications, (ii) the utility of blast thresholds to define specific AML subsets, and (iii) the relevance of former AML subtypes in view of recent genetic acquisitions. All of this was addressed by the 2022 ICC and WHO classifications, which adopted similar (yet not overlapping), genetically oriented approaches [13].
The ICC stratifies AML into five molecular subgroups: (i) AML with recurrent genetic abnormalities (both gene rearrangements and gene mutations); (ii) AML with mutated TP53; (iii) AML with myelodysplasia (MDS)-related gene mutations; (iv) AML with MDS-related cytogenetic abnormalities; and (v) AML, not otherwise specified (NOS) (Table 1; Figure 1) [9]. To simplify the classification and avoid confusion among entities with overlapping genetic features, the ICC no longer considers therapy-related AML and AML following MDS, myeloproliferative neoplasms (MPNs) or MDS/MPN as distinct disease subcategories. These, instead, are regarded as diagnostic qualifiers to be added to any AML subtype, whenever appropriate [9,14].

Table 1.
The 2022 ICC and WHO Classification of AML.
AML with recurrent genetic abnormalities has been expanded to include several newly identified gene rearrangements, as well as AML with NPM1 mutations and with in-frame bZIP CEBPA mutations (Table 1). This substitutes the former AML with biallelic CEPBA mutations, since in-frame bZIP derangements have been recently associated with favorable outcome, irrespective of their allelic status [15,16]. No other type of CEPBA mutation is included in this category [9]. For all AML with recurrent genetic abnormalities, the minimal blast threshold is now lowered to 10% of nucleated cells, with the notable exception of AML with t(9;22)(q34.1;q11.2), which still requires at least 20% blasts to facilitate its distinction from progression of CML. The 20% threshold also applies to all other AML subgroups (Table 1) [9,14].
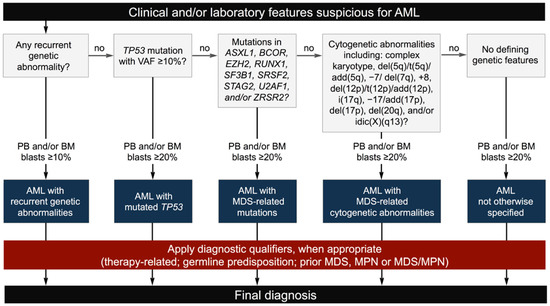
Figure 1.
The ICC diagnostic flow-chart for AML (modified from [17]).
The newly introduced AML with TP53 mutation encompasses AML with ≥20% blasts and mono/biallelic mutations of TP53, irrespective of blast differentiation and/or phenotype [9]. In all published series, this entity is characterized by particularly poor outcome, transcending both blast count and disease ontogeny (e.g., secondary evolution from prior myeloid neoplasms; therapy relatedness; presence of MDS-related genetic changes) [18,19,20]. Of note, since most cases of formerly defined pure erythroid leukemia bear TP53 derangements [21,22], this entity should now be diagnosed as AML with TP53 mutation, with an optional comment on its erythroid differentiation.
AML with MDS-related cytogenetic abnormalities and AML with MDS-related gene mutations (Table 1) substitute the former category of AML with myelodysplasia-related changes (MRCs) [14]. These new subtypes both require ≥20% blasts. Morphologic dysplasia no longer qualifies an AML as MDS-related, given its poor reproducibility, its limited prognostic value and the occurrence of bone marrow (BM) dysplasia even in cases with no history of MDS (e.g., AML with NPM1 or CEPBA mutations) [23,24]. These subtypes account for about 10% of all AMLs and bear a poor prognosis as previously shown for AML-MRC [25]. Grouping together these molecularly homogenous disorders will likely contribute to a better prognostication and management of patients.
When genetic studies have excluded any of the previously defined molecular abnormalities, a diagnosis of AML, NOS is warranted. In such cases, the type of blast differentiation/morphologic subtype can be specified according to the guidelines outlined in the 4th edition of the WHO Classification. Of note, the number of AML cases diagnosed as NOS is expected to decrease in the near future, as a result of further acquisitions on the molecular bases of these disorders.
In summary, the ICC provides a hierarchical classification of AML, based on genetic determinants with clinical and prognostic relevance. Predisposing factors and/or clinical determinants are downgraded to diagnostic qualifiers that should not impact on the molecular classification (Figure 1).
The 2022 WHO Classification has adopted the same classification framework with a few relevant differences. Unlike the ICC, the WHO does not recognize TP53 mutations as genetic classifiers and does not consider AML with TP53 mutations as a separate diagnostic category [10,13]. Nevertheless, TP53 derangements are acknowledged as markers of poor prognosis that may impact on therapy decisions. Furthermore, the 2022 WHO Classification (i) includes fewer gene rearrangements in the category of AML with defining genetic abnormalities, (ii) allows any biallelic mutation (not only in-frame bZIP derangements) as well as monoallelic bZIP mutations for the diagnosis of AML with CEBPA mutation, (iii) contains slight differences in the list of chromosomal changes and gene mutations defining MDS-related AML and (iv) does not require any blast threshold for the diagnosis of AML with defining genetic abnormalities (with the exception of AML with BCR::ABL1 fusion and with CEBPA mutation) [10] (Figure 2; Table 1).
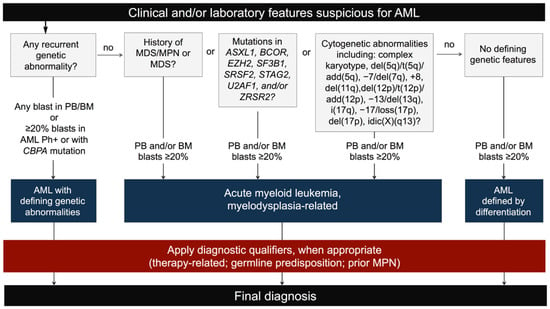
Figure 2.
The 2022 WHO Classification diagnostic flow-chart for AML.
Therefore, both the ICC and 2022 WHO Classification have significantly revised blast thresholds in genetically defined AML. In detail, while the 2022 WHO Classification regards blast cutoffs as largely arbitrary in this context, the ICC adopts a 10% threshold to support the differential diagnosis with MDS and other myeloid neoplasms, which are still regarded as aggressive AML precursors [9]. Despite these differences, both classifications acknowledge the blurred boundaries between AML and MDS, as illustrated by the extensive revisions that have also been made on MDS classification (Table 2). These include (i) a greater emphasis on genetic derangements, (ii) an update of genetically defined entities (MDS with isolated 5q deletion, with SF3B1 mutation and with biallelic TP53 mutations) and (iii) a revised MDS nomenclature, limiting the relevance of morphology. In such a context, the ICC also emphasizes the close relationship between AML and MDS with high blasts (i.e., MDS with 5–19% blasts in PB and/or 10–19% blasts in BM), by renaming these forms MDS/AML and by proposing their molecular stratification according to the AML framework.

Table 2.
Classification of MDS according to the ICC and 2022 WHO Classification.
2.2. Novelties in the Therapy of Acute Myeloid Leukemia (AML)
In the second half of the twentieth century, the management of AML sailed on a dead calm sea. Virtually all attempts to identify new drugs failed and treatment of AML basically relied on one-size-fits-all approaches. The only curative option for young and fit patients was induction chemotherapy with cytarabine plus an anthracycline (“3 + 7” regimen), followed by consolidation with high-dose cytarabine and, if possible, allogeneic stem cell transplantation (alloSCT) [26]. Older or unfit patients were treated with non-curative regimens, including low-dose cytarabine or, more recently, hypomethylating agents (HMAs) [27,28].
In the last few decades, genetic studies have demonstrated the heterogeneity of AML, laying the foundations for new tailored therapies [6]. In this rapidly evolving scenario, AML risk classification has also changed and will likely keep on changing, due to new molecular markers and to treatment options aimed at neutralizing the poor prognosis of some genetic variants. A paradigmatic example of such developments is the reclassification of FLT3-mutated AML as intermediate-risk disease by the 2022 recommendations of the European Leukemia Network (ELN) [17].
In this new era, key issues in AML treatment include: (i) the identification of targetable mutations at diagnosis and relapse; (ii) the choice of appropriate drug combinations to maximize treatment response and to overcome drug resistance; (iii) the choice of low-intensity vs. intensive therapy; (iv) the monitoring of minimal residual disease (MRD) to assess deep remission and relapse risk; (v) the management of new drug-related side effects; and (vi) the appropriate selection of patients for alloSCT to improve transplant outcomes.
Since 2017, several compounds have been approved and many others are being intensively investigated. These comprise both gene mutation-targeting drugs (the FLT3 inhibitors, midostaurin and gilteritinib; the IDH1/IDH2 inhibitors, enasidenib, ivosidenib and olutasidenib) and broadly active compounds (the antibody–drug conjugate, gentuzumab ozogamicin; the liposomal formulation of cytarabine and daunorubicin, CPX-351; the Sonic Hedgehog pathway inhibitor, glasdegib; the BCL2 inhibitor, venetoclax; the oral azacitidine formulation, CC-486, for maintenance therapy). These can be used as frontline treatment or at relapse [29,30], considering the clinical setting and genomic features of the disease. Gentuzumab ozogamicin is indicated in core binding factor (CBF) AML, CPX-351 in MDS-related and therapy-related AML, midostaurin in FLT3-mutated AML and IDH inhibitors in IDH1/2-mutated AML [28].
Besides these new compounds, significant advances in so-called low-intensity therapies have been granted by combining HMAs with venetoclax (HMA/V) [31,32]. As an alternative to intensive therapy, HMA/V is characterized by better survival rates and quality of life, especially in old/unfit patients and in relapsing/refractory (R/R) disease [33]. Given its favorable profile, HMA/V has also been investigated as frontline therapy for young adults with high-risk AML, who would otherwise be fit for intensive chemotherapy. Finally, the introduction of CC-486 ushered in a new season for maintenance therapy of AML, as it was associated with improved clinical outcome in patients ineligible for alloSCT, who are initially treated with intensive therapy [34].
Despite this progress, unmet needs remain in the treatment of R/R disease and of AML with adverse genetic features (e.g., AML with TP53 mutation or KTM2A rearrangements) [35,36]. Promising approaches include doublet or triplet drug regimens combining HMA/intensive chemotherapy with venetoclax [31], APR-246 (a mutant p53 reactivator) [37], FLT3 [38], IDH1/2 [39] or Menin inhibitors [40], or with monoclonal antibodies against CD47 (magrolimab) [41] and TIM3 (sabatolimab) [42]. Ongoing trials are testing these combinations and will hopefully disclose new ways to treat such aggressive AMLs.
3. Updates on the Classification, Diagnosis and Therapy of Chronic Myelomonocytic Leukemia (CMML)
MDS/MPNs are a heterogeneous group of myeloid neoplasms, characterized by both myelodysplastic and myeloproliferative features [43,44]. Over the past decade, significant progress has been made on the molecular bases and natural history of MDS/MPNs. This prompted some relevant changes in the classification and nomenclature of such disorders.
One of the most relevant updates in the ICC classification regards juvenile myelomonocytic leukemia (JMML), a pediatric-age myeloid neoplasm that has been removed from the group of MDS/MPN to be included in a newly created category termed “pediatric and/or germline mutation-associated disorders” [45]. In the 2022 WHO Classification, instead, JMML has been included among the MPNs [10]. Other significnat changes regard MDS/MPN with ring sideroblasts (RSs) and thrombocytosis (MDS/MPN-RS-T). In both the ICC and 2022 WHO Classification, this entity has been renamed MDS/MPN with SF3B1 mutation and thrombocytosis. The ICC allows diagnosing this condition even in the absence of RSs, provided that SF3B1 mutations with variant allele frequency (VAF) ≥ 10% are documented [9,44]. The 2022 WHO Classification, instead, requires the documentation of RS in all cases and does not define a minimum VAF for SF3B1 mutations [10]. Finally, both classifications recognize rare MPD/MPN-RS-T lacking SF3B1 mutations. The ICC identifies a distinct diagnostic category for such cases (i.e., MDS/MPN-RS-T, NOS) [9], while the 2022 WHO Classification retains them among the MDS/MPNs with SF3B1 mutation and thrombocytosis, provided that RSs account for ≥15% of erythroid precursors [10].
Besides these differences, the 2022 ICC and WHO Classifications acknowledge recurrent genetic changes, precursor conditions and early disease phases of MDS/MPN that were not accounted for by prior classifications. All of this holds particularly true for CMML, whose diagnostic criteria have been extensively revised to include this new information and to support the differential diagnosis with other entities.
3.1. New Classification and Updates in the Diagnosis of Chronic Myelomonocytic Leukemia (CMML)
The ICC proposed a significant revision of CMML diagnostic criteria, specifically focused on (i) lowering the threshold of peripheral blood (PB) monocytosis, (ii) adding cytopenia as a new diagnostic requirement and (iii) emphasizing the role of genetic abnormalities to assess disease clonality [9]. Furthermore, both the ICC and 2022 WHO Classifications acknowledge the relevance of phenoypic studies to distinguish CMML from other causes of monocytosis (i.e., ≥94% CD14+/CD16− classical monocytes in PB are strongly associated with CMML as compared to any other condition) [9,10]. In this context, however, it is worth noting that the specificity and (more importantly) sensitivity of flow cytometry assays in CMML have been challenged by recent studies [46,47]. As such, the diagnosis of CMML must always rely on the integration of clinical–pathological, phenotypic and molecular data.
The threshold of PB monocytosis is now set at 0.5 × 109/L with monocytes being ≥10% of the WBC [9]. This lower value is justified by the results of multi-institutional studies, showing genetic and clinical overlap between conventional CMML (absolute monocytes ≥ 1.0 × 109/L) and so-called “oligomonocytic” CMML (absolute monocytes 0.5–1.0 × 109/L) [48,49]. To ensure diagnostic specificity, monocytosis must be accompanied by cytopenia, clonal genetic abnormalities and consistent BM findings (Figure 3). AML and potential CMML mimickers (i.e., MPN, MDS, CML with p190 fusion; myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions (M/LN-Eo-TKs)) must also be excluded. In the absence of genetically defined clonality, the ICC allows a diagnosis of CMML with higher monocyte count (monocytes ≥ 1 × 109/L and ≥10% of the WBCs) together with morphologic/phenotypic findings (increased PB/BM blasts, morphologic dysplasia and/or CMML-specific phenotypic aberrances by flow cytometry) (Table 3) [9]. In the presence of clonality, the ICC limits the value of morphologic dysplasia, which is now replaced by cytopenia as a marker of ineffective hematopoiesis [44]. This is justified by the poor reproducibility of morphology and by the often-subtle dysplastic changes of most CMMLs. The ICC defines cytopenia as hemoglobin levels <130 g/L (for males) and <120 g/L (for females), absolute PMN count <1.8 × 109/L and/or platelet count <150 × 109/L [9]. While anemia (with or without thrombocytopenia) is very common in CMML, a small subset of cases lacks cytopenia at diagnosis. These are usually early phase CMML that become cytopenic later in the course of the disease [50].
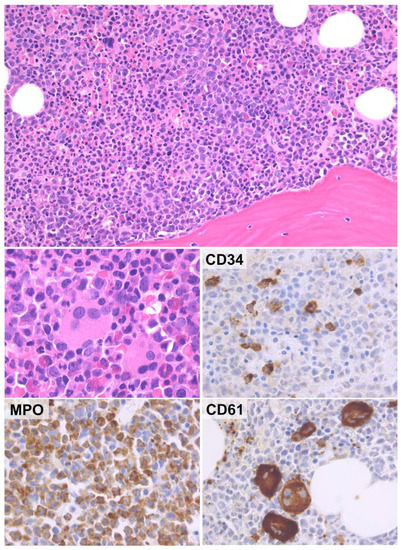
Figure 3.
Histological features of CMML. The BM of CMML is usually hypercellular with increased, left-shifted granulopoiesis (upper panel). Megakaryocytes are often increased and have MDS-like features (middle left panel). Immunohistochemical analysis discloses variable CD34-positive myeloid precursors and markedly increased MPO-positive myeloid cells. CD61 highlights both atypical megakaryocytes and micromegakaryocytes (H&E and immunoperoxidase stains; original magnification ×20, ×40 and ×63).

Table 3.
The ICC diagnostic criteria for CMML.
Based on WBC counts, two disease variants are recognized: CMML with MDS-like phenotype (WBC < 13 × 109/L) and CMML with MPN-like phenotype (WBC ≥ 13 × 109/L) [44]. Distinction between these forms is biologically and clinically relevant, as CMML with MPN-like phenotype is enriched in RAS pathway, JAKV617F and SETBP1 mutations and has a more aggressive clinical course [51]. Subgrouping of CMML according to PB/BM blasts is also simplified, by re-adopting a two-tiered system (CMML-1: <5% in PB and <10% in BM; CMML-2: 5–19% in PB and 10–19% in BM). This is justified by the limited prognostic value of distinguishing between prior CMML-0 and CMML-1 subgroups [52,53].
Finally, the ICC recognizes molecular derangements, which correlate with specific clinical–prognostic features (e.g., NPM1 and SF3B1 mutations). CMMLs with NPM1 mutations are associated with particularly poor prognosis and with high risk of AML progression [54,55]. Despite this, NPM1 mutations should not prompt a diagnosis of AML, if the diagnostic criteria of CMML are met [44]. By contrast, SF3B1 mutations in cases of otherwise typical CMML are associated with a more favorable outcome with low rates of AML progression. These rare cases present with MDS-like phenotype and are morphologically characterized by numerous RS in the BM [56].
Of note, the ICC also introduced clonal monocytosis of undetermined significance (CMUS) as a precursor condition of CMML. CMUS is defined by persistent monocytosis (monocytes ≥ 0.5 × 109/L and ≥10% of the WBC) with at least one myeloid neoplasm-associated mutation (variant allele frequency ≥2%) and with BM findings inconsistent with CMML or other types of myeloid malignancy (Table 4) [9]. If cytopenia is also present, a diagnosis of clonal cytopenia and monocytosis of undetermined significance (CCMUS) is warranted [9]. CMUS and CCMUS must be distinguished from reactive monocytosis, since they frequently progress to overt CMML and other myeloid malignancies [57]. Thus, a thorough diagnostic workup is always recommended before making a diagnosis of CMUS or CCMUS.

Table 4.
The ICC diagnostic criteria for CMUS.
The 2022 WHO Classification introduces several changes also adopted by the ICC (Table 5), but it proposes a hierarchical approach for the diagnosis of CMML with slight differences from the ICC scheme. In particular, the 2022 WHO Classification does not include cytopenia as a diagnostic requirement for CMML and retains morphologic dysplasia as a necessary diagnostic criterion for cases with low monocyte counts (0.5–1 × 109/L) [10]. Moreover (in contrast to the ICC approach), the WHO 2022 Classification mandates to diagnose all NPM1-mutated myeloid neoplasms with CMML features as AML with NPM1 mutation, given their high rate of acute leukemic progression. Finally, the 2022 WHO Classification does not recognize CMUS and CCMUS as separate diagnostic categories [10].

Table 5.
The 2022 WHO diagnostic criteria for CMML.
3.2. Novelties in the Therapy of Chronic Myelomonocytic Leukemia (CMML)
Unlike other myeloid malignancies, the treatment of CMML remains challenging. This is largely due to the biological heterogeneity and intrinsic chemoresistance of the disease and to the lack of dedicated clinical trials for such a rare entity [58]. To date, the only curative approach for CMML is alloSCT, but this is indicated in only a minority of cases, due to the advanced age and comorbidities of many patients [58,59]. As such, treatment options are largely confined to controlling myeloid proliferation in cases with MPN-like phenotype and to compensating cytopenia in cases with MDS-like phenotype.
As for the control of myeloid proliferation, seminal studies in the late 1990s reported disappointing results for chemotherapy, with minimal (if any) clinical response and high toxicity rates [60]. Since then, hydroxyurea (HU) has been increasingly used to control symptoms and disease burden. More recently, HMAs have also been associated with satisfactory response and good safety profiles. The recommended HMAs in CMML include 5-azacitidine (5-AZA), decitabine and the oral formulation ASTX727 (decitabine/cedazuridine). Of note, these recommendations are based on clinical trials designed mostly for MDS and including only small subsets of CMML [61,62,63,64].
Later real-world studies and small non-randomized clinical trials on HMA-treated CMML reported 40–50% overall response (OR) and 7–17% complete response (CR) rates [65,66,67]. However, HMA had no effect on mutational allele burden and on acute leukemic progression, even in patients undergoing clinical response [68,69]. Furthermore, a large retrospective multicenter study suggested a survival advantage for HMA-treated high-risk myeloproliferative CMML, with minimal (if any) effect on lower-risk disease [70]. Finally, a recent phase 3 clinical trial (DACOTA trial) failed to demonstrate any survival advantage for HMAs over HU [71]. As such, no conclusion can be drawn on the best treatment for myeloid proliferation in CMML and both HMAs and HU are the standard of care in this setting.
In recent years, new therapeutic opportunities for CMML have been identified. These include (i) combination therapies with HMA (e.g., 5-AZA plus venetoclax; 5-AZA plus IDH1/2 inhibitors), (ii) new compounds for CMML-related cytopenias (e.g., sotatercept or luspatercept for anemia; eltrombopag for thrombocytopenia) and (iii) therapies targeting CMML-associated molecular derangements (e.g., inhibitors of GM-CSF, RAS-MAPK or Sonic Hedgehog pathways) [58]. Unfortunately, none of these is curative and further research is needed to address the many unmet needs of CMML therapy.
4. Updates on the Classification, Diagnosis and Therapy of Chronic Myeloid Leukemia (CML)
Since the publication of the 2016 WHO Classification, clinical and molecular advances have prompted relevant changes in phase stratification of CML. Significant updates have specifically focused on accelerated phase (AP) and blast phase (BP) disease. This has been paralleled by the advent of new therapeutic strategies, aimed at overcoming therapy resistance to conventional tyrosine kinase inhibitors (TKIs).
4.1. New Classification and Updates in the Diagnosis of Chronic Myeloid Leukemia (CML)
The ICC retains the traditional partition of CML into three phases and proposes simplified criteria for the diagnosis of AP and BP disease [9]. AP is now defined by (i) 10–19% blasts in PB and/or BM, (ii) ≥20% basophils in PB and/or (iii) additional clonal cytogenetic abnormalities in the neoplastic clone (i.e., second Philadelphia chromosome; trisomy 8; isochromosome 17q; trisomy 19; abnormalities of 3q16.2; complex karyotype, defined as ≥3 cytogenetic abnormalities) (Table 6) [9,72]. The 2016 criteria of worsening thrombocytopenia and increasing leukocytosis, thrombocytosis or splenomegaly unresponsive to therapy have been dropped from the definition of AP. Likewise, the provisional criteria of response to TKIs are now excluded from the defining features of AP [8,9]. In line with the 2016 WHO Classification, the ICC acknowledges that BM fibrosis often associated with increased, dysplastic megakaryopoiesis is common in AP [73] (Figure 4), but this is not a stand-alone criterion for the diagnosis of disease progression [72]. The ICC also better specifies the diagnostic criteria of BP, which now include (i) ≥20% blasts in PB and/or BM, (ii) myeloid sarcoma (i.e., extra-medullary blast proliferation) or (iii) >5% lymphoid blasts in PB and/or BM (i.e., lymphoblastic crisis) (Table 6) [72,74].

Table 6.
ICC diagnostic criteria of accelerated and blast phase CML.
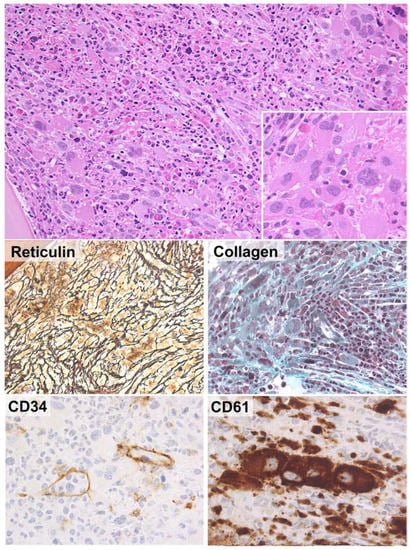
Figure 4.
BM histological features correlating with high-risk CML. In rare cases, the BM of CML shows markedly increased megakaryocytopoiesis with clusters of atypical (MDS-like or myelofibrosis-like) megakaryocytes (upper panel, insert). These findings are usually associated with severe reticulin and collagen fibrosis (middle panels). Immunohistochemical analysis for CD34 reveals patent sinusoids with intra-vascular hematopoiesis. Increased CD34-positive myeloid precursors may or may not be documented. CD61 immunostain is positive in tight clusters of megakaryocytes. These histological features are associated with a more aggressive clinical course and poorer response to TKIs (H&E and immunoperoxidase stains; original magnification ×20, ×40 and ×63).
The 2022 WHO Classification adopts an alternative, biphasic scheme for the natural history of CML, only recognizing chronic phase (CP) and BP disease. AP is no longer considered an independent step in the biological continuum of CML and is regarded as “high-risk CP” [10]. The removal of AP as a diagnostic category is paralleled by the introduction of a list of features that correlate with high-risk CP (Table 7) [10,75]. All the ICC defining parameters of AP are included in it [9,72]. Finally, the 2022 WHO criteria of BP largely correspond to those of the ICC, although no actual threshold for lymphoblastsis provided by the WHO [10]. In conclusion, despite differences in terminology, the 2022 ICC and WHO Classifications provide roughly similar risk stratifications for CML, which are in keeping with clinical observations following the introduction of TKIs.

Table 7.
Features associated with increased risk of progression in chronic phase CML according to the 2022 WHO Classification.
4.2. Novelties in the Therapy of Chronic Myeloid Leukemia (CML)
The discovery of the Philadelphia chromosome paved the way to TKI-based therapies for CML [76]. This has dramatically changed the paradigm of CML management, prompting deep and fast molecular responses, decreasing disease progression and prolonging life expectancy to nearly that of the general population. Long-term outcome studies specifically indicate that TKIs have increased the 10-year OS of CML from roughly 20% to 80–90% [77,78].
Several TKIs are available for the treatment of CML, including imatinib (first-generation TKIs, 1G-TKIs), dasatinib, nilotinib and bosutinib (second-generation TKIs, 2G-TKIs), ponatinib, olverembatinib and vodobatinib (third-generation TKIs) and PF-114 (fourth-generation TKI). While 1G-TKI, 2G-TKI and ponatinib have entered clinical practice [79,80,81,82], olverembatinib, vodobatinib and PF-114 are still under investigation in dedicated trials [83]. In October 2021, a new allosteric inhibitor of BCR-ABL1 (asciminib) joined the TKI family [84]. Unlike other compounds, asciminib binds the myristoyl pocket of BCR-ABL1, blocking its kinase activtabity. This unique mechanism may prove particularly useful to overcome resistance to conventional TKIs.
To date, key points in the management of CML include: (i) the choice of first-line TKI; (ii) the management of TKI resistance and side effects; (iii) the prevention and treatment of AP/BP disease; (iv) therapy withdrawal following remission (aka therapy-free remission, TFR).
As for the choice of upfront therapy, current guidelines endorse any 1G/2G-TKI as a therapeutic option [85]. The choice usually relies on several parameters, including disease risk stratification, patient comorbidities, treatment goals and costs and physician and patient preference [86]. Patients with intolerance/resistance to 1G/2G-TKIs may receive a further 2G-TKI or ponatinib/asciminib, after careful re-assessment of disease phase and clonal evolution. AlloSCT still represents the only curative treatment for CP disease resistant to ≥2 TKIs. As for BP disease, the therapeutic options include AML-like or B-ALL-like regimens (depending on blast phenotype) in addition to TKIs. AlloSCT also represents a valuable therapeutic option in all elegible patients, with 5-year overall survival rates of about 60% [87].
Another major issue in the therapy of CML is when and how to consider TKI discontinuation. Starting from the STIM1 study [88] and from reports on interferon (IFN)-treated CML [89], several trials and real-world studies have demonstrated the safety of TFR in selected patients with CP CML. This has led to incorporation of TFR as a goal of treatment by the 2020 ELN and National Comprehensive Cancer Network (NCCN) recommendations [90,91].
Factors that are associated with TFR include (i) the baseline disease risk category, (ii) the duration of TKI treatment, (iii) the response to TKI and (iv) the depth and duration of molecular response [92]. Since only a minority of patients achieves durable TFR, many strategies are under investigation to increase these figures. Possible options include asciminib as frontline therapy (NCT05143840 trial), combination therapies with ruxolitinib and TKIs (NCT03610971 trial) and further administration of TKIs following TFR failure (NCT04838041 trial). The results of these studies will hopefully improve TFR rates, marking further progress in the management of CML.
5. Updates on the Classification, Diagnosis and Therapy of Myeloid Neoplasms (MNs) with Eosinophilia
MNs with eosinophilia are a heterogeneous group of myeloid disorders that mainly include chronic eosinophilic leukemia, NOS (CEL, NOS) and the renamed, genetically assigned group of M/LN-Eo-TK. The diagnostic criteria for CEL, NOS have been updated and its boundaries with other eosinophilic disorders have been refined, plus new entities have been added to the group of M/LN-Eo-TK. These changes have a direct impact on the management of patients.
5.1. New Classification and Updates in the Diagnosis of Myeloid Neoplasms (MNs) with Eosinophilia
The 2022 ICC and WHO Classifications introduced significant changes to the diagnostic criteria of CEL, NOS (Table 8) [9]. In particular, the ICC adopts a more strict definition of hypereosinophila (HE) and introduces BM morphology as a supportive diagnostic criterion. According to the ICC, HE is now defined by high absolute eosinophil counts (≥1.5 × 109/L) with relative eosinophilia (≥10% of the WBCs). This is in line with the diagnostic approach to other MNs (e.g., monocyte count in CMML) and avoids ambiguities in cases with extreme leukocytosis and high absolute eosinophil counts but low percentage of eosinophils [93]. In recent years, BM morphology has emerged as a robust diagnostic marker of CEL, NOS, since marrow dysplasia and/or reticulin fibrosis are typical of this condition and almost never observed in non-clonal HE [94]. As such, in the presence of persistent eosinophilia, diagnostically appropriate BM findings can be usedto diagnose CEL, NOS even in the absence of clonal abnormalities and/or increased blasts, after other possible causes of eosinophila have been excluded (Table 8) [9].

Table 8.
Diagnostic criteria for CEL, NOS according to the ICC and 2022 WHO Classification.
The 2022 WHO Classification adopts slightly different diagnostic criteria, as it (i) requires only high absolute eosinophil counts (≥1.5 × 109/L) with no relative eosinophilia, (ii) considers both clonal genetic derangements and abnormal BM findings as mandatory features and (iii) dismisses high blast counts from the diagnostic checklist (Table 8) [10].
As for other MNs with eosinophilia, both the 2022 ICC and WHO Classifications have renamed the category of M/LN-Eo-TK to emphasize the relevance of TK gene fusions in the pathogenesis and therapy of such conditions. Furthermore, the former list of M/LN-Eo-TKs (i.e., M/LN with PDGFRA, PDGFRB and FGFR1 rearrangements) has been expanded to also include M/LN with FLT3 and ETV6::ABL1 gene fusions as well as M/LN with JAK2 rearrangements other than those with PCM1 [9,10,93] (Table 9). The WHO Classification has introduced a further diagnostic category of “M/LN with other TK rearrangements” for entities presenting as M/LN-Eo-TK with rare or unconventional genetic features [10] (Table 9).

Table 9.
Classification of M/LN-Eo-TK according to the ICC and 2022 WHO Classification.
M/LN-Eo-TKs have very heterogeneous clinical–pathological presentation, resembling either MPN, MDS/MPN, AML or B-cell and T-cell acute lymphoblastic leukemia (ALL). In this latter case, to avoid confusion with conventional ALL with TK rearrangements, a diagnosis of M/LN-Eo-TK is appropriate if (i) there is concurrent or previous evidence of an MN, (ii) a residual MN is documented after ALL therapy or (iii) TK rearrangements are found in myeloid cells [93]. Likewise, since mast cell clones resembling systemic mastocytosis (SM) can be documented in M/LN-Eo-TK, assessment for TK rearrangements is recommended in all SM with HE or lacking KIT mutations. If TK rearrangements are documented, the diagnosis of M/LN-Eo-TK supersedes that of SM [93].
5.2. Novelties in the Therapy of Myeloid Neoplasms (MNs) with Eosinophilia
The treatment of MNs with eosinophilia is challenging, given the limited number of studies on this topic and the overall rarity of such conditions. CEL, NOS remains a very aggressive disease with limited and non-standardized therapeutic options. HU or IFN can be used to control CEL-associated myeloproliferative features and steroids can reduce PB eosinophilia. In young and fit patients, alloSCT remains the only curative option [95].
In M/LN-Eo-TK, constitutive TK signaling is a potential target for TKIs, yet the efficacy of these compounds is highly variable. Of note, evidence on TKI therapy in such disorders relies only on single case reports and small case series. Despite this, the 2021 NCCN guidelines recommend TKIs as frontline treatment for both CP and BP disease, especially in the absence of clinical trials [96]. The type of gene fusion dictates response to TKIs. PDGFRA and PDGFRB translocations have shown exquisite sensitivity to imatinib [97,98]. 2G/3G-TKIs or FLT3 inhibitors should be restricted to cases with primary/secondary imatinib resistance, as a bridge to alloSCT [99,100]. By contrast, FGFR1, JAK2, FLT3 and ETV6::ABL1 rearrangements have more aggressive clinical course and benefit from other therapies (especially alloSCT) [101].
In M/LN with FGFR1 rearrangement, sustained molecular remissions can be obtained with alloSCT or (more recently) with the selective FGFR inhibitor, pemigatinib [102,103]. In M/LN with ETV6::ABL1 rearrangement, durable remissions have been observed with 2G/3G-TKIs in CP disease, with minimal (if any) effect in BP disease [101]. In M/LN with FLT3 rearrangement, FLT3 inhibitors (sunitinib and sorafenib) have shown at best short-lived remissions and alloSCT represents the only curative option [104,105,106]. The same holds true for M/LN with JAK2 rearrangement, for which only short-term remissions or a role in maintenance therapy have been reported for JAK2 inhibitors (e.g., ruxolitinib) [101,107,108]. In conclusion, the treatment of M/LN-Eo-TK is still largely empirical and anecdotal. Further studies will hopefully disclose new molecular targets for such aggressive MNs.
6. Conclusions
In addition to morphology, immunophenotyping and cytogenetics (which have been the mainstay of diagnostic assessment over the past several decades and remain relevant today), genomic characterization and advances in sequencing technology have improved our understanding of MNs, opening novel therapeutic opportunities [109]. The need for genomic integration has prompted the formulation of new disease classification proposals, some of which have been discussed in this review. All of this certainly represents a challenge for pathologists and hematologists dealing with myeloid leukemias. Nevertheless, the sheer amount of data at our disposal represents an unprecedented opportunity to understand these neoplasms and, ultimately, our role in diagnosing and treating them.
Although the relevance of molecular tests cannot be overestimated, the 2022 ICC and WHO Classifications clearly demonstrate that integration of clinical, morphologic, immunophenotypic and genetic data remains the mainstay for the diagnosis of MNs [9,10]. This is in keeping with the decade-long approach to the diagnosis of hematopoietic disorders [94,110,111], which has shaped current classifications and treatment options. On the other hand, the discovery of recurrent genetic abnormalities and molecular determinants remains crucial for providing new therapeutic options.
In the foreseeable future, clinical–pathological and genetic studies will shed light on the still many questions concerning myeloid leukemias. The most compelling issues specifically regard (i) the relevance of blast thresholds for the diagnosis and prognosis of some MNs, (ii) the hierarchy of clinical, morphological and genetic data to define specific disease entities, (iii) the nature and type of molecular derangements to be included in genetically based classifications, (iv) the management of MNs characterized by poor response to conventional therapies and (v) the treatment of newly discovered entities. These problems can be solved only by the close collaboration of experts from different fields, in keeping with the inspiring principles of current hematology practice.
Author Contributions
M.P. and C.G.: writing of the paper; A.O.: review conceptualization, manuscript supervision and final editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kampen, K.R. The discovery and early understanding of leukemia. Leuk. Res. 2012, 36, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Geary, C.G. The story of chronic myeloid leukaemia. Br. J. Haematol. 2000, 110, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, A. Ueber acute Leukamie. Dtsch Med. Wochenschr. 1895, 21, 639–642. [Google Scholar] [CrossRef]
- Reschad, H.; Schilling-Torgau, V. Ueber eine neue Leukämie durch echte Uebergangsformen (Splenozytenleukämie) und ihre Bedeutung für die Selbständigkeit dieser Zellen. Münchener Med. Wchnschrift 1913, 60, 1981–1984. [Google Scholar]
- Neame, P.B.; Soamboonsrup, P.; Browman, G.P.; Meyer, R.M.; Benger, A.; Wilson, W.E.; McBride, J.A. Classifying Acute Leukemia by Immunophenotyping: A Combined FAB-Immunologic Classification of AML. Blood 1986, 68, 1355–1362. [Google Scholar] [CrossRef] [PubMed]
- Papaemmanuil, E.; Gerstung, M.; Bullinger, L.; Gaidzik, V.I.; Paschka, P.; Roberts, N.D.; Potter, N.E.; Heuser, M.; Thol, F.; Bolli, N.; et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016, 374, 2209–2221. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.-M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef]
- Vardiman, J.W.; Harris, N.L.; Brunning, R.D. The World Health Organization (WHO) classification of the myeloid neoplasms. Blood 2002, 100, 2292–2302. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Hasserjian, R.P.; Orazi, A.; Mathews, V.; Roberts, A.W.; Schiffer, C.A.; Roug, A.S.; Cazzola, M.; Döhner, H.; Tefferi, A. Classification of myeloid neoplasms/acute leukemia: Global perspectives and the international consensus classification approach. Am. J. Hematol. 2022, 97, 514–518. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Martelli, M.P. Comparison of the International Consensus and 5th WHO edition classifications of adult myelodysplastic syndromes and acute myeloid leukemia. Am. J. Hematol. 2023, 98, 481–492. [Google Scholar] [CrossRef]
- Weinberg, O.K.; Porwit, A.; Orazi, A.; Hasserjian, R.P.; Foucar, K.; Duncavage, E.J.; Arber, D.A. The International Consensus Classification of acute myeloid leukemia. Virchows Arch. 2022, 482, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Tarlock, K.; Lamble, A.J.; Wang, Y.C.; Gerbing, R.B.; Ries, R.E.; Loken, M.R.; Brodersen, L.E.; Pardo, L.; Leonti, A.; Smith, J.L.; et al. CEBPA-bZip mutations are associated with favorable prognosis in de novo AML: A report from the Children’s Oncology Group. Blood 2021, 138, 1137–1147. [Google Scholar] [CrossRef]
- Taube, F.; Georgi, J.A.; Kramer, M.; Stasik, S.; Middeke, J.M.; Röllig, C.; Krug, U.; Krämer, A.; Scholl, S.; Hochhaus, A.; et al. CEBPA mutations in 4708 patients with acute myeloid leukemia: Differential impact of bZIP and TAD mutations on outcome. Blood 2022, 139, 87–103. [Google Scholar] [CrossRef]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Ok, C.Y.; Patel, K.P.; Garcia-Manero, G.; Routbort, M.J.; Peng, J.; Tang, G.; Goswami, M.; Young, O.C.; Singh, R.; Medeiros, L.J.; et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J. Hematol. Oncol. 2015, 8, 45. [Google Scholar] [CrossRef]
- Grob, T.; Al Hinai, A.S.A.; Sanders, M.A.; Kavelaars, F.G.; Rijken, M.; Gradowska, P.L.; Biemond, B.J.; Breems, D.A.; Maertens, J.; Kooy, M.V.M.; et al. Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood 2022, 139, 2347–2354. [Google Scholar] [CrossRef]
- Niparuck, P.; Police, P.; Noikongdee, P.; Siriputtanapong, K.; Limsuwanachot, N.; Rerkamnuaychoke, B.; Chuncharunee, S.; Siriboonpiputtana, T. TP53 mutation in newly diagnosed acute myeloid leukemia and myelodysplastic syndrome. Diagn. Pathol. 2021, 16, 100. [Google Scholar] [CrossRef]
- Fang, H.; Wang, S.A.; Khoury, J.D.; El Hussein, S.; Kim, D.H.; Tashakori, M.; Tang, Z.; Li, S.; Hu, Z.; Jelloul, F.Z.; et al. Pure erythroid leukemia is characterized by biallelic TP53 inactivation and abnormal p53 expression patterns in de novo and secondary cases. Haematologica 2022, 107, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- Pizzi, M.; Sbaraglia, M.; De Bartolo, D.; Santo, L.D.; Binotto, G.; Tosato, F.; Pravato, S.; Scapinello, G.; Martines, A.; Bonaldi, L.; et al. Relevance of bone marrow histology in challenging cases of Acute Myeloid Leukemia. Int. J. Lab. Hematol. 2021, 44, e107–e110. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Macijewski, K.; Weiss, T.; Bacher, U.; Schnittger, S.; Kern, W.; Kohlmann, A.; Klein, H.-U.; Vignetti, M.; Piciocchi, A.; et al. Multilineage dysplasia has no impact on biologic, clinicopathologic, and prognostic features of AML with mutated nucleophosmin (NPM1). Blood 2010, 115, 3776–3786. [Google Scholar] [CrossRef] [PubMed]
- Haferlach, T.; Bennett, J.; Löffler, H.; Gassmann, W.; Andersen, J.W.; Tuzuner, N.; BÜChner, T. Acute myeloid leukemia with translocation (8;21). Cytomorphology, dysplasia and prognostic factors in 41 cases. AML Cooperative Group and ECOG. Leuk. Lymphoma 1996, 23, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Baer, C.; Walter, W.; Stengel, A.; Hutter, S.; Meggendorfer, M.; Kern, W.; Haferlach, C.; Haferlach, T. Molecular Classification of AML-MRC Reveals a Distinct Profile and Identifies MRC-like Patients with Poor Overall Survival. Blood 2019, 134, 2735. [Google Scholar] [CrossRef]
- Yates, J.W.; Wallace, H.J., Jr.; Ellison, R.R.; Holland, J.F. Cytosine arabinoside (NSC-63878) and daunorubicin (NSC-83142) therapy in acute nonlymphocytic leukemia. Cancer Chemother. Rep. 1973, 57, 485–488. [Google Scholar]
- Dombret, H.; Seymour, J.F.; Butrym, A.; Wierzbowska, A.; Selleslag, D.; Jang, J.H.; Kumar, R.; Cavenagh, J.; Schuh, A.C.; Candoni, A.; et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood 2015, 126, 291–299. [Google Scholar] [CrossRef]
- Kantarjian, H.M.; Thomas, X.G.; Dmoszynska, A.; Wierzbowska, A.; Mazur, G.; Mayer, J.; Gau, J.-P.; Chou, W.-C.; Buckstein, R.; Cermak, J.; et al. Multicenter, Randomized, Open-Label, Phase III Trial of Decitabine Versus Patient Choice, with Physician Advice, of Either Supportive Care or Low-Dose Cytarabine for the Treatment of Older Patients with Newly Diagnosed Acute Myeloid Leukemia. J. Clin. Oncol. 2012, 30, 2670–2677. [Google Scholar] [CrossRef]
- Liu, H. Emerging agents and regimens for AML. J. Hematol. Oncol. 2021, 14, 49. [Google Scholar] [CrossRef]
- Kayser, S.; Levis, M.J. Updates on targeted therapies for acute myeloid leukaemia. Br. J. Haematol. 2021, 196, 316–328. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Pratz, K.; Pullarkat, V.; Jonas, B.A.; Arellano, M.; Becker, P.S.; Frankfurt, O.; Konopleva, M.; Wei, A.H.; Kantarjian, H.M.; et al. Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood 2018, 133, 7–17. [Google Scholar] [CrossRef]
- Dinardo, C.D.; Jonas, B.A.; Pullarkat, V.; Thirman, M.J.; Garcia, J.S.; Wei, A.H.; Konopleva, M.; Döhner, H.; Letai, A.; Fenaux, P.; et al. Azacitidine and Venetoclax in Previously Untreated Acute Myeloid Leukemia. N. Engl. J. Med. 2020, 383, 617–629. [Google Scholar] [CrossRef] [PubMed]
- Samra, B.; Konopleva, M.; Isidori, A.; Daver, N.; Dinardo, C. Venetoclax-Based Combinations in Acute Myeloid Leukemia: Current Evidence and Future Directions. Front. Oncol. 2020, 10, 562558. [Google Scholar] [CrossRef] [PubMed]
- Roboz, G.J.; Döhner, H.; Pocock, C.; Dombret, H.; Ravandi, F.; Jang, J.H.; Selleslag, D.; Mayer, J.; Martens, U.M.; Liesveld, J.; et al. Oral azacitidine preserves favorable level of fatigue and health-related quality of life for patients with acute myeloid leukemia in remission: Results from the phase 3, placebo-controlled QUAZAR AML-001 trial. Haematologica 2021, 106, 3240–3244. [Google Scholar] [CrossRef] [PubMed]
- Hunter, A.M.; Sallman, D.A. Current status and new treatment approaches in TP53 mutated AML. Best Pract. Res. Clin. Haematol. 2019, 32, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Rücker, F.G.; Schlenk, R.F.; Bullinger, L.; Kayser, S.; Teleanu, V.; Kett, H.; Habdank, M.; Kugler, C.-M.; Holzmann, K.; Gaidzik, V.I.; et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood 2012, 119, 2114–2121. [Google Scholar] [CrossRef]
- Lehmann, S.; Bykov, V.J.; Ali, D.; Andrén, O.; Cherif, H.; Tidefelt, U.; Uggla, B.; Yachnin, J.; Juliusson, G.; Moshfegh, A.; et al. Targeting p53 in Vivo: A First-in-Human Study with p53-Targeting Compound APR-246 in Refractory Hematologic Malignancies and Prostate Cancer. J. Clin. Oncol. 2012, 30, 3633–3639. [Google Scholar] [CrossRef]
- Maiti, A.; DiNardo, C.D.; Daver, N.G.; Rausch, C.R.; Ravandi, F.; Kadia, T.M.; Pemmaraju, N.; Borthakur, G.; Bose, P.; Issa, G.C.; et al. Triplet therapy with venetoclax, FLT3 inhibitor and decitabine for FLT3-mutated acute myeloid leukemia. Blood Cancer J. 2021, 11, 25. [Google Scholar] [CrossRef]
- Lachowiez, C.; Borthakur, G.; Loghavi, S.; Zeng, Z.; Kadia, T.M.; Masarova, L.; Dinardo, C.D. A phase Ib/II study of ivosidenib with venetoclax +/− azacitidine in IDH1-mutated myeloid malignancies. J. Clin. Oncol. 2021, 39, 7012. [Google Scholar] [CrossRef]
- Swaminathan, M.; Bourgeois, W.; Armstrong, S.; Wang, E.S. Menin Inhibitors in Acute Myeloid Leukemia-What Does the Future Hold? Cancer J. 2022, 28, 62–66. [Google Scholar] [CrossRef]
- Daver, N.G.; Vyas, P.; Kambhampati, S.; Al Malki, M.M.; Larson, R.; Asch, A.; Mannis, G.; Chai-Ho, W.; Tanaka, T.; Bradley, T.; et al. Tolerability and efficacy of the first-in-class anti-cd47 antibody magrolimab combined with azacitidine in frontline patient with tp53-mutated acute myeloid leukemia: Phase 1b results. Hemasphere 2022, 6, 33–34. [Google Scholar] [CrossRef]
- Brunner, A.M.; Esteve, J.; Porkka, K.; Knapper, S.; Traer, E.; Scholl, S.; Tovar, N. Efficacy and Safety of Sabatolimab (MBG453) in Combination with Hypomethylating Agents (HMAs) in Patients (Pts) with Very High/High-Risk Myelodysplastic Syndrome (vHR/HR-MDS) and Acute Myeloid Leukemia (AML): Final Analysis from a Phase Ib Study. Blood 2021, 138, 244. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T. Evidence-Based Minireview: Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: A focused review. Hematology 2020, 2020, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Arber, D.A.; Bueso-Ramos, C.; Hasserjian, R.P.; Orazi, A. Advances in myelodysplastic/myeloproliferative neoplasms. Virchows Arch. 2022, 482, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Rudelius, M.; Weinberg, O.K.; Niemeyer, C.M.; Shimamura, A.; Calvo, K.R. The International Consensus Classification (ICC) of hematologic neoplasms with germline predisposition, pediatric myelodysplastic syndrome, and juvenile myelomonocytic leukemia. Virchows Arch. 2022, 482, 113–130. [Google Scholar] [CrossRef] [PubMed]
- Pophali, P.A.; Timm, M.M.; Mangaonkar, A.A.; Shi, M.; Reichard, K.; Tefferi, A.; Pavelko, K.; Villasboas, J.C.; Jevremovic, D.; Patnaik, M.M. Practical limitations of monocyte subset repartitioning by multiparametric flow cytometry in chronic myelomonocytic leukemia. Blood Cancer J. 2019, 9, 65. [Google Scholar] [CrossRef]
- Murali, A.; Cross, D.; Mollee, P. The use of monocyte subset repartitioning by flow cytometry for diagnosis of chronic myelomonocytic leukaemia. Blood Cancer J. 2021, 11, 6. [Google Scholar] [CrossRef]
- Geyer, J.T.; Tam, W.; Liu, Y.-C.; Chen, Z.; Wang, S.A.; Bueso-Ramos, C.; Oak, J.; Arber, D.A.; Hsi, E.; Rogers, H.J.; et al. Oligomonocytic chronic myelomonocytic leukemia (chronic myelomonocytic leukemia without absolute monocytosis) displays a similar clinicopathologic and mutational profile to classical chronic myelomonocytic leukemia. Mod. Pathol. 2017, 30, 1213–1222. [Google Scholar] [CrossRef]
- Calvo, X.; Garcia-Gisbert, N.; Parraga, I.; Gibert, J.; Florensa, L.; Andrade-Campos, M.; Merchan, B.; Garcia-Avila, S.; Montesdeoca, S.; Fernández-Rodríguez, C.; et al. Oligomonocytic and overt chronic myelomonocytic leukemia show similar clinical, genomic, and immunophenotypic features. Blood Adv. 2020, 4, 5285–5296. [Google Scholar] [CrossRef]
- Itzykson, R.; Fenaux, P.; Bowen, D.; Cross, N.C.P.; Cortes, J.; De Witte, T.; Germing, U.; Onida, F.; Padron, E.; Platzbecker, U.; et al. Diagnosis and Treatment of Chronic Myelomonocytic Leukemias in Adults: Recommendations from the European Hematology Association and the European LeukemiaNet. Hemasphere 2018, 2, e150. [Google Scholar] [CrossRef]
- Carr, R.M.; Vorobyev, D.; Lasho, T.; Marks, D.L.; Tolosa, E.J.; Vedder, A.; Almada, L.L.; Yurcheko, A.; Padioleau, I.; Alver, B.; et al. RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis. Nat. Commun. 2021, 12, 2901. [Google Scholar] [CrossRef]
- Loghavi, S.; Sui, D.; Wei, P.; Garcia-Manero, G.; Pierce, S.; Routbort, M.J.; Jabbour, E.J.; Pemmaraju, N.; Kanagal-Shamanna, R.; Gur, H.D.; et al. Validation of the 2017 revision of the WHO chronic myelomonocytic leukemia categories. Blood Adv. 2018, 2, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Xicoy, B.; Triguero, A.; Such, E.; García, O.; Jiménez, M.-J.; Arnán, M.; Bernal, T.; Diaz-Beya, M.; Valcárcel, D.; Pedro, C.; et al. The division of chronic myelomonocytic leukemia (CMML)-1 into CMML-0 and CMML-1 according to 2016 World Health Organization (WHO) classification has no impact in outcome in a large series of patients from the Spanish group of MDS. Leuk. Res. 2018, 70, 34–36. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zuo, Z.; Fu, B.; Oki, Y.; Tang, G.; Goswami, M.; Priyanka, P.; Muzzafar, T.; Medeiros, L.J.; Luthra, R.; et al. Chronic myelomonocytic leukemia with nucleophosmin (NPM1) mutation. Eur. J. Haematol. 2015, 96, 65–71. [Google Scholar] [CrossRef]
- Vallapureddy, R.; Lasho, T.L.; Hoversten, K.; Finke, C.M.; Ketterling, R.; Hanson, C.; Gangat, N.; Tefferi, A.; Patnaik, M.M. Nucleophosmin 1 (NPM1) mutations in chronic myelomonocytic leukemia and their prognostic relevance. Am. J. Hematol. 2017, 92, E614–E618. [Google Scholar] [CrossRef] [PubMed]
- Wudhikarn, K.; Loghavi, S.; Mangaonkar, A.A.; Al-Kali, A.; Binder, M.; Carr, R.; Reichard, K.; Finke, C.; Howard, M.; Gangat, N.; et al. SF3B1-mutant CMML defines a predominantly dysplastic CMML subtype with a superior acute leukemia-free survival. Blood Adv. 2020, 4, 5716–5721. [Google Scholar] [PubMed]
- Valent, P.; Orazi, A.; Savona, M.R.; Patnaik, M.M.; Onida, F.; Van de Loosdrecht, A.A.; Haase, D.; Haferlach, T.; Elena, C.; Pleyer, L.; et al. Proposed diagnostic criteria for classical chronic myelomonocytic leukemia (CMML), CMML variants and pre-CMML conditions. Haematologica 2019, 104, 1935–1949. [Google Scholar] [CrossRef]
- Renneville, A.; Patnaik, M.M.; Chan, O.; Padron, E.; Solary, E. Increasing recognition and emerging therapies argue for dedicated clinical trials in chronic myelomonocytic leukemia. Leukemia 2021, 35, 2739–2751. [Google Scholar] [CrossRef]
- De Witte, T.; Bowen, D.; Robin, M.; Malcovati, L.; Niederwieser, D.; Yakoub-Agha, I.; Kröger, N. Allogeneic hematopoietic stem cell transplantation for MDS and CMML: Recommendations from an international expert panel. Blood 2017, 129, 1753–1762. [Google Scholar] [CrossRef]
- Wattel, E.; Guerci, A.; Hecquet, B.; Economopoulos, T.; Copplestone, A.; Mahé, B.; Couteaux, M.E.; Resegotti, L.; Voglova, V.; Foussard, C.; et al. A randomized trial of hydroxyurea versus VP16 in adult chronic myelomonocytic leukemia. Groupe Français des Myélodysplasies and European CMML Group. Blood 1996, 88, 2480–2487. [Google Scholar] [CrossRef]
- Silverman, L.R.; Demakos, E.P.; Peterson, B.L.; Kornblith, A.B.; Holland, J.C.; Odchimar-Reissig, R.; Stone, R.M.; Nelson, D.; Powell, B.L.; DeCastro, C.M.; et al. Randomized Controlled Trial of Azacitidine in Patients with the Myelodysplastic Syndrome: A Study of the Cancer and Leukemia Group B. J. Clin. Oncol. 2002, 20, 2429–2440. [Google Scholar] [CrossRef] [PubMed]
- Fenaux, P.; Mufti, G.J.; Hellstrom-Lindberg, E.; Santini, V.; Finelli, C.; Giagounidis, A.; Schoch, R.; Gattermann, N.; Sanz, G.; List, A.; et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: A randomised, open-label, phase III study. Lancet Oncol. 2009, 10, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Kantarjian, H.; Kadia, T.; DiNardo, C.; Daver, N.; Borthakur, G.; Jabbour, E.; Garcia-Manero, G.; Konopleva, M.; Ravandi, F. Acute myeloid leukemia: Current progress and future directions. Blood Cancer J. 2021, 11, 41. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manero, G.; Griffiths, E.A.; Steensma, D.P.; Roboz, G.J.; Wells, R.A.; McCloskey, J.; Odenike, O.; DeZern, A.E.; Yee, K.; Busque, L.; et al. Oral cedazuridine/decitabine for MDS and CMML: A phase 2 pharmacokinetic/pharmacodynamic randomized crossover study. Blood 2020, 136, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Braun, T.; Itzykson, R.; Renneville, A.; De Renzis, B.; Dreyfus, F.; Laribi, K.; Bouabdallah, K.; Vey, N.; Toma, A.; Recher, C.; et al. Molecular predictors of response to decitabine in advanced chronic myelomonocytic leukemia: A phase 2 trial. Blood 2011, 118, 3824–3831. [Google Scholar] [CrossRef] [PubMed]
- Wijermans, P.; Rüter, B.; Baer, M.; Slack, J.; Saba, H.; Lübbert, M. Efficacy of decitabine in the treatment of patients with chronic myelomonocytic leukemia (CMML). Leuk. Res. 2008, 32, 587–591. [Google Scholar] [CrossRef]
- Santini, V.; Allione, B.; Zini, G.; Gioia, D.; Lunghi, M.; Poloni, A.; Cilloni, D.; Sanna, A.; Masiera, E.; Ceccarelli, M.; et al. A phase II, multicentre trial of decitabine in higher-risk chronic myelomonocytic leukemia. Leukemia 2017, 32, 413–418. [Google Scholar] [CrossRef]
- Unnikrishnan, A.; Papaemmanuil, E.; Beck, D.; Deshpande, N.P.; Verma, A.; Kumari, A.; Woll, P.S.; Richards, L.A.; Knezevic, K.; Chandrakanthan, V.; et al. Integrative Genomics Identifies the Molecular Basis of Resistance to Azacitidine Therapy in Myelodysplastic Syndromes. Cell Rep. 2017, 20, 572–585. [Google Scholar] [CrossRef]
- Merlevede, J.; Droin, N.; Qin, T.; Meldi, K.; Yoshida, K.; Morabito, M.; Chautard, E.; Auboeuf, D.; Fenaux, P.; Braun, T.; et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat. Commun. 2016, 7, 10767. [Google Scholar] [CrossRef]
- Pleyer, L.; Leisch, M.; Kourakli, A.; Padron, E.; Maciejewski, J.P.; Cirici, B.X.; Kaivers, J.; Ungerstedt, J.; Heibl, S.; Patiou, P.; et al. Outcomes of patients with chronic myelomonocytic leukaemia treated with non-curative therapies: A retrospective cohort study. Lancet Haematol. 2021, 8, e135–e148. [Google Scholar] [CrossRef]
- Itzykson, R.; Santini, V.; Thepot, S.; Ades, L.; Chaffaut, C.; Giagounidis, A.; Morabito, M.; Droin, N.; Lübbert, M.; Sapena, R.; et al. Decitabine Versus Hydroxyurea for Advanced Proliferative Chronic Myelomonocytic Leukemia: Results of a Randomized Phase III Trial within the EMSCO Network. J. Clin. Oncol. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Gianelli, U.; Thiele, J.; Orazi, A.; Gangat, N.; Vannucchi, A.M.; Tefferi, A.; Kvasnicka, H.M. International Consensus Classification of myeloid and lymphoid neoplasms: Myeloproliferative neoplasms. Virchows Arch. 2022, 482, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Binotto, G.; Bertorelle, R.; Bonaldi, L.; Frison, L.; Vianello, F.; Pizzi, M. Chronic Myeloid Leukemia with Myelofibrosis-Like Features. Clues of Accelerated Phase? Int. J. Surg. Pathol. 2019, 27, 771–772. [Google Scholar] [CrossRef]
- How, J.; Venkataraman, V.; Hobbs, G.S. Blast and accelerated phase CML: Room for improvement. Hematology 2021, 2021, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Sato, E.; Iriyama, N.; Tokuhira, M.; Takaku, T.; Ishikawa, M.; Nakazato, T.; Sugimoto, K.; Fujita, H.; Kimura, Y.; Fujioka, I.; et al. The EUTOS long-term survival score predicts disease-specific mortality and molecular responses among patients with chronic myeloid leukemia in a practice-based cohort. Cancer Med. 2020, 9, 8931–8939. [Google Scholar] [CrossRef]
- Rowley, J.D. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature 1973, 243, 290–293. [Google Scholar] [CrossRef]
- Huang, X.; Cortes, J.; Kantarjian, H. Estimations of the increasing prevalence and plateau prevalence of chronic myeloid leukemia in the era of tyrosine kinase inhibitor therapy. Cancer 2012, 118, 3123–3127. [Google Scholar] [CrossRef] [PubMed]
- Hochhaus, A.; Larson, R.A.; Guilhot, F.; Radich, J.P.; Branford, S.; Hughes, T.P.; Baccarani, M.; Deininger, M.W.; Cervantes, F.; Fujihara, S.; et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N. Engl. J. Med. 2017, 376, 917–927. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, S.G.; Guilhot, F.; Larson, R.A.; Gathmann, I.; Baccarani, M.; Cervantes, F.; Cornelissen, J.J.; Fischer, T.; Hochhaus, A.; Hughes, T.; et al. Imatinib Compared with Interferon and Low-Dose Cytarabine for Newly Diagnosed Chronic-Phase Chronic Myeloid Leukemia. N. Engl. J. Med. 2003, 348, 994–1004. [Google Scholar] [CrossRef]
- Tokarski, J.S.; Newitt, J.A.; Chang, C.Y.J.; Cheng, J.D.; Wittekind, M.; Kiefer, S.E.; Kish, K.; Lee, F.Y.; Borzillerri, R.; Lombardo, L.J.; et al. The Structure of Dasatinib (BMS-354825) Bound to Activated ABL Kinase Domain Elucidates Its Inhibitory Activity against Imatinib-Resistant ABL Mutants. Cancer Res. 2006, 66, 5790–5797. [Google Scholar] [CrossRef]
- Weisberg, E.; Manley, P.; Breitenstein, W.; Brüggen, J.; Cowan-Jacob, S.W.; Ray, A.; Griffin, J.D. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell 2005, 7, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Gambacorti-Passerini, C.; Deininger, M.W.; Mauro, M.J.; Chuah, C.; Kim, D.-W.; Dyagil, I.; Glushko, N.; Milojkovic, D.; Le Coutre, P.; et al. Bosutinib versus Imatinib for Newly Diagnosed Chronic Myeloid Leukemia: Results from the Randomized BFORE Trial. J. Clin. Oncol. 2018, 36, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Scalzulli, E.; Carmosino, I.; Bisegna, M.L.; Martelli, M.; Breccia, M. CML Resistant to 2nd-Generation TKIs: Mechanisms, Next Steps, and New Directions. Curr. Hematol. Malign-Rep. 2022, 17, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Hughes, T.P.; Mauro, M.J.; Cortes, J.E.; Minami, H.; Rea, D.; DeAngelo, D.J.; Breccia, M.; Goh, Y.-T.; Talpaz, M.; Hochhaus, A.; et al. Asciminib in Chronic Myeloid Leukemia after ABL Kinase Inhibitor Failure. N. Engl. J. Med. 2019, 381, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Narlı Özdemir, Z.; Kılıçaslan, N.A.; Yılmaz, M.; Eşkazan, A.E. Guidelines for the treatment of chronic myeloid leukemia from the NCCN and ELN: Differences and similarities. Int. J. Hematol. 2023, 117, 3–15. [Google Scholar] [CrossRef]
- Ono, T. Which Tyrosine Kinase Inhibitors Should Be Selected as the First-Line Treatment for Chronic Myelogenous Leukemia in Chronic Phase? Cancers 2021, 13, 5116. [Google Scholar] [CrossRef] [PubMed]
- Yohanan, B.; George, B. Current Management of Chronic Myeloid Leukemia Myeloid Blast Phase. Clin. Med. Insights Oncol. 2022, 16, 1–9. [Google Scholar] [CrossRef]
- Mahon, F.-X.; Réa, D.; Guilhot, J.; Guilhot, F.; Huguet, F.; Nicolini, F.; Legros, L.; Charbonnier, A.; Guerci, A.; Varet, B.; et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: The prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010, 11, 1029–1035. [Google Scholar] [CrossRef]
- Latagliata, R.; Romano, A.; Mancini, M.; Breccia, M.; Carmosino, I.; Vozella, F.; Montagna, C.; Volpicelli, P.; De Angelis, F.; Petrucci, L.; et al. Discontinuation of alpha-interferon treatment in patients with chronic myeloid leukemia in long-lasting complete molecular response. Leuk. Lymphoma 2015, 57, 99–102. [Google Scholar] [CrossRef]
- Hochhaus, A.; Baccarani, M.; Silver, R.T.; Schiffer, C.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Deininger, M.W.; Guilhot, F.; et al. European LeukemiaNet 2020 recommendations for treating chronic myeloid leukemia. Leukemia 2020, 34, 966–984. [Google Scholar] [CrossRef]
- Deininger, M.W.; Shah, N.P.; Altman, J.K.; Berman, E.; Bhatia, R.; Bhatnagar, B.; Sundar, H. Chronic Myeloid Leukemia, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2020, 18, 1385–1415. [Google Scholar] [CrossRef]
- Stuckey, R.; López-Rodríguez, J.F.; Sánchez-Sosa, S.; Segura-Díaz, A.; Sánchez-Farías, N.; Bilbao-Sieyro, C.; Gómez-Casares, M.T. Predictive indicators of successful tyrosine kinase inhibitor discontinuation in patients with chronic myeloid leukemia. World J. Clin. Oncol. 2020, 11, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Tzankov, A.; Reichard, K.K.; Hasserjian, R.P.; Arber, D.A.; Orazi, A.; Wang, S.A. Updates on eosinophilic disorders. Virchows Arch. 2022, 482, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Hasserjian, R.P.; Tam, W.; Tsai, A.; Geyer, J.T.; George, T.; Foucar, K.; Rogers, H.J.; Hsi, E.D.; Rea, B.A.; et al. Bone marrow morphology is a strong discriminator between chronic eosinophilic leukemia, not otherwise specified and reactive idiopathic hypereosinophilic syndrome. Haematologica 2017, 102, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Morsia, E.; Reichard, K.; Pardanani, A.; Tefferi, A.; Gangat, N. WHO defined chronic eosinophilic leukemia, not otherwise specified (CEL, NOS): A contemporary series from the Mayo Clinic. Am. J. Hematol. 2020, 95, E172–E174. [Google Scholar] [CrossRef]
- Gerds, A.T.; Gotlib, J.; Bose, P.; Deininger, M.W.; Dunbar, A.; Elshoury, A.; George, T.I.; Gojo, I.; Gundabolu, K.; Hexner, E.; et al. Myeloid/Lymphoid Neoplasms with Eosinophilia and TK Fusion Genes, Version 3.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2020, 18, 1248–1269. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.-Q.; Qin, T.-J.; Xu, Z.-F.; Zhang, Y.; Ai, X.F.; Li, B.; Xiao, Z.J. Long-term outcomes of imatinib in patients with FIP1L1/PDGFRA associated chronic eosinophilic leukemia: Experience of a single center in China. Oncotarget 2016, 7, 33229. [Google Scholar] [CrossRef] [PubMed]
- Jawhar, M.; Naumann, N.; Schwaab, J.; Baurmann, H.; Casper, J.; Dang, T.-A.; Dietze, L.; Döhner, K.; Hänel, A.; Lathan, B.; et al. Imatinib in myeloid/lymphoid neoplasms with eosinophilia and rearrangement of PDGFRB in chronic or blast phase. Ann. Hematol. 2017, 96, 1463–1470. [Google Scholar] [CrossRef]
- Lierman, E.; Michaux, L.; Beullens, E.; Pierre, P.; Marynen, P.; Cools, J.; Vandenberghe, P. FIP1L1-PDGFRα D842V, a novel panresistant mutant, emerging after treatment of FIP1L1-PDGFRα T674I eosinophilic leukemia with single agent sorafenib. Leukemia 2009, 23, 845–851. [Google Scholar] [CrossRef]
- Sadovnik, I.; Lierman, E.; Peter, B.; Herrmann, H.; Suppan, V.; Stefanzl, G.; Haas, O.; Lion, T.; Pickl, W.; Cools, J.; et al. Identification of Ponatinib as a potent inhibitor of growth, migration, and activation of neoplastic eosinophils carrying FIP1L1-PDGFRA. Exp. Hematol. 2014, 42, 282–293.e4. [Google Scholar] [CrossRef]
- Schwaab, J.; Naumann, N.; Luebke, J.; Jawhar, M.; Somervaille, T.C.P.; Williams, M.S.; Frewin, R.; Jost, P.J.; Lichtenegger, F.S.; La Rosée, P.; et al. Response to tyrosine kinase inhibitors in myeloid neoplasms associated with PCM1-JAK2, BCR-JAK2 and ETV6-ABL1 fusion genes. Am. J. Hematol. 2020, 95, 824–833. [Google Scholar] [CrossRef]
- Verstovsek, S.; Vannucchi, A.M.; Rambaldi, A.; Gotlib, M.J.R.; Mead, A.J.; Hochhaus, A.; Kiladjian, J.-J.; Boluda, J.C.H.; Asatiani, E.; Lihou, B.C.; et al. Interim Results from Fight-203, a Phase 2, Open-Label, Multicenter Study Evaluating the Efficacy and Safety of Pemigatinib (INCB054828) in Patients with Myeloid/Lymphoid Neoplasms with Rearrangement of Fibroblast Growth Factor Receptor 1 (FGFR1). Blood 2018, 132, 690. [Google Scholar] [CrossRef]
- Hernández-Boluda, J.-C.; Pereira, A.; Zinger, N.; Gras, L.; Martino, R.; Nikolousis, E.; Finke, J.; Chinea, A.; Rambaldi, A.; Robin, M.; et al. Allogeneic hematopoietic cell transplantation in patients with myeloid/lymphoid neoplasm with FGFR1-rearrangement: A study of the Chronic Malignancies Working Party of EBMT. Bone Marrow Transplant. 2022, 57, 416–422. [Google Scholar] [CrossRef]
- Yao, J.; Xu, L.; Aypar, U.; Meyerson, H.J.; Londono, D.; Gao, Q.; Baik, J.; Dietz, J.; Benayed, R.; Sigler, A.; et al. Myeloid/lymphoid neoplasms with eosinophilia/basophilia and ETV6-ABL1 fusion: Cell-of-origin and response to tyrosine kinase inhibition. Haematologica 2020, 106, 614–618. [Google Scholar] [CrossRef]
- Falchi, L.; Mehrotra, M.; Newberry, K.J.; Lyle, L.M.; Lu, G.; Patel, K.P.; Luthra, R.; Popat, U.; Verstovsek, S. Professor of Medicine ETV6–FLT3 fusion gene-positive, eosinophilia-associated myeloproliferative neoplasm successfully treated with sorafenib and allogeneic stem cell transplant. Leukemia 2014, 28, 2090–2092. [Google Scholar] [CrossRef] [PubMed]
- Munthe-Kaas, M.C.M.; Forthun, R.B.; Brendehaug, A.M.; Eek, A.K.M.; Høysæter, T.B.; Osnes, L.T.M.; Prescott, T.M.; Spetalen, S.M.; Hovland, R. Partial Response to Sorafenib in a Child with a Myeloid/Lymphoid Neoplasm, Eosinophilia, and a ZMYM2-FLT3 Fusion. J. Pediatr. Hematol. 2020, 43, e508–e511. [Google Scholar] [CrossRef] [PubMed]
- Rumi, E.; Milosevic, J.D.; Selleslag, D.; Casetti, I.; Lierman, E.; Pietra, D.; Cavalloni, C.; Bellini, M.; Milanesi, C.; Dambruoso, I.; et al. Efficacy of ruxolitinib in myeloid neoplasms with PCM1-JAK2 fusion gene. Ann. Hematol. 2015, 94, 1927–1928. [Google Scholar] [CrossRef]
- Rizzuto, G.; Leoncin, M.; Imbergamo, S.; Taurino, D.; Mico, M.C.; Tosi, M.; Michelato, A.; Buklijas, K.; Spinelli, O.; Lussana, F.; et al. Sequential allogeneic transplantation and ruxolitinib maintenance for a synchronous PCM1-JAK2 positive myeloid sarcoma and acute B-lymphoblastic leukemia. Clin. Case Rep. 2022, 10, e05212. [Google Scholar] [CrossRef]
- Duncavage, E.J.; Bagg, A.; Hasserjian, R.P.; DiNardo, C.D.; Godley, L.A.; Iacobucci, I.; Jaiswal, S.; Malcovati, L.; Vannucchi, A.M.; Patel, K.P.; et al. Genomic profiling for clinical decision making in myeloid neoplasms and acute leukemia. Blood 2022, 140, 2228–2247. [Google Scholar] [CrossRef]
- Pizzi, M. Crossing the Borders: An Integrated Approach to Myeloproliferative Neoplasms and Mastocytoses. Cancers 2021, 13, 1492. [Google Scholar] [CrossRef]
- Orazi, A. Histopathology in the Diagnosis and Classification of Acute Myeloid Leukemia, Myelodysplastic Syndromes, and Myelodysplastic/Myeloproliferative Diseases. Pathobiology 2007, 74, 97–114. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).